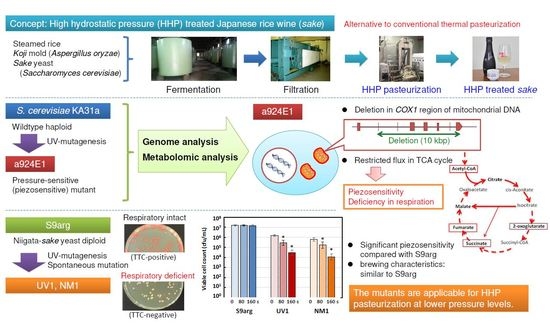
Genomic and Metabolomic Analyses of a Piezosensitive Mutant of Saccharomyces cerevisiae and Application for Generation of Piezosensitive Niigata-Sake Yeast Strains
Abstract
:1. Introduction
2. Materials and Methods
2.1. Yeast Strains and Growth Conditions
2.2. Screening of Aerobic Respiration-Deficient Mutants
2.3. High Hydrostatic Pressure (HHP) Treatment
2.4. Genome Analysis
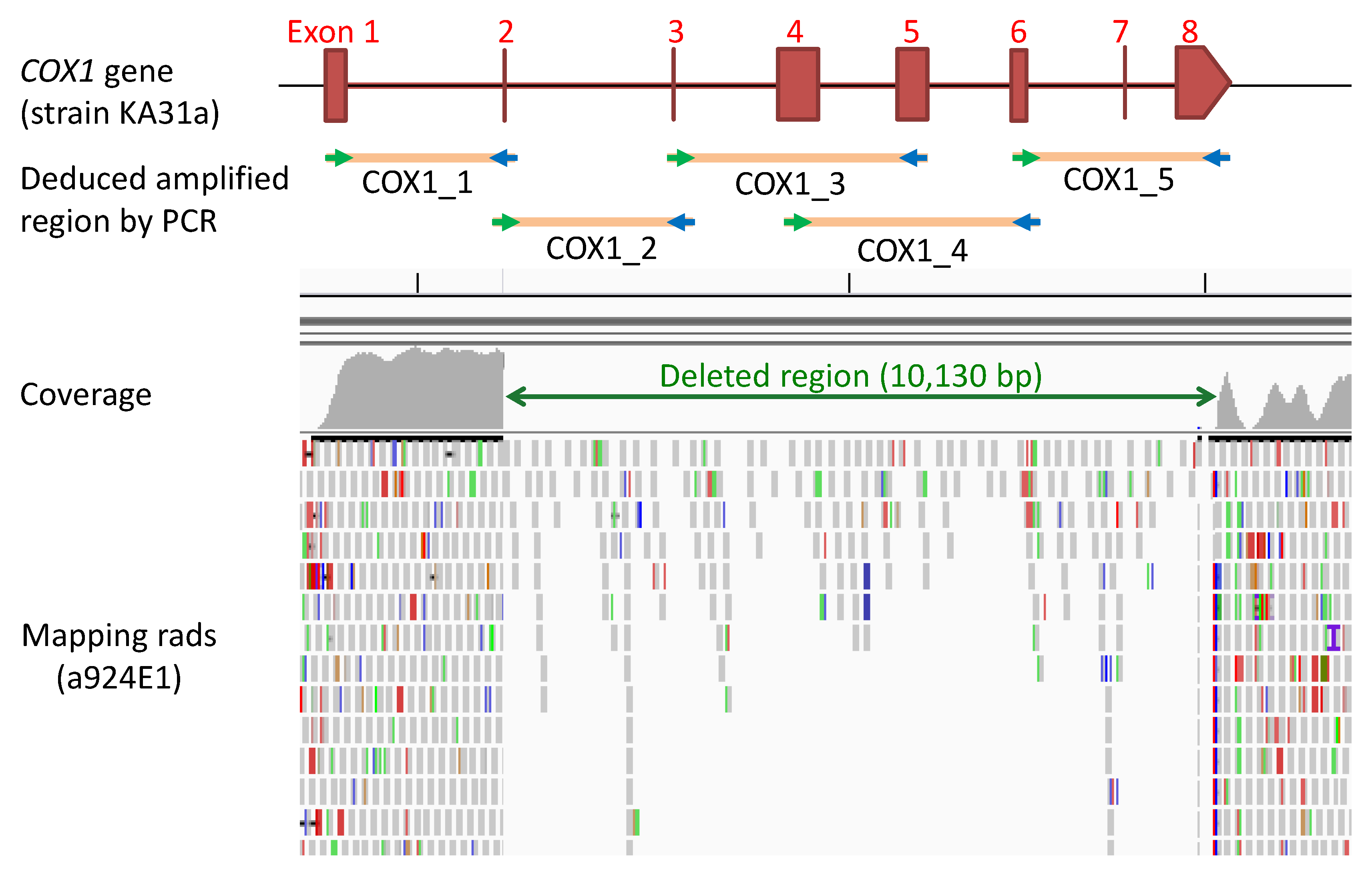
2.5. PCR Analysis
2.6. Extraction and Analyses of Metabolites
2.7. Laboratory-Scale Sake Brewing Test
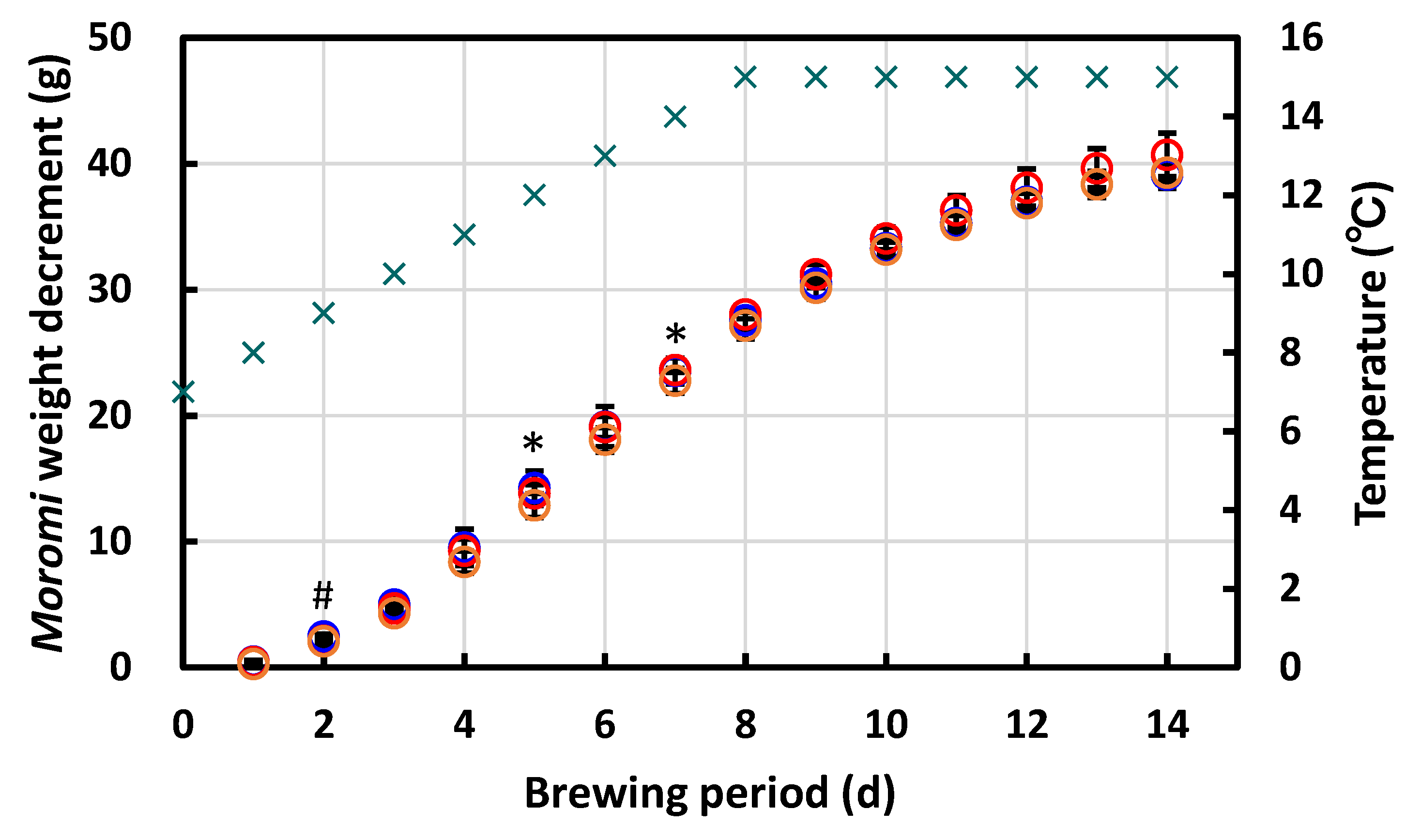
2.8. Characterization of Flavor Components in the Moromi
3. Results and Discussion
3.1. Growth and Pressure Inactivation Characteristics of Strains KA31a and a924E1
3.2. Genome Analysis of Strains KA31a and a924E1
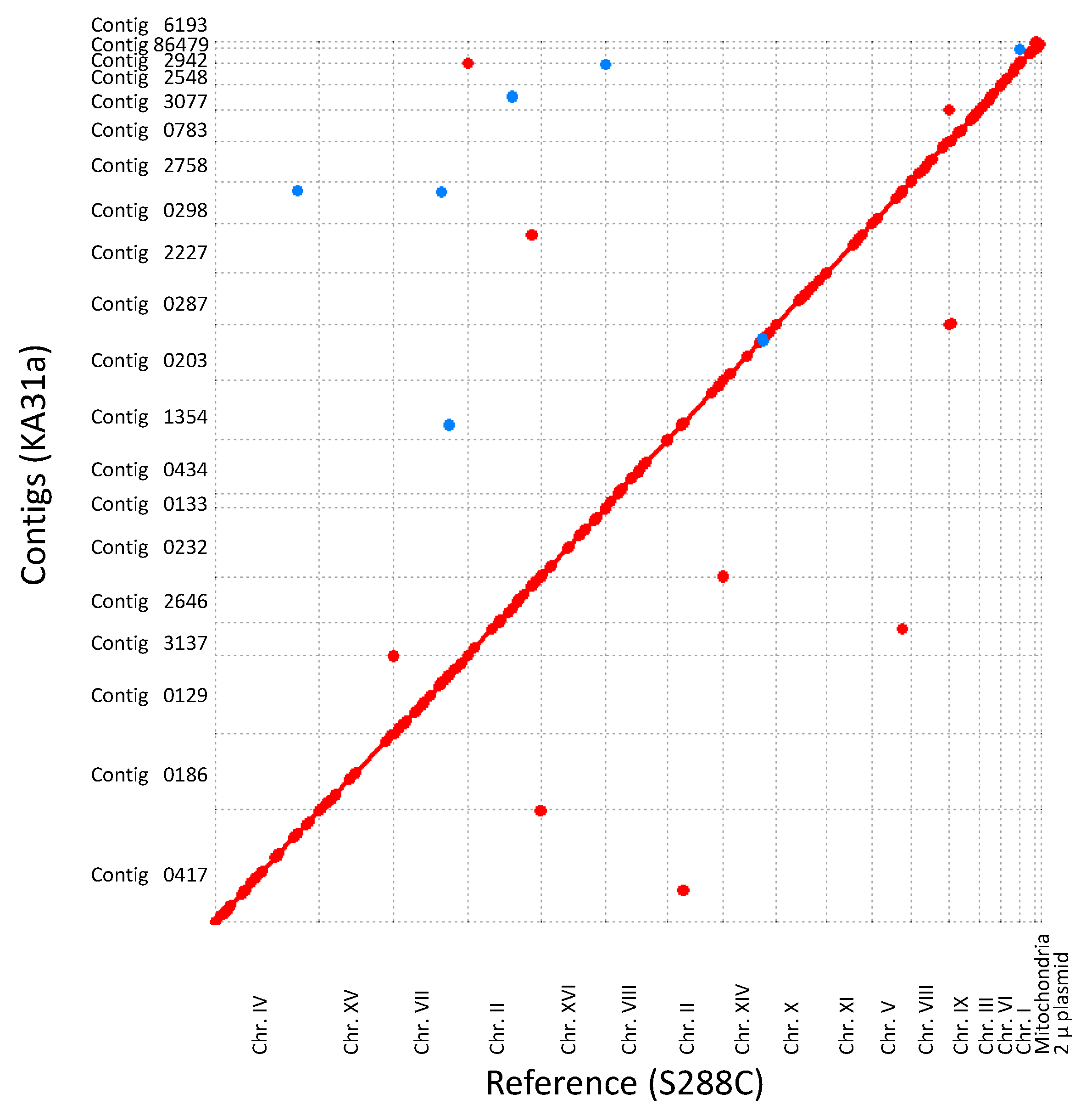
3.2.1. Genome Analysis of Strain KA31a
3.2.2. Genome Analysis of Strain a924E1
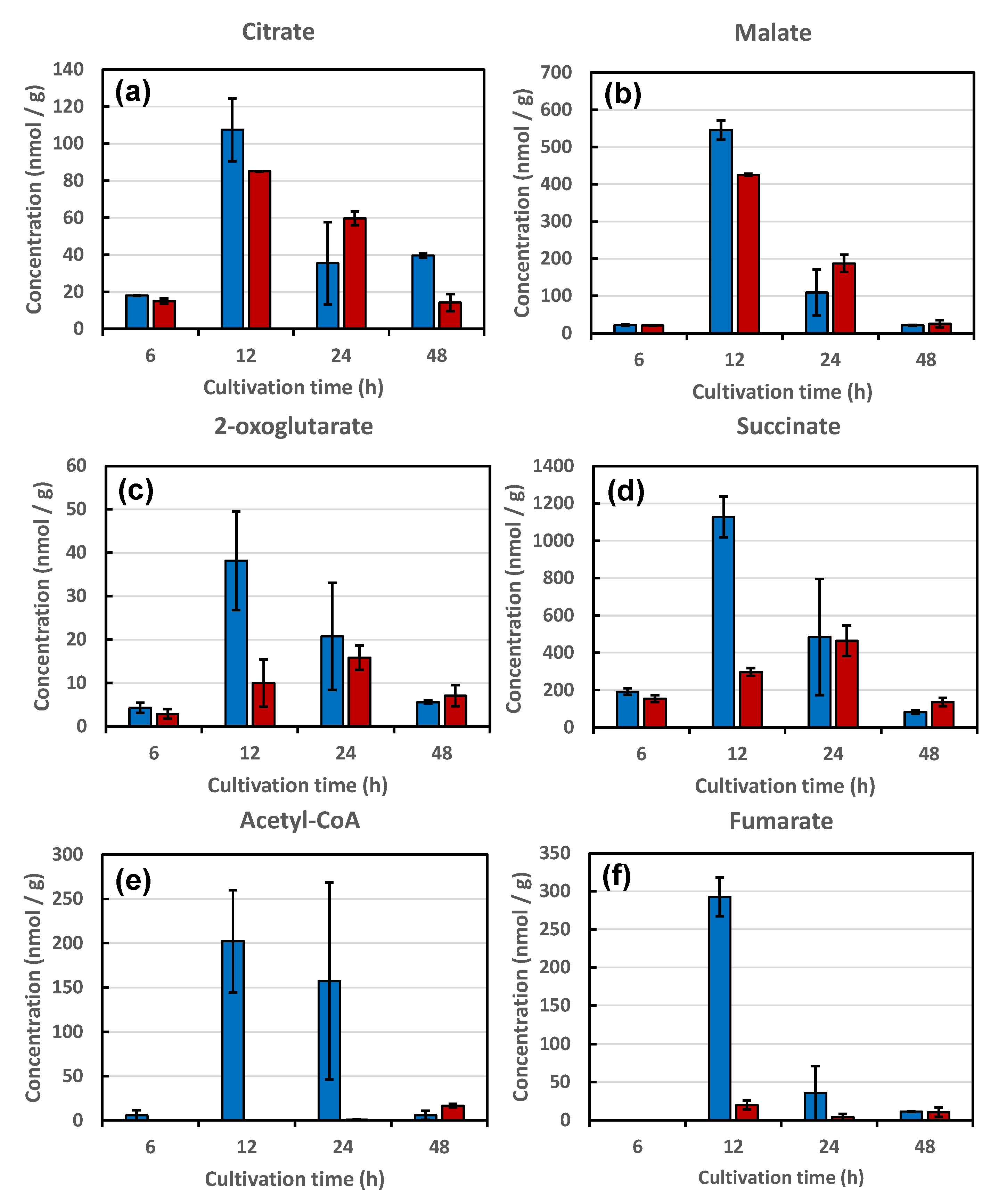
3.3. Metabolites Analysis of Strains KA31a and a924E1
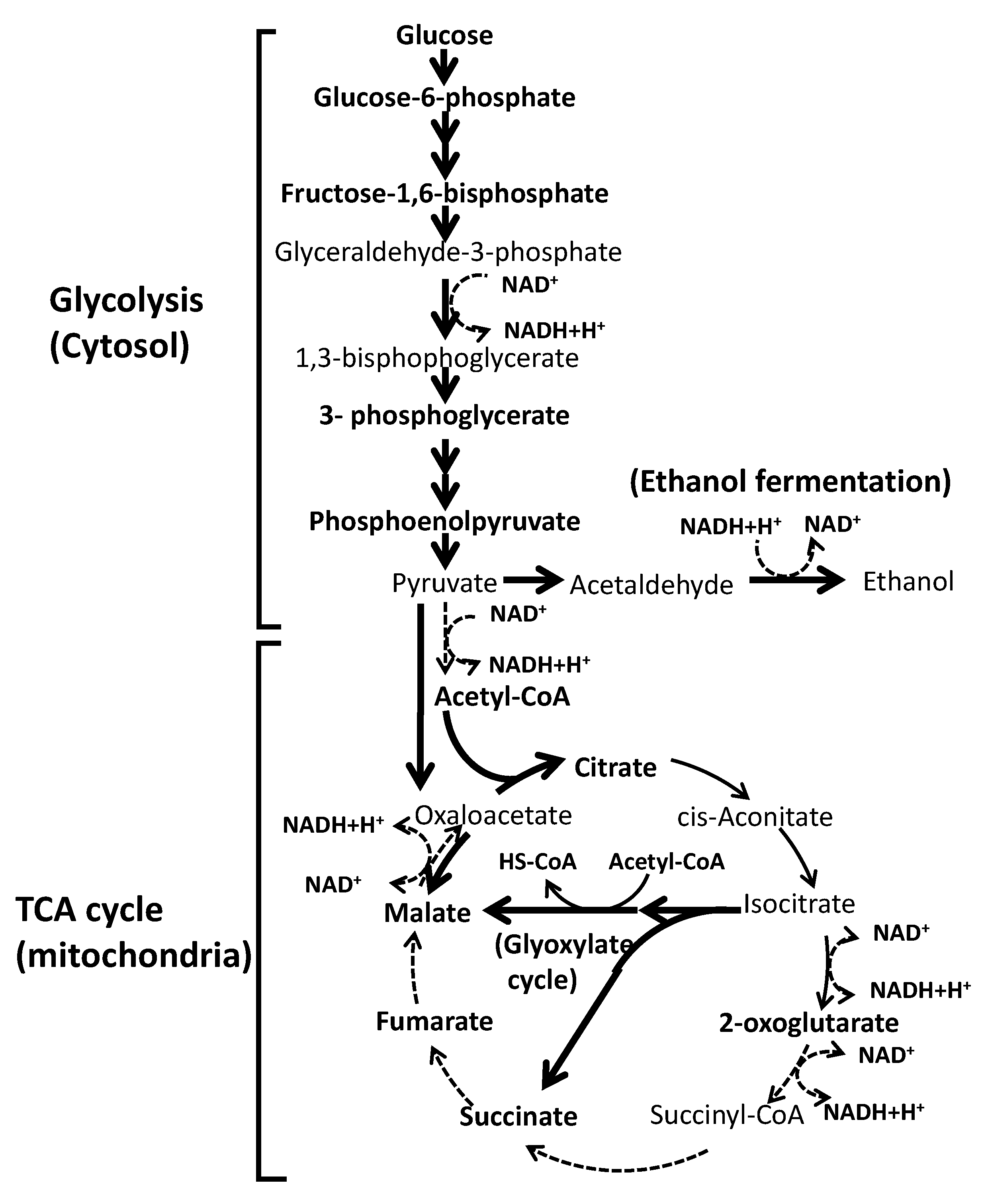
3.3.1. Metabolites of Glycolysis
3.3.2. Metabolites in the TCA Cycle
3.3.3. Relationship between the Restricted Metabolism and Piezosensitivity
3.4. Generation of Piezosensitive Mutants from Niigata-Sake Yeast Strain S9arg
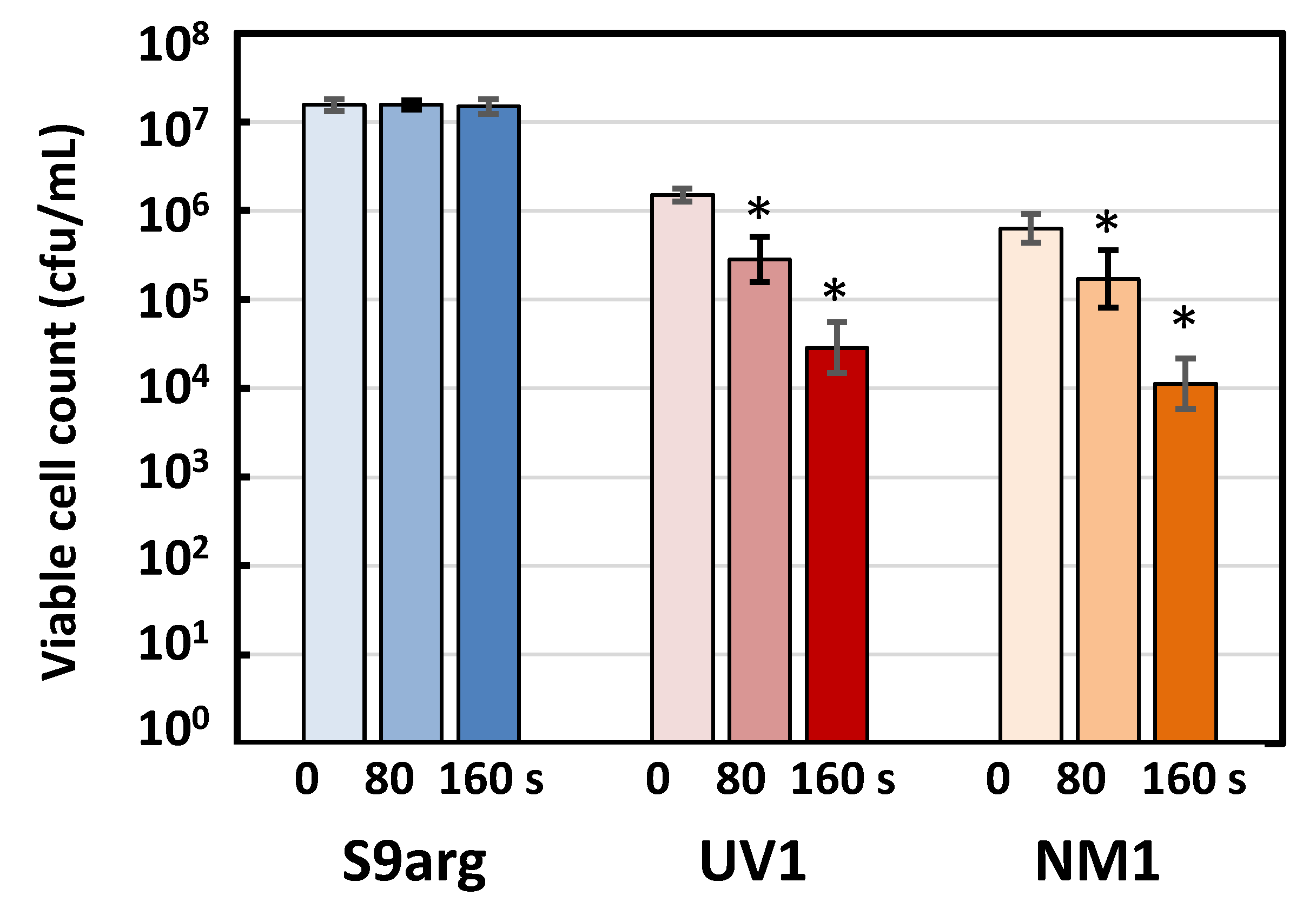
3.4.1. Screening and Obtain the Piezosensitive Mutants Derived from Strain S9arg
3.4.2. Pressure Inactivation Characteristics of Strains UV1 and NM1
3.4.3. Laboratory-Scale Sake Brewing Test
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Shigematsu, T.; Nasuhara, Y.; Nagai, G.; Nomura, K.; Ikarashi, K.; Hirayama, M.; Hayashi, M.; Ueno, S.; Fujii, T. Isolation and characterization of barosensitive mutants of Saccharomyces cerevisiae obtained by UV mutagenesis. J. Food Sci. 2010, 75, M509–M514. [Google Scholar] [CrossRef] [PubMed]
- Shigematsu, T.; Nomura, K.; Nasuhara, Y.; Ikarashi, K.; Nagai, G.; Hirayama, M.; Hayashi, M.; Ueno, S.; Fujii, T. Thermosensitivity of a barosensitive Saccharomyces cerevisiae mutant obtained by UV mutagenesis. High Press. Res. 2010, 30, 524–529. [Google Scholar] [CrossRef]
- Nanba, M.; Nomura, K.; Nasuhara, Y.; Hayashi, M.; Kido, M.; Hayashi, M.; Iguchi, A.; Shigematsu, T. Importance of cell-damage causing growth delay for high-pressure inactivation of Saccharomyces cerevisiae. High Press. Res. 2013, 33, 299–307. [Google Scholar] [CrossRef]
- Nomura, K.; Iwahashi, H. Pressure-regulated fermentation: A revolutionary approach that utilizes hydrostatic pressure. Rev. Agric. Sci. 2014, 2, 1–10. [Google Scholar] [CrossRef]
- Nomura, K.; Ikezaki, M.; Kataoka, C.; Hori, S.; Aoki, T.; Kuribayashi, T.; Kaneoke, M.; Iguchi, A.; Shigematsu, T. High hydrostatic pressure pasteurization of a draft sake brewed using a Niigata-sake yeast. High Press. Res. 2019, 39, 301–312. [Google Scholar] [CrossRef]
- Shigematsu, T.; Takahashi, I.; Aoki, T.; Kaneoke, M.; Sekine, A.; Miyawaki, T.; Maeda, S.; Ito, M. Development of a sparkling cloudy draft sake “AWANAMA” using high hydrostatic pressure pasteurization technology. Rev. High Press. Sci. Technol. 2019, 29, 22–30. [Google Scholar] [CrossRef]
- Nomura, K.; Kuwabara, Y.; Kuwabara, W.; Takahashi, H.; Nakajima, K.; Hayashi, M.; Iguchi, A.; Shigematsu, T. Comparative analysis on inactivation kinetics of between piezotolerant and piezosensitive mutant strains of Saccharomyces cerevisiae under combinations of high hydrostatic pressure and temperature. Biophys. Chem. 2017, 231, 87–94. [Google Scholar] [CrossRef] [PubMed]
- Nomura, K.; Iwahashi, H.; Iguchi, A.; Shigematsu, T. Barosensitivity in Saccharomyces cerevisiae is closely associated with a deletion of the COX1 gene. J. Food Sci. 2015, 80, M1051–M1059. [Google Scholar] [CrossRef] [PubMed]
- Kuribayashi, T.; Sato, K.; Joh, T.; Kaneoke, M. Breeding of a non-urea-producing sake yeast carrying a FAS2 mutation. Mycoscience 2017, 58, 302–306. [Google Scholar] [CrossRef]
- Nagai, S. Induction of the respiration-deficient mutation in yeast by various synthetic dyes. Science 1959, 130, 1188–1189. [Google Scholar] [CrossRef] [PubMed]
- Huxley, C.; Green, E.D.; Dunham, I. Rapid assessment of S. cerevisiae mating type by PCR. Trends Genet. 1990, 6, 236. [Google Scholar] [CrossRef] [PubMed]
- Hasunuma, T.; Sanda, T.; Yamada, R.; Yoshimura, K.; Ishii, J.; Kondo, A. Metabolic pathway engineering based on metabolomics confers acetic and formic acid tolerance to a recombinant xylose-fermenting strain of Saccharomyces cerevisiae. Microbial. Cell Fact. 2011, 10, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harada, K.; Ohyama, Y.; Tabushi, T.; Kobayashi, A.; Fukusaki, E. Quantitative analysis of anionic metabolites for Catharanthus roseus by capillary electrophoresis using sulfonated capillary coupled with electrospray ionization-tandem mass. J. Biosci. Bioeng. 2008, 105, 249–260. [Google Scholar] [CrossRef] [PubMed]
- Nomura, K.; Hoshino, H.; Igoshi, K.; Onozuka, H.; Tanaka, E.; Hayashi, M.; Yamazaki, H.; Takaku, H.; Iguchi, A.; Shigematsu, T. Brewing chajracteristics of piezosensitive sake yeasts. High Press. Res. 2018, 38, 165–176. [Google Scholar] [CrossRef]
- Merz, S.; Wstermann, B. Genome-wide deletion mutant analysis reveals genes required for respiratory growth, mitochondrial genome maintenance and mitochondrial protein synthesis in Saccharomyces cerevisiae. Genome Biol. 2009, 10, R95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Osumi, M. Effects of hydrostatic pressure to ultrastructure of yeast cells. In Pressure-Processed Food–Research and Development; Hayashi, R., Ed.; Sanei Shuppan: Kyoto, Japan, 1990; pp. 157–164. (In Japanese) [Google Scholar]
- Abe, F.; Horikoshi, K. Tryptophan permease gene TAT2 confers high-pressure growth in Saccharomyces cerevisiae. Mol. Cell. Biol. 2000, 20, 8093–8102. [Google Scholar] [CrossRef] [PubMed]
- Abe, F. Effect of growth-permissive pressures on the physiology of Saccharomyces cerevisiae. In High-Pressure Microbiology; Michiels, C., Bartlett, D.H., Aertsen, A., Eds.; ASM Press: Washington, DC, USA, 2008; pp. 167–179. [Google Scholar] [CrossRef]
- Abe, F.; Minegishi, H. Global screening of genes essential for growth in high pressure and cold environments: Searching for basic adaptive strategies using a yeast deletion library. Genetics 2008, 178, 851–872. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iwahashi, H.; Nwaka, S.; Obuchi, K. Evidence for the contribution of neutral trehalase in barotolerance of Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2000, 66, 5182−5185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kobori, H.; Sato, M.; Tameike, A.; Hamada, K.; Shimada, S.; Osumi, M. Ultrastructural effects of pressure stress to the nucleus in Saccharomyces cerevisiae: A study by immunoelectron microscopy using frozen thin sections. FEMS Microbiol. Lett. 1995, 132, 253−258. [Google Scholar] [CrossRef]
- Nomura, K.; Iwahashi, H.; Iguchi, A.; Shigematsu, T. Depletion of arginine in yeast cells decreases the resistance to hydrostatic pressure. High Press. Res. 2015, 35, 310–316. [Google Scholar] [CrossRef]


| Primer | Sequence | Product Size (bp) |
|---|---|---|
| COX1_1F | 5’-TTTTTAGTGGTATGGCAGGAACAG-3’ | 2576 |
| COX1_1R | 5’-TAATACAGCATGACCAACTACTAA-3’ | |
| COX1_2F | 5’-TTAGTAGTTGGTCATGCTGTATTA-3’ | 2546 |
| COX1_2R | 5’-ACCTCCAATTAAAGCAGGCATTAC-3’ | |
| COX1_3F | 5’-GTAATGCCTGCTTTAATTGGAGGT-3’ | 3225 |
| COX1_3R | 5’-TGCTCTAAGATCTGCATCTAATCC-3’ | |
| COX1_4F | 5’-GCTACAGATACAGCATTTCCAAGA-3’ | 3461 |
| COX1_4R | 5’-GTTAGCTAAGGCAACACCAGTTAAA-3’ | |
| COX1_5F | 5’-TTTAACTGGTGTTGCCTTAGCTAAC-3’ | 2978 |
| COX1_5R | 5’-AGTGTACAGCTGGTGGAGAAGTTA-3’ |
| Compound | Q1 (m/z) | Q3 (m/z) |
|---|---|---|
| Glucose (and Fructose) 1 | 178.9 | 88.8 |
| Glucose-6-phosphate (and Glucose-1-phosphate) 2 | 258.8 | 78.9 |
| Fructose-1,6-bisphophate | 339.0 | 78.9 |
| 3-phosphoglycerate | 184.7 | 79.0 |
| Phosphoenolpyruvate | 166.7 | 79.0 |
| Acetyl coenzyme A (Acetyl-CoA) | 807.6 | 79.0 |
| Citrate | 190.9 | 86.8 |
| 2-oxoglutarate | 144.8 | 101.1 |
| Succinate | 116.9 | 72.9 |
| Fumarate | 114.2 | 72.9 |
| Malate | 132.8 | 114.9 |
| Nicotinamide adenine dinucleotide (NAD+) | 661.7 | 539.9 |
| Reduced nicotinamide adenine dinucleotide (NADH) | 663.4 | 78.8 |
| 1,4-piperazinediethanesulfonate (PIPES) | 300.7 | 192.9 |
| S9arg | UV1 | NM1 | K7 | |
|---|---|---|---|---|
| Ethanol (%) | 12.6 ± 2.3 | 14.6± 3.5 | 11.6 ± 2.0 | 14.5 ± 1.5 |
| Flavor components (mg/L) | ||||
| Isoamyl acetate | 0.82 ± 0.06 | 0.70 ± 0.12 | 0.72 ± 0.12 | 0.74 ± 0.63 |
| Isoamyl alcohol | 68.4 ± 13.6 | 71.4 ± 7.5 | 78.0± 12.0 | 120.7 ± 31.6 * |
| Ethyl caproate | 7.35 ± 3.95 | 6.94 ± 3.57 | 4.01 ± 1.22 | 0.41 ± 0.21 * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shigematsu, T.; Kaneko, Y.; Ikezaki, M.; Kataoka, C.; Nomura, K.; Nakano, A.; Aii, J.; Aoki, T.; Kuribayashi, T.; Kaneoke, M.; et al. Genomic and Metabolomic Analyses of a Piezosensitive Mutant of Saccharomyces cerevisiae and Application for Generation of Piezosensitive Niigata-Sake Yeast Strains. Foods 2021, 10, 2247. https://doi.org/10.3390/foods10102247
Shigematsu T, Kaneko Y, Ikezaki M, Kataoka C, Nomura K, Nakano A, Aii J, Aoki T, Kuribayashi T, Kaneoke M, et al. Genomic and Metabolomic Analyses of a Piezosensitive Mutant of Saccharomyces cerevisiae and Application for Generation of Piezosensitive Niigata-Sake Yeast Strains. Foods. 2021; 10(10):2247. https://doi.org/10.3390/foods10102247
Chicago/Turabian StyleShigematsu, Toru, Yuta Kaneko, Minami Ikezaki, Chihiro Kataoka, Kazuki Nomura, Ayana Nakano, Jotaro Aii, Toshio Aoki, Takashi Kuribayashi, Mitsuoki Kaneoke, and et al. 2021. "Genomic and Metabolomic Analyses of a Piezosensitive Mutant of Saccharomyces cerevisiae and Application for Generation of Piezosensitive Niigata-Sake Yeast Strains" Foods 10, no. 10: 2247. https://doi.org/10.3390/foods10102247
APA StyleShigematsu, T., Kaneko, Y., Ikezaki, M., Kataoka, C., Nomura, K., Nakano, A., Aii, J., Aoki, T., Kuribayashi, T., Kaneoke, M., Hori, S., & Iguchi, A. (2021). Genomic and Metabolomic Analyses of a Piezosensitive Mutant of Saccharomyces cerevisiae and Application for Generation of Piezosensitive Niigata-Sake Yeast Strains. Foods, 10(10), 2247. https://doi.org/10.3390/foods10102247