Determination of Technological Parameters and Characterization of Microbiota of the Spontaneous Sourdough Fermentation of Hull-Less Barley
Abstract
:1. Introduction
2. Materials and Methods
2.1. Raw Material
2.2. Experimental Design for Sourdough Preparation and Statistical Analysis
2.3. Determination of Number of Lactic Acid Bacteria (LAB) and Yeasts and Statistical Analysis
2.4. Determination of pH
2.5. Statistical Analysis
2.6. Microorganisms Identification
3. Results and Discussion
3.1. The Influence of Technological Parameters to the Development of LAB, Yeasts
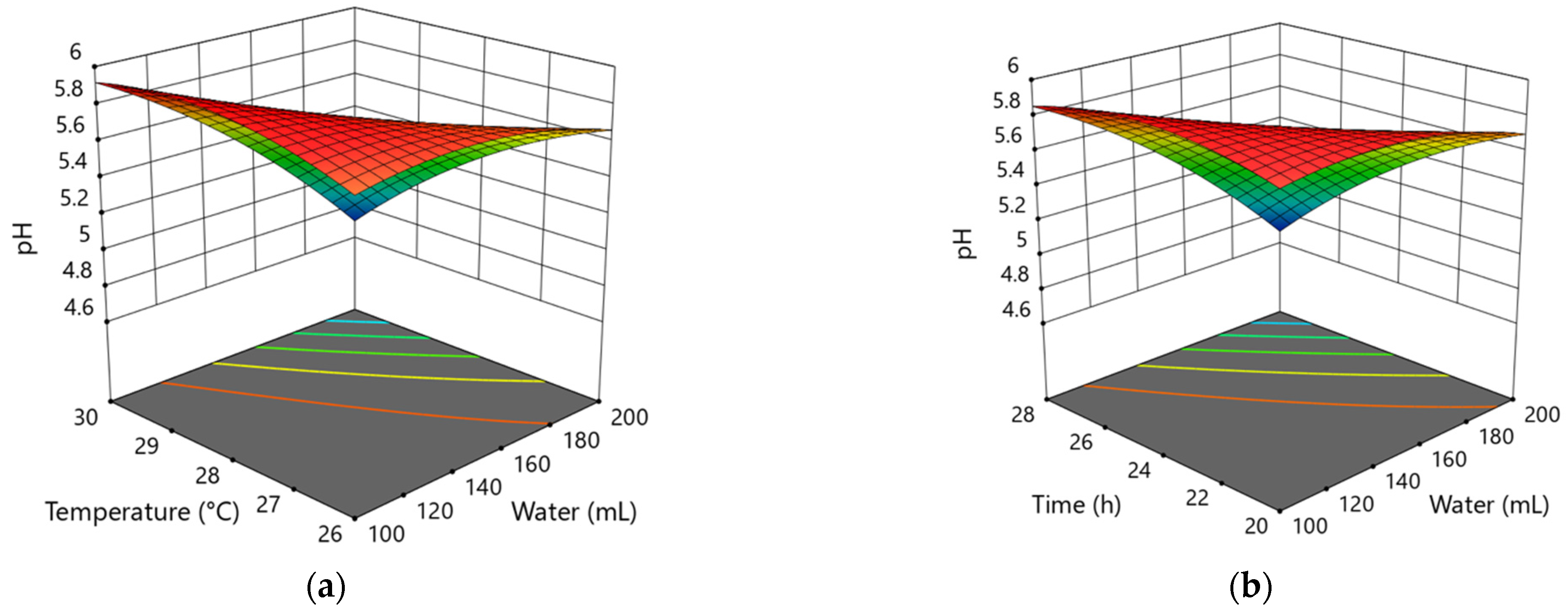
3.1.1. Modeling of the Influence of Variable Factors in the 1st Step
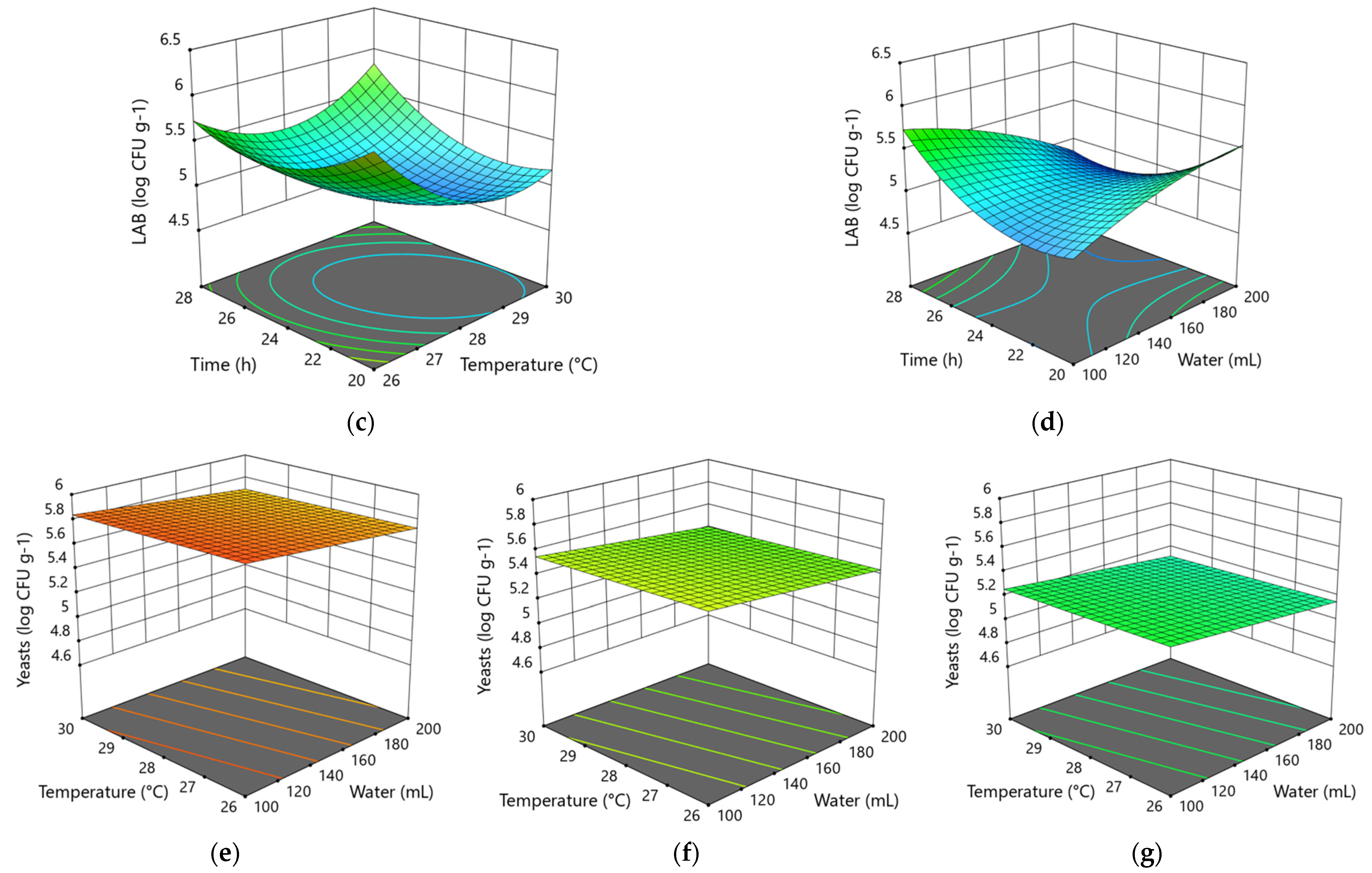
3.1.2. Modeling of the Influence of Variable Factors in the 2nd Backslopping Step
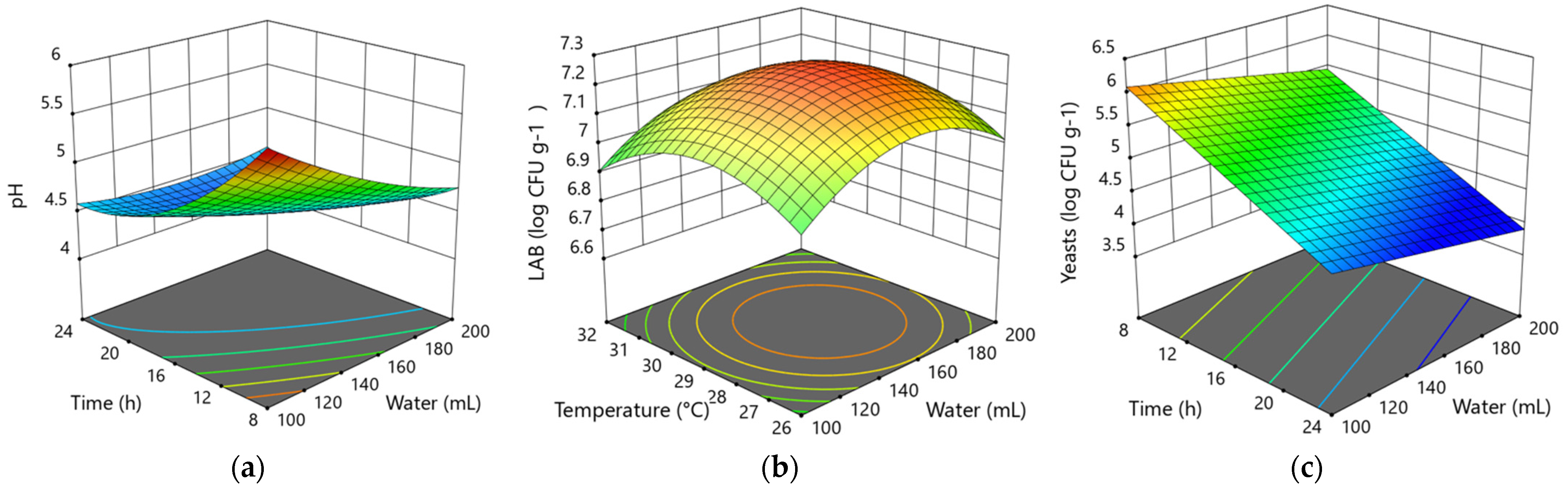
3.1.3. Modeling of the Influence of Variable Factors in the 3rd Backslopping Step
3.2. The Dynamics of the Development of Lactic Acid Bacteria and Yeasts in Three-Step Fermentation According to the Optimized Factors
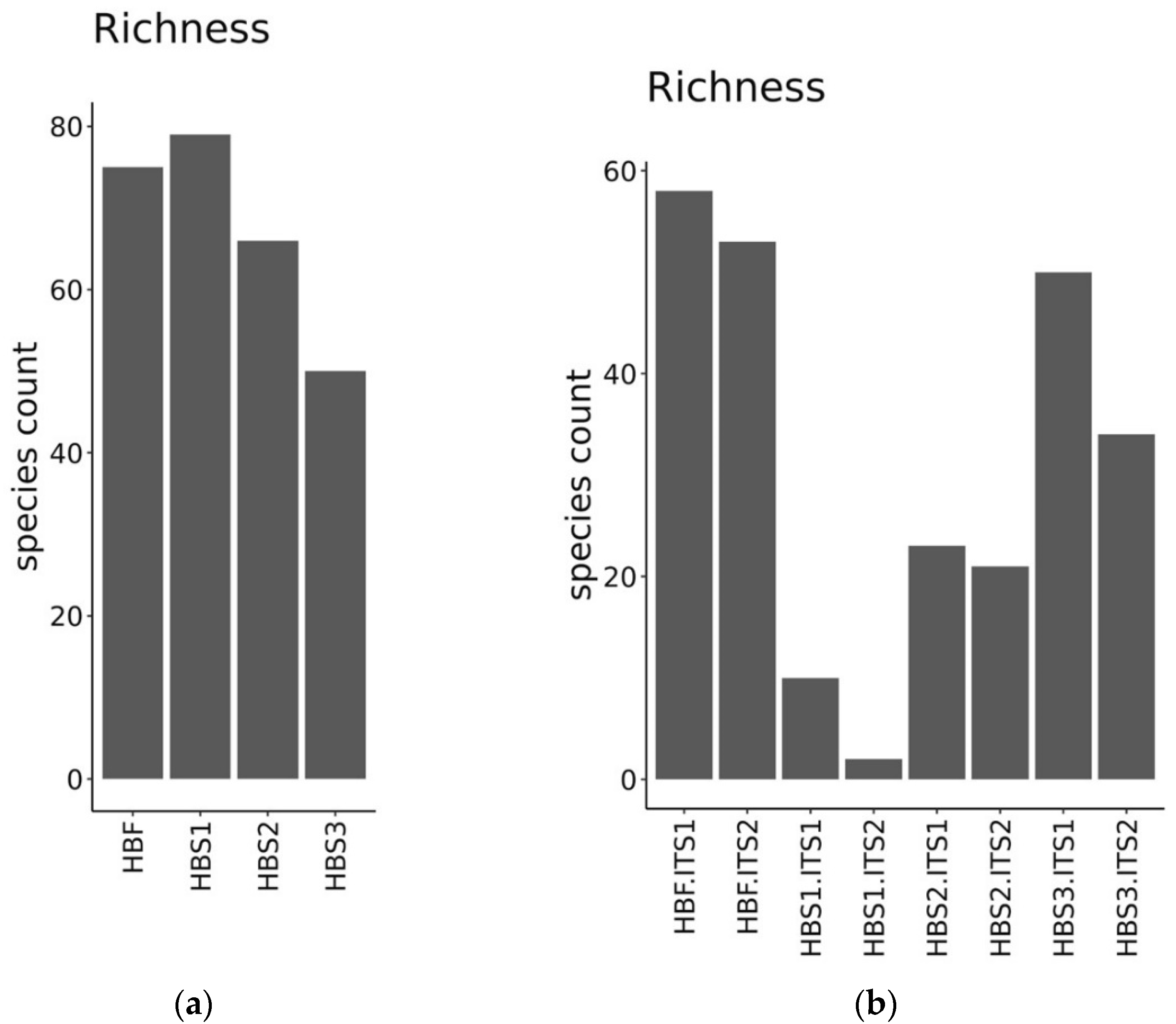
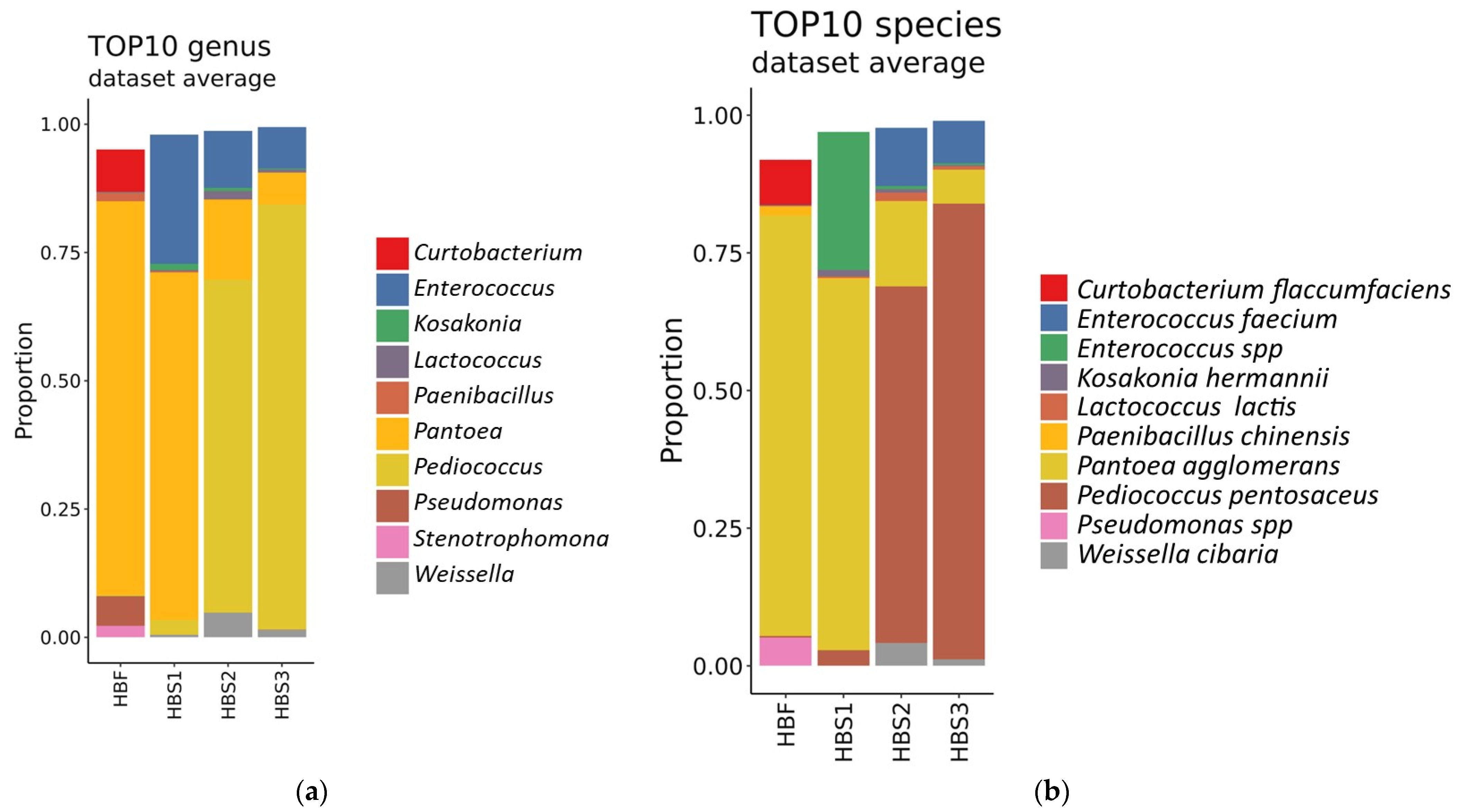
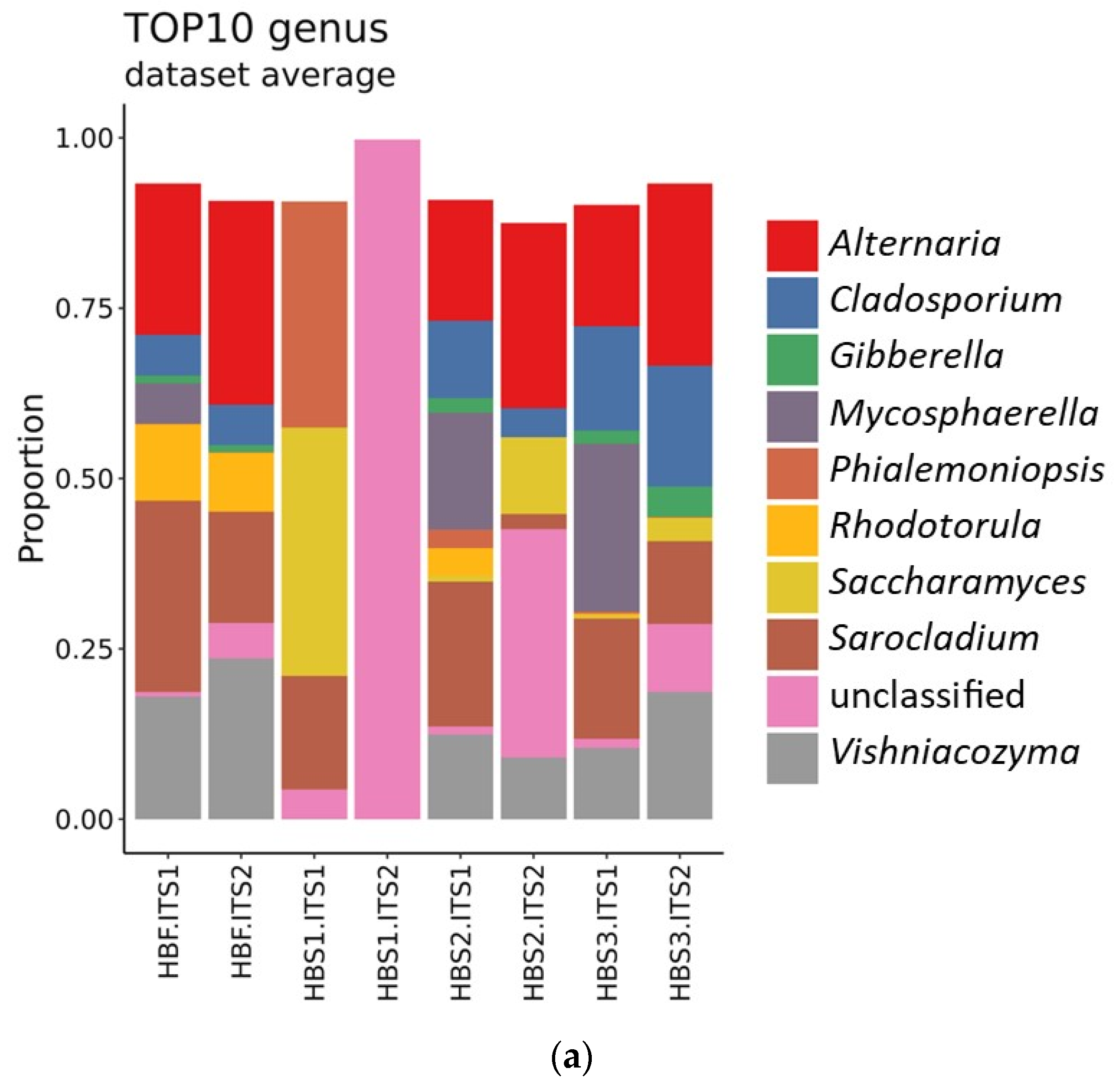
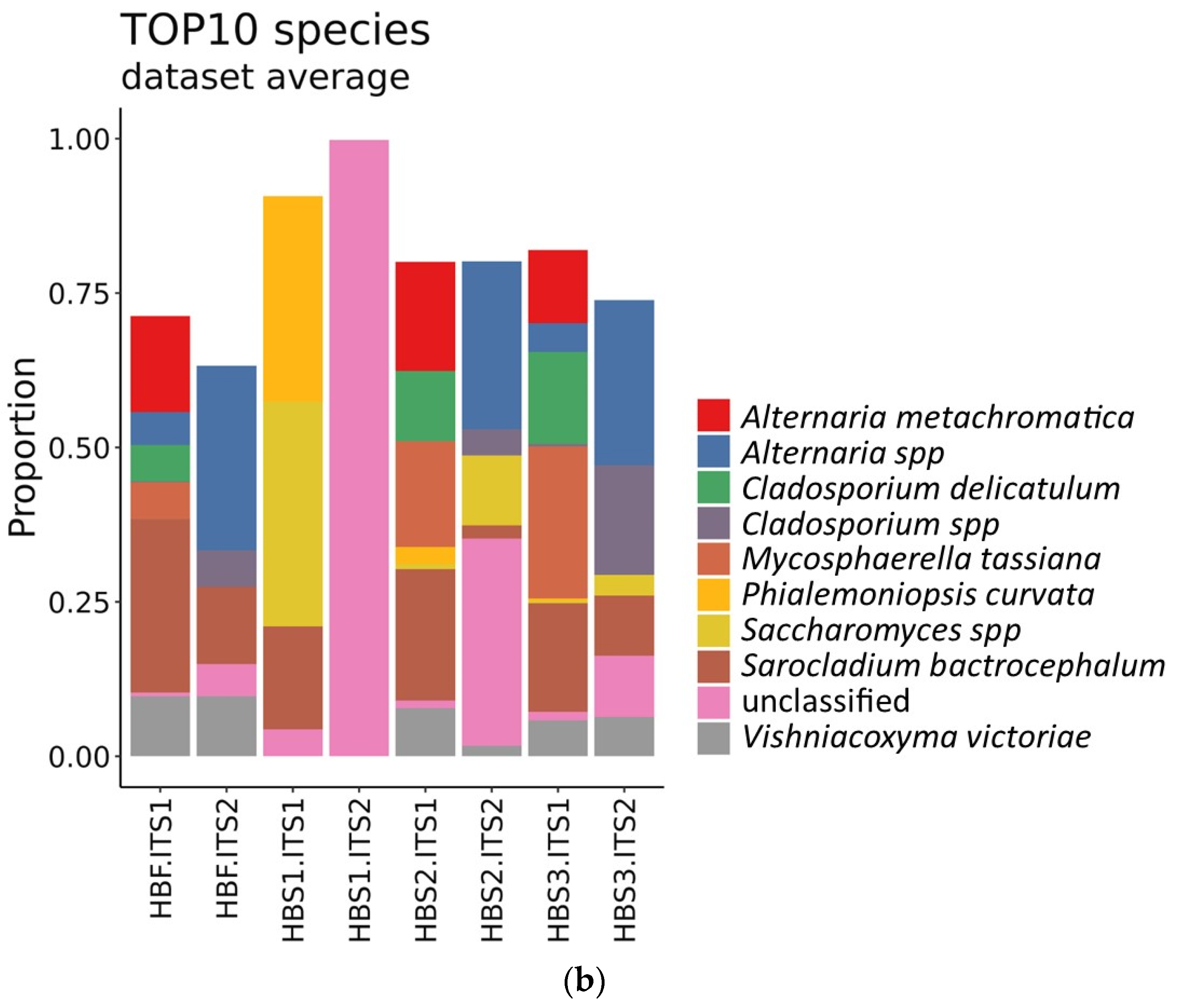
3.3. Identification of Microorganisms in the Hull-Less Barley Sourdough in the Three Backslopping Steps of Fermentation
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Šterna, V.; Zute, S.; Jakobsone, I. Grain composition and functional ingredients of barley varieties created in Latvia. Proc. Latv. Acad. Sci. Sect. B Nat. Exact Appl. Sci. 2015, 69, 158–162. [Google Scholar] [CrossRef] [Green Version]
- Baik, B.K.; Ullrich, S.E. Barley for food: Characteristics, improvement, and renewed interest. J. Cereal Sci. 2008, 48, 233–242. [Google Scholar] [CrossRef]
- Pejcz, E.; Czaja, A.; Wojciechowicz-Budzisz, A.; Gil, Z.; Spychaj, R. The potential of naked barley sourdough to improve the quality and dietary fibre content of barley enriched wheat bread. J. Cereal Sci. 2017, 77, 97–101. [Google Scholar] [CrossRef]
- Ma, S.; Wang, Z.; Guo, X.; Wang, F.; Huang, J.; Sun, B.; Wang, X. Sourdough improves the quality of whole-wheat flour products: Mechanisms and challenges—A review. Food Chem. 2021, 360, 130038. [Google Scholar] [CrossRef]
- Pejcz, E.; Gil, Z.; Wojciechowicz-Budzisz, A.; Półtorak, M.; Romanowska, A. Effect of technological process on the nutritional quality of naked barley enriched rye bread. J. Cereal Sci. 2015, 65, 215–219. [Google Scholar] [CrossRef]
- De Angelis, M.; Minervini, F.; Siragusa, S.; Rizzello, C.G.; Gobbetti, M. Wholemeal wheat flours drive the microbiome and functional features of wheat sourdoughs. Int. J. Food Microbiol. 2019, 302, 35–46. [Google Scholar] [CrossRef] [PubMed]
- Gobbetti, M.; Gänzle, M. Handbook on Sourdough Biotechnology; Springer: Boston, MA, USA, 2013; ISBN 9781461454250. [Google Scholar]
- De Vuyst, L.; Van Kerrebroeck, S.; Leroy, F. Microbial Ecology and Process Technology of Sourdough Fermentation. Adv. Appl. Microbiol. 2017, 100, 49–160. [Google Scholar] [PubMed]
- De Vuyst, L.; Van Kerrebroeck, S.; Harth, H.; Huys, G.; Daniel, H.M.; Weckx, S. Microbial ecology of sourdough fermentations: Diverse or uniform? Food Microbiol. 2014, 37, 11–29. [Google Scholar] [CrossRef] [PubMed]
- Harth, H.; Van Kerrebroeck, S.; De Vuyst, L. Community dynamics and metabolite target analysis of spontaneous, backslopped barley sourdough fermentations under laboratory and bakery conditions. Int. J. Food Microbiol. 2016, 228, 22–32. [Google Scholar] [CrossRef]
- Van Kerrebroeck, S.; Maes, D.; De Vuyst, L. Sourdoughs as a function of their species diversity and process conditions, a meta-analysis. Trends Food Sci. Technol. 2017, 68, 152–159. [Google Scholar] [CrossRef]
- Kozlinskis, E.; Skudra, L.; Rakcejeva, T.; Kunkulberga, D. Changes in the chemical and microbiological properties of spontaneous rye sourdough during fermentation. Spontano Rudzu Maizes Ieraugu Kim. Mikrobiol. Rad. Izmainas Ferment. Laik. 2010, 25, 67–77. [Google Scholar]
- Spicher, G.; Stephan, H. Handbuch Sauerteig: Biology, Biochemie, Technologie; B Behrš Verlag GMBH & Co.: Humburg, Germany, 1993. [Google Scholar]
- Brandt, M.J.; Gänzle, M.G. Handbuch Sauerteig; Behr’s Verlag DE: Humburg, Germany, 2006; ISBN 3-86022-076-4. [Google Scholar]
- Demirkesen-Bicak, H.; Arici, M.; Yaman, M.; Karasu, S.; Sagdic, O. Effect of different fermentation condition on estimated glycemic index, in vitro starch digestibility, and textural and sensory properties of sourdough bread. Foods 2021, 10, 514. [Google Scholar] [CrossRef]
- Zheng, J.; Wittouck, S.; Salvetti, E.; Franz, C.M.A.P.; Harris, H.M.B.; Mattarelli, P.; O’toole, P.W.; Pot, B.; Vandamme, P.; Walter, J.; et al. A taxonomic note on the genus Lactobacillus: Description of 23 novel genera, emended description of the genus Lactobacillus beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 2020, 70, 2782–2858. [Google Scholar] [CrossRef]
- Çakır, E.; Arıcı, M.; Durak, M.Z. Biodiversity and techno-functional properties of lactic acid bacteria in fermented hull-less barley sourdough. J. Biosci. Bioeng. 2020, 130, 450–456. [Google Scholar] [CrossRef]
- Oshiro, M.; Zendo, T.; Nakayama, J. Diversity and dynamics of sourdough lactic acid bacteriota created by a slow food fermentation system. J. Biosci. Bioeng. 2021, 131, 333–340. [Google Scholar] [CrossRef] [PubMed]
- De Vuyst, L.; Harth, H.; Van Kerrebroeck, S.; Leroy, F. Yeast diversity of sourdoughs and associated metabolic properties and functionalities. Int. J. Food Microbiol. 2016, 239, 26–34. [Google Scholar] [CrossRef]
- Abedfar, A.; Sadeghi, A. Response surface methodology for investigating the effects of sourdough fermentation conditions on Iranian cup bread properties. Heliyon 2019, 5, e02608. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katina, K.; Heiniö, R.L.; Autio, K.; Poutanen, K. Optimization of sourdough process for improved sensory profile and texture of wheat bread. LWT Food Sci. Technol. 2006, 39, 1189–1202. [Google Scholar] [CrossRef]
- ISO 6887-1:1999. Microbiology of Food and Feeding Staffs—Preparation of Test Samples, Initial Suspension and Decimal Dilutions for Microbiological Examination—Part 1: General Rules for the Preparation of the Initial Suspension and Decimals Dilutions; ISO: Geneva, Switzerland, 1999. [Google Scholar]
- ISO 15214. Microbiology of Food and Animal Feeding Stuffs—Horizontal Method for the Emumeration of Mesophilic Lactic Acid Bacteria—Colony-Count Technique at 30 °C; ISO: Geneva, Switzerland, 1998. [Google Scholar]
- ISO 21527-1. Microbiology of Food and Animal Feeding Stuffs. Horizontal Method for the Enumeration of Yeasts and Molds; ISO: Geneva, Switzerland, 2008. [Google Scholar]
- AACC 02-52:1999. Hydrogen-Ion Activity (pH)—Electrometric Method. Approved Methods of the American Association of Cereal Chemists; AACC: St. Paul, MN, USA, 1999. [Google Scholar]
- Kazantseva, J.; Malv, E.; Kaleda, A.; Kallastu, A.; Meikas, A. Optimisation of sample storage and DNA extraction for human gut microbiota studies. BMC Microbiol. 2021, 21, 158. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Lozupone, C.A.; Turnbaugh, P.J.; Fierer, N.; Knight, R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. USA 2011, 108, 4516–4522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kõljalg, U.; Nilsson, R.H.; Abarenkov, K.; Tedersoo, L.; Taylor, A.F.S.; Bahram, M.; Bates, S.T.; Bruns, T.D.; Bengtsson-Palme, J.; Callaghan, T.M.; et al. Towards a unified paradigm for sequence-based identification of fungi. Mol. Ecol. 2013, 22, 5271–5277. [Google Scholar] [CrossRef] [Green Version]
- Lee, T.D.; Walters, W.A.; Lennon, N.J.; Bochicchio, J.; Krohn, A.; Caporaso, J.G.; Pennanen, T. Accurate Estimation of Fungal Diversity and Abundance through Improved Lineage-Specific Primers Optimized for Illumina Amplicon Sequencing. Appl. Environ. Microbiol. 2016, 82, 7217–7226. [Google Scholar]
- BION-Meta Program. Available online: https://github.com/nielsl/mcdonald-et-al (accessed on 16 May 2016).
- Katsi, P.; Kosma, I.; Michailidou, S.; Argiriou, A.; Badeka, A.; Kontominas, M. Characterization of Artisanal Spontaneous Sourdough Wheat Bread from Central Greece: Evaluation of Physico-Chemical, Microbiological, and Sensory Properties in Relation to Conventional Yeast Leavened Wheat Bread. Foods 2021, 10, 635. [Google Scholar] [CrossRef] [PubMed]
- Minervini, F.; Lattanzi, A.; De Angelis, M.; Celano, G.; Gobbetti, M. House microbiotas as sources of lactic acid bacteria and yeasts in traditional Italian sourdoughs. Food Microbiol. 2015, 52, 66–76. [Google Scholar] [CrossRef] [PubMed]
- Weckx, S.; Van der Meulen, R.; Maes, D.; Scheirlinck, I.; Huys, G.; Vandamme, P.; De Vuyst, L. Lactic acid bacteria community dynamics and metabolite production of rye sourdough fermentations share characteristics of wheat and spelt sourdough fermentations. Food Microbiol. 2010, 27, 1000–1008. [Google Scholar] [CrossRef] [PubMed]
- Corsetti, A.; Settanni, L.; Valmorri, S.; Mastrangelo, M.; Suzzi, G. Identification of subdominant sourdough lactic acid bacteria and their evolution during laboratory-scale fermentations. Food Microbiol. 2007, 24, 592–600. [Google Scholar] [CrossRef]
- Huys, G.; Daniel, H.M.; De Vuyst, L. Taxonomy and Biodiversity of Sourdough Yeasts and Lactic Acid Bacteria. In Handbook on Sourdough Biotechnology; Gobbetti, M., Ganzle, M., Eds.; Springer: New York, NY, USA, 2013; p. 298. [Google Scholar]
- Porto, M.C.W.; Kuniyoshi, T.M.; Azevedo, P.O.S.; Vitolo, M.; Oliveira, R.P.S. Pediococcus spp.: An important genus of lactic acid bacteria and pediocin producers. Biotechnol. Adv. 2017, 35, 361–374. [Google Scholar] [CrossRef] [PubMed]
- Scherf, K.A.; Wieser, H.; Koehler, P. Novel approaches for enzymatic gluten degradation to create high-quality gluten-free products. Food Res. Int. 2018, 110, 62–72. [Google Scholar] [CrossRef]

| Independent Variables | Units | Symbols | Coded Levels | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1st Backslopping Step | 2nd Backslopping Step | 3rd Backslopping Step | |||||||||
| −1 | 0 | 1 | −1 | 0 | 1 | −1 | 0 | 1 | |||
| Amount of added water | mL | X1 | 100 | 150 | 200 | 100 | 150 | 200 | 100 | 150 | 200 |
| Temperature of fermentation | °C | X2 | 26 | 28 | 30 | 29 | 32 | 35 | 26 | 29 | 32 |
| Time of fermentation | h | X3 | 20 | 24 | 28 | 8 | 16 | 24 | 8 | 14 | 20 |
| 1st Backslopping Step | ||||||
|---|---|---|---|---|---|---|
| Independent Variables | Responses | |||||
| Run | X1 | X2 | X3 | pH | LAB | Yeasts |
| Water, mL | Temperature, °C | Time, h | log CFU g−1 | log CFU g−1 | ||
| 1 | 100 | 26 | 24 | 5.84 ± 0.09 | 5.50 ± 0.31 | 5.89 ± 0.09 |
| 2 | 200 | 26 | 24 | 5.72 ± 0.00 | 5.29 ± 0.05 | 5.34 ± 0.33 |
| 3 | 100 | 30 | 24 | 5.86 ± 0.02 | 5.27 ± 0.31 | 5.74 ± 0.22 |
| 4 | 200 | 30 | 24 | 4.62 ± 0.09 | 5.02 ± 0.02 | 5.04 ± 0.04 |
| 5 | 100 | 28 | 20 | 5.82 ± 0.05 | 4.93 ± 0.14 | 5.80 ± 0.08 |
| 6 | 200 | 28 | 20 | 5.72 ± 0.00 | 5.58 ± 0.41 | 5.86 ± 0.09 |
| 7 | 100 | 28 | 28 | 5.83 ± 0.02 | 5.48 ± 0.27 | 4.70 ± 0.11 |
| 8 | 200 | 28 | 28 | 4.68 ± 0.00 | 4.69 ± 0.41 | 5.33 ± 0.32 |
| 9 | 150 | 26 | 20 | 5.77 ± 0.00 | 6.50 ± 0.26 | 5.58 ± 0.25 |
| 10 | 150 | 30 | 20 | 5.72 ± 0.02 | 5.48 ± 0.23 | 5.95 ± 0.04 |
| 11 | 150 | 26 | 28 | 5.50 ± 0.13 | 5.45 ± 0.22 | 5.45 ± 0.28 |
| 12 | 150 | 30 | 28 | 5.03 ± 0.04 | 5.39 ± 0.08 | 5.39 ± 0.23 |
| 13 | 150 | 28 | 24 | 5.70 ± 0.07 | 4.97 ± 0.13 | 5.41 ± 0.03 |
| 14 | 150 | 28 | 24 | 5.75 ± 0.00 | 4.86 ± 0.54 | 5.54 ± 0.06 |
| 15 | 150 | 28 | 24 | 5.70 ± 0.09 | 5.01 ± 0.21 | 5.42 ± 0.09 |
| Responses | Effect of Factors in the Models | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Source | Model | X1 | X2 | X3 | X1X2 | X1X3 | X2X3 | X12 | X22 | X32 | |
| 1st step | |||||||||||
| pH | p-value | 0.0007 | 0.0002 | 0.0017 | 0.0006 | 0.0018 | 0.0023 | 0.0720 | 0.0850 | 0.0699 | 0.0746 |
| Coef | 5.72 | −0.3263 | −0.2000 | −0.2487 | −0.2800 | −0.2625 | −0.1050 | −0.1029 | −0.1104 | −0.1079 | |
| R2 | 0.9826 | ||||||||||
| LAB | p-value | 0.0119 | 0.2708 | 0.0225 | 0.0283 | 0.9117 | 0.0085 | 0.0380 | 0.2888 | 0.0048 | 0.0141 |
| Coef | 4.95 | −0.0750 | −0.1975 | −0.1850 | −0.0100 | −0.3600 | 0.2400 | −0.1058 | 0.4292 | 0.3292 | |
| R2 | 0.9441 | ||||||||||
| Yeasts | p-value | 0.0743 | 0.4937 | 0.8627 | 0.0136 | ||||||
| Coef | 5.50 | 0.4937 | 0.8627 | 0.0136 | |||||||
| R2 | 0.4537 | ||||||||||
| 2nd step | |||||||||||
| pH | p-value | 0.0009 | 0.0004 | 0.5801 | 0.0002 | 0.6013 | 0.0025 | 0.0818 | 0.0636 | 0.0953 | 0.0010 |
| Coef | 4.47 | −0.2687 | −0.0188 | −0.3150 | −0.0250 | 0.2525 | 0.0975 | 0.1108 | 0.0958 | 0.3233 | |
| R2 | 0.9812 | ||||||||||
| LAB | p-value | 0.0445 | 0.6672 | 0.2348 | 0.3794 | 0.6056 | 0.0131 | 0.0902 | |||
| Coef | 5.45 | 0.0972 | 0.1262 | 0.0844 | 0.0431 | 0.1909 | 0.0772 | ||||
| R2 | 0.7376 | ||||||||||
| Yeasts | p-value | 0.0140 | 0.1612 | 0.3647 | 0.0035 | ||||||
| Coef | 5.00 | −0.3150 | −0.1988 | −0.7763 | |||||||
| R2 | 0.6039 | ||||||||||
| 3rd step | |||||||||||
| pH | p-value | 0.1163 | 0.0441 | 0.4796 | 0.2170 | ||||||
| Coef | 4.12 | −0.0737 | −0.0237 | −0.0425 | |||||||
| R2 | 0.4026 | ||||||||||
| LAB | p-value | 0.1908 | 0.3008 | 0.7747 | 0.0323 | 1.0000 | 0.4724 | 0.8536 | 0.0821 | 0.0946 | 0.2037 |
| Coef | 7.21 | 0.0525 | −0.0138 | −0.1338 | 0.0000 | 0.0500 | 0.0125 | −0.1454 | −0.1379 | −0.0979 | |
| R2 | 0.8029 | ||||||||||
| Yeasts | p-value | 0.2109 | 0.7649 | 0.1490 | 0.9053 | 0.1876 | 0.4292 | 0.8290 | 0.6004 | 0.2224 | 0.0191 |
| Coef | 7.24 | −0.0663 | −0.3575 | −0.0263 | 0.4525 | −0.2550 | −0.0675 | −0.1725 | −0.4300 | −1.05 | |
| R2 | 0.7925 | ||||||||||
| 2nd Backslopping Step | ||||||
|---|---|---|---|---|---|---|
| Independent Variables | Responses | |||||
| Run | X1 | X2 | X3 | pH | LAB | Yeasts |
| Water, mL | Temperature, °C | Time, h | log CFU g−1 | log CFU g−1 | ||
| 1 | 100 | 29 | 16 | 5.02 ± 0.02 | 5.98 ± 0.39 | 6.26 ± 0.08 |
| 2 | 200 | 29 | 16 | 4.44 ± 0.02 | 5.48 ± 0.08 | 4.27 ± 0.07 |
| 3 | 100 | 35 | 16 | 4.97 ± 0.03 | 5.79 ± 0.04 | 4.57 ± 0.29 |
| 4 | 200 | 35 | 16 | 4.29 ± 0.01 | 5.55 ± 0.06 | 4.35 ± 0.09 |
| 5 | 100 | 32 | 8 | 5.71 ± 0.02 | 5.78 ± 0.07 | 6.11 ± 0.04 |
| 6 | 200 | 32 | 8 | 4.76 ± 0.01 | 4.60 ± 0.07 | 5.62 ± 0.12 |
| 7 | 100 | 32 | 24 | 4.55 ± 0.03 | 5.58 ± 0.13 | 4.53 ± 0.22 |
| 8 | 200 | 32 | 24 | 4.61 ± 0.01 | 5.93 ± 0.10 | 4.71 ± 0.08 |
| 9 | 150 | 29 | 8 | 5.28 ± 0.04 | 5.96 ± 0.27 | 6.35 ± 0.26 |
| 10 | 150 | 35 | 8 | 5.11 ± 0.01 | 5.39 ± 0.05 | 6.21 ± 0.04 |
| 11 | 150 | 29 | 24 | 4.48 ± 0.01 | 5.49 ± 0.08 | 4.34 ± 0.09 |
| 12 | 150 | 35 | 24 | 4.70 ± 0.02 | 5.84 ± 0.09 | 4.50 ± 0.04 |
| 13 | 150 | 32 | 16 | 4.43 ± 0.03 | 5.32 ± 0.16 | 4.41 ± 0.07 |
| 14 | 150 | 32 | 16 | 4.42 ± 0.02 | 5.29 ± 0.11 | 4.30 ± 0.09 |
| 15 | 150 | 32 | 16 | 4.57 ± 0.02 | 5.46 ± 0.09 | 4.40 ± 0.11 |
| 3rd Backslopping Step | ||||||
|---|---|---|---|---|---|---|
| Independent Variables | Responses | |||||
| Run | X1 | X2 | X3 | pH | LAB | Yeasts |
| Water, mL | Temperature, °C | Time, h | log CFU g−1 | log CFU g−1 | ||
| 1 | 100 | 26 | 14 | 4.27 ± 0.01 | 6.98 ± 0.08 | 7.05 ± 0.21 |
| 2 | 200 | 26 | 14 | 4.13 ± 0.01 | 7.03 ±0.39 | 6.45 ± 0.26 |
| 3 | 100 | 32 | 14 | 4.24 ± 0.02 | 6.83 ± 0.18 | 5.92 ± 0.50 |
| 4 | 200 | 32 | 14 | 4.06 ± 0.03 | 6.88 ± 0.08 | 7.13 ± 0.07 |
| 5 | 100 | 29 | 8 | 4.22 ± 0.02 | 7.14 ± 0.14 | 6.17 ± 0.14 |
| 6 | 200 | 29 | 8 | 4.09 ± 0.00 | 7.20 ± 0.10 | 6.11 ± 0.10 |
| 7 | 100 | 29 | 20 | 4.01 ± 0.04 | 6.64 ± 0.32 | 6.43 ± 0.10 |
| 8 | 200 | 29 | 20 | 3.87 ± 0.03 | 6.90 ± 0.13 | 5.35 ± 0.13 |
| 9 | 150 | 26 | 8 | 4.15 ± 0.03 | 7.01 ± 0.08 | 6.22 ± 0.27 |
| 10 | 150 | 32 | 8 | 4.08 ± 0.00 | 7.08 ± 0.15 | 5.15 ± 0.09 |
| 11 | 150 | 26 | 20 | 4.17 ± 0.01 | 6.85 ± 0.07 | 6.50 ± 0.11 |
| 12 | 150 | 32 | 20 | 4.15 ± 0.03 | 6.97 ± 0.15 | 5.16 ± 0.08 |
| 13 | 150 | 29 | 14 | 4.25 ± 0.02 | 7.13 ± 0.09 | 7.92 ± 0.07 |
| 14 | 150 | 29 | 14 | 4.13 ± 0.01 | 7.23 ± 0.05 | 7.15 ± 0.09 |
| 15 | 150 | 29 | 14 | 4.04 ± 0.01 | 7.80 ± 0.06 | 6.65 ± 0.14 |
| Backslopping Step | Parameters | Predicted Values | Experimental Values | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Water | Temperature | Time | pH | LAB | Yeasts | pH | LAB | Yeasts | |
| mL | °C | h | log CFU g−1 | log CFU g−1 | |||||
| 1st | 113 | 30 | 24 | 5.7 | 5.3 | 5.5 | 5.44 ± 0.04a | 5.85 ± 0.50b | 6.75 ± 0.47a |
| 2nd | 137 | 31 | 14 | 4.7 | 5.6 | 5.3 | 4.62 ± 0.05b | 5.99 ± 0.44b | 6.86 ± 0.04a |
| 3rd | 150 | 28.5 | 12 | 4.1 | 7.2 | 6.8 | 3.80 ± 0.05c | 8.22 ± 0.14a | 7.34 ± 0.68a |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reidzane, S.; Kruma, Z.; Kazantseva, J.; Traksmaa, A.; Klava, D. Determination of Technological Parameters and Characterization of Microbiota of the Spontaneous Sourdough Fermentation of Hull-Less Barley. Foods 2021, 10, 2253. https://doi.org/10.3390/foods10102253
Reidzane S, Kruma Z, Kazantseva J, Traksmaa A, Klava D. Determination of Technological Parameters and Characterization of Microbiota of the Spontaneous Sourdough Fermentation of Hull-Less Barley. Foods. 2021; 10(10):2253. https://doi.org/10.3390/foods10102253
Chicago/Turabian StyleReidzane, Sanita, Zanda Kruma, Jekaterina Kazantseva, Anna Traksmaa, and Dace Klava. 2021. "Determination of Technological Parameters and Characterization of Microbiota of the Spontaneous Sourdough Fermentation of Hull-Less Barley" Foods 10, no. 10: 2253. https://doi.org/10.3390/foods10102253
APA StyleReidzane, S., Kruma, Z., Kazantseva, J., Traksmaa, A., & Klava, D. (2021). Determination of Technological Parameters and Characterization of Microbiota of the Spontaneous Sourdough Fermentation of Hull-Less Barley. Foods, 10(10), 2253. https://doi.org/10.3390/foods10102253







