Substantial Differences in the Subgingival Microbiome Measured by 16S Metagenomics According to Periodontitis Status in Older Women
Abstract
1. Introduction
2. Results
2.1. Characteristics of Study Group
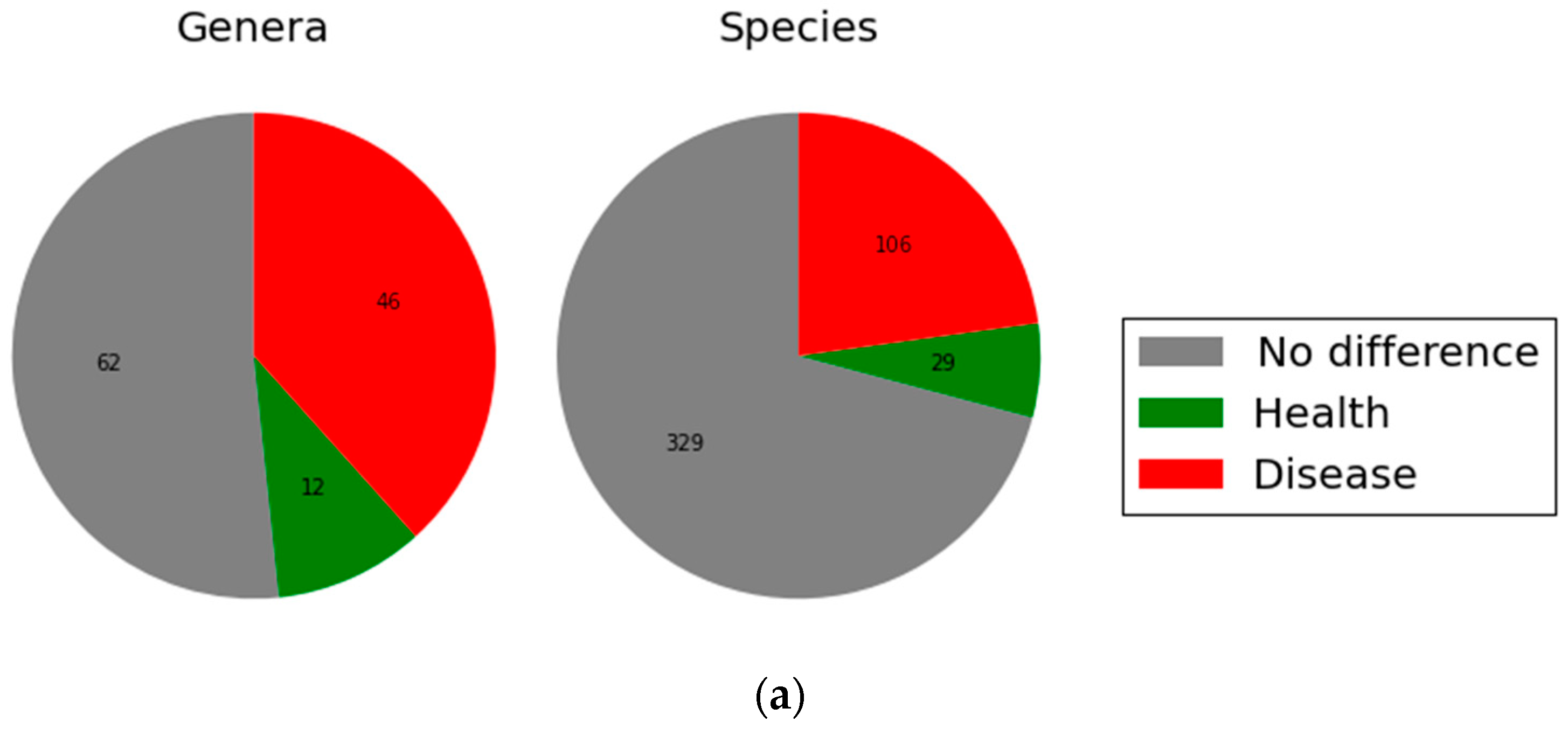
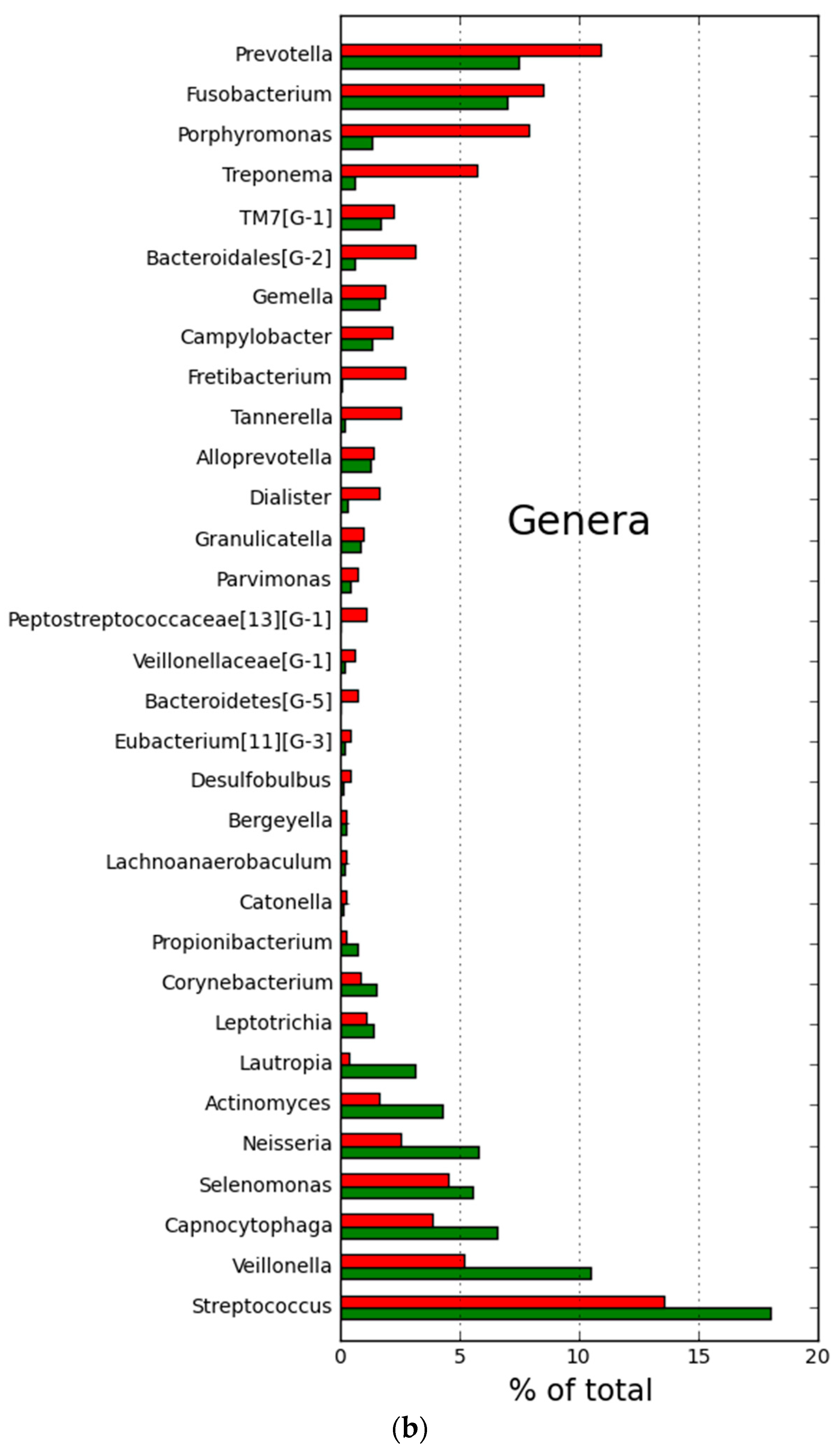
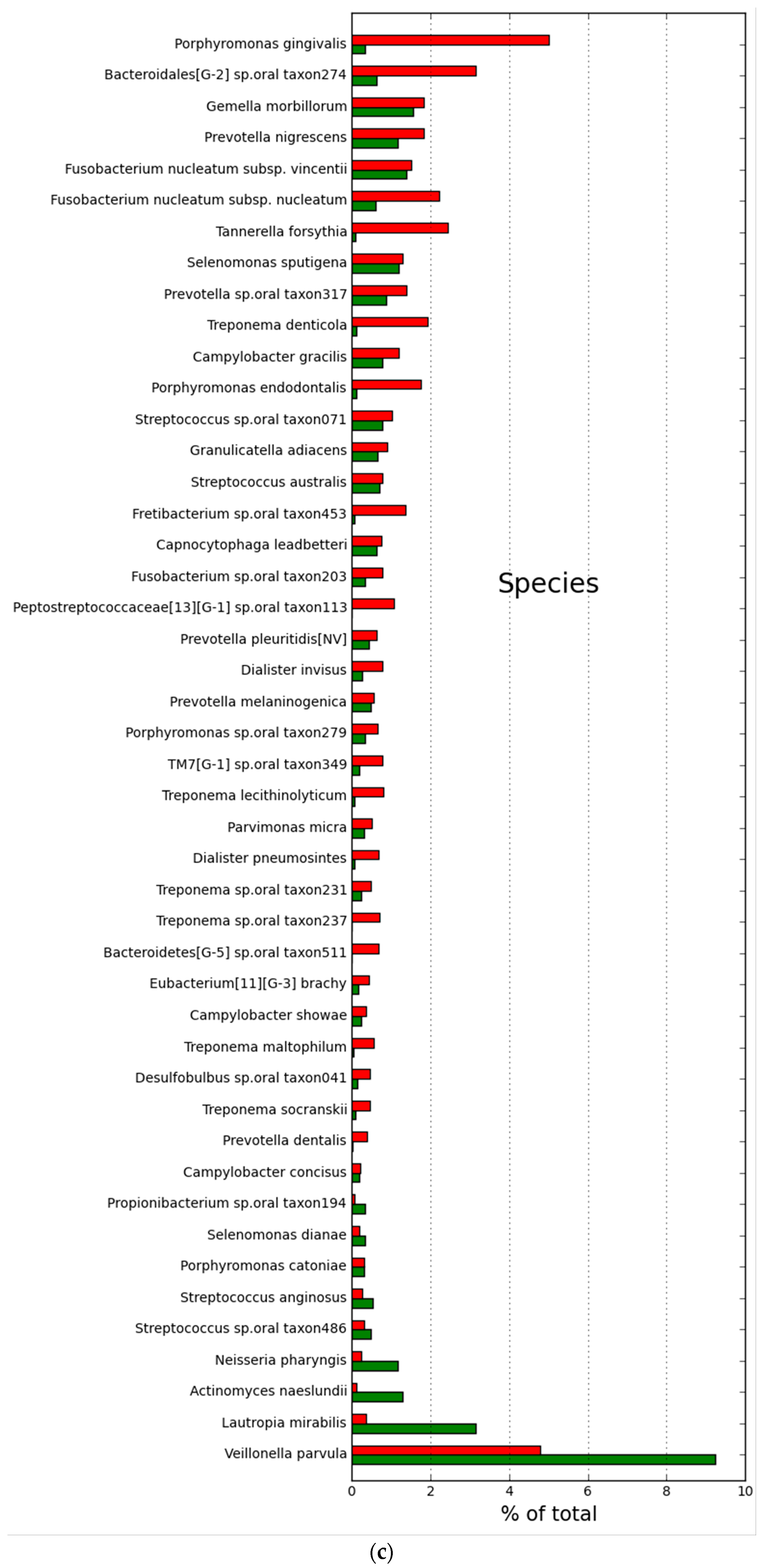
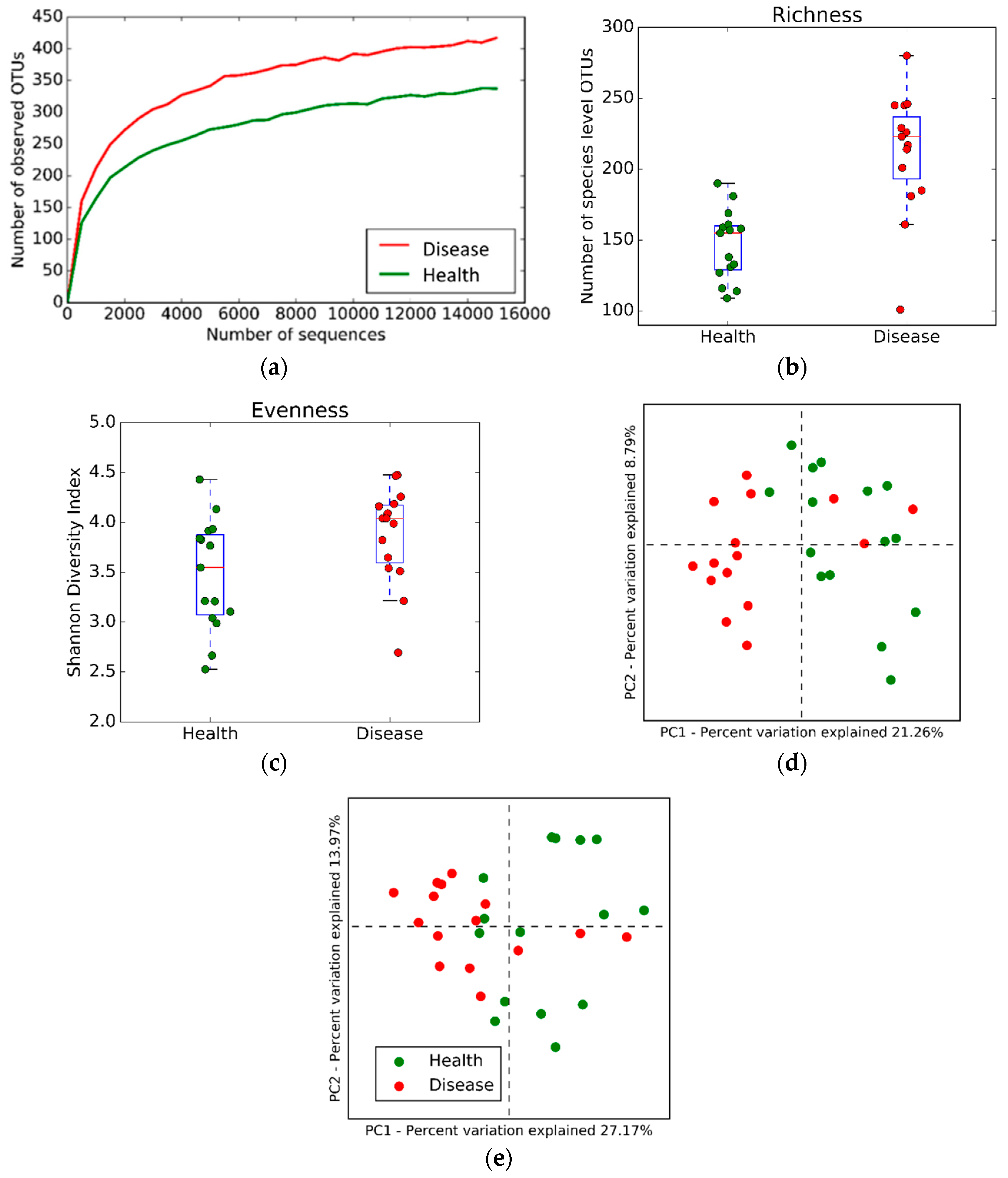
2.2. Microbial Community Structure and Composition
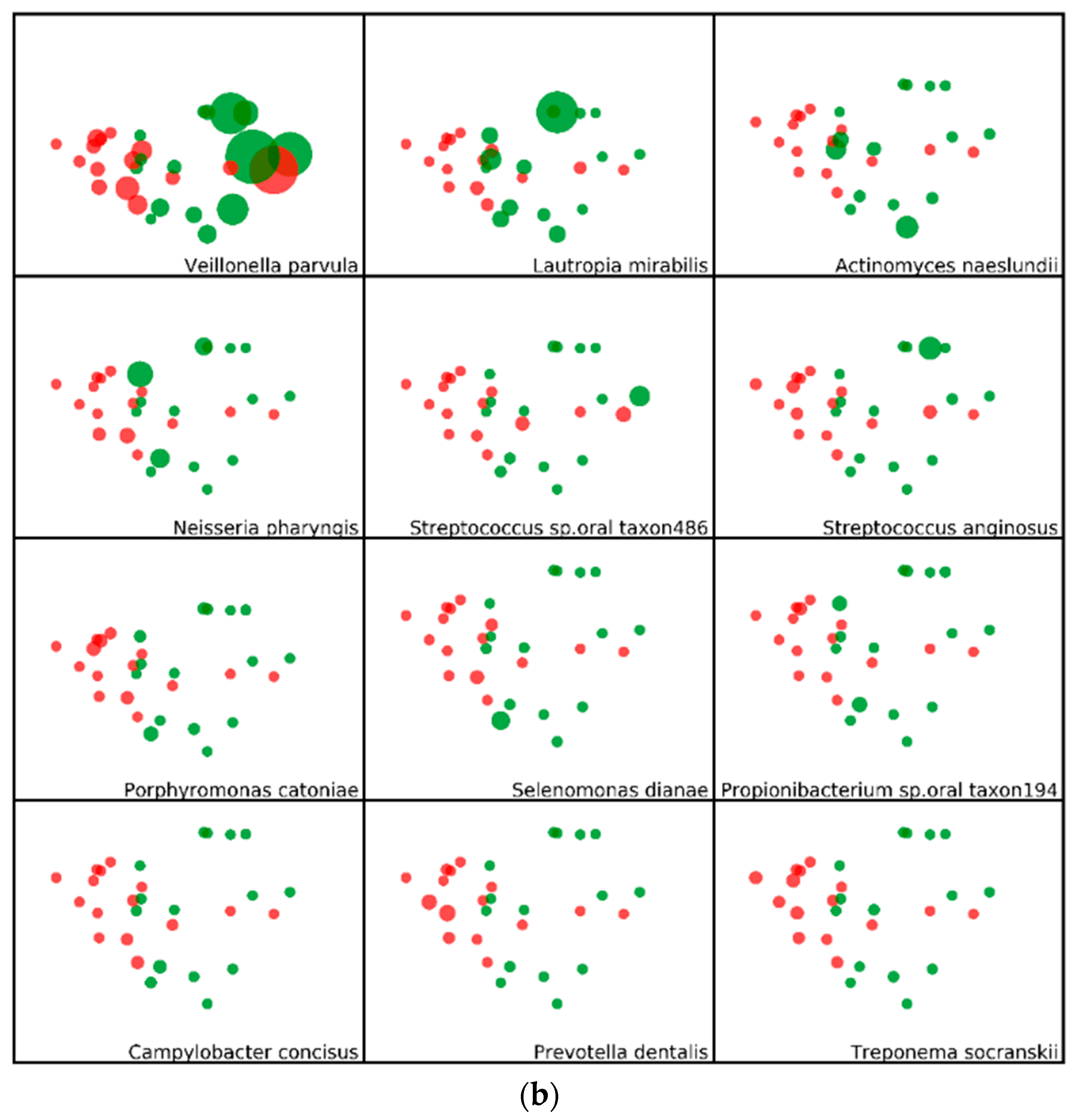
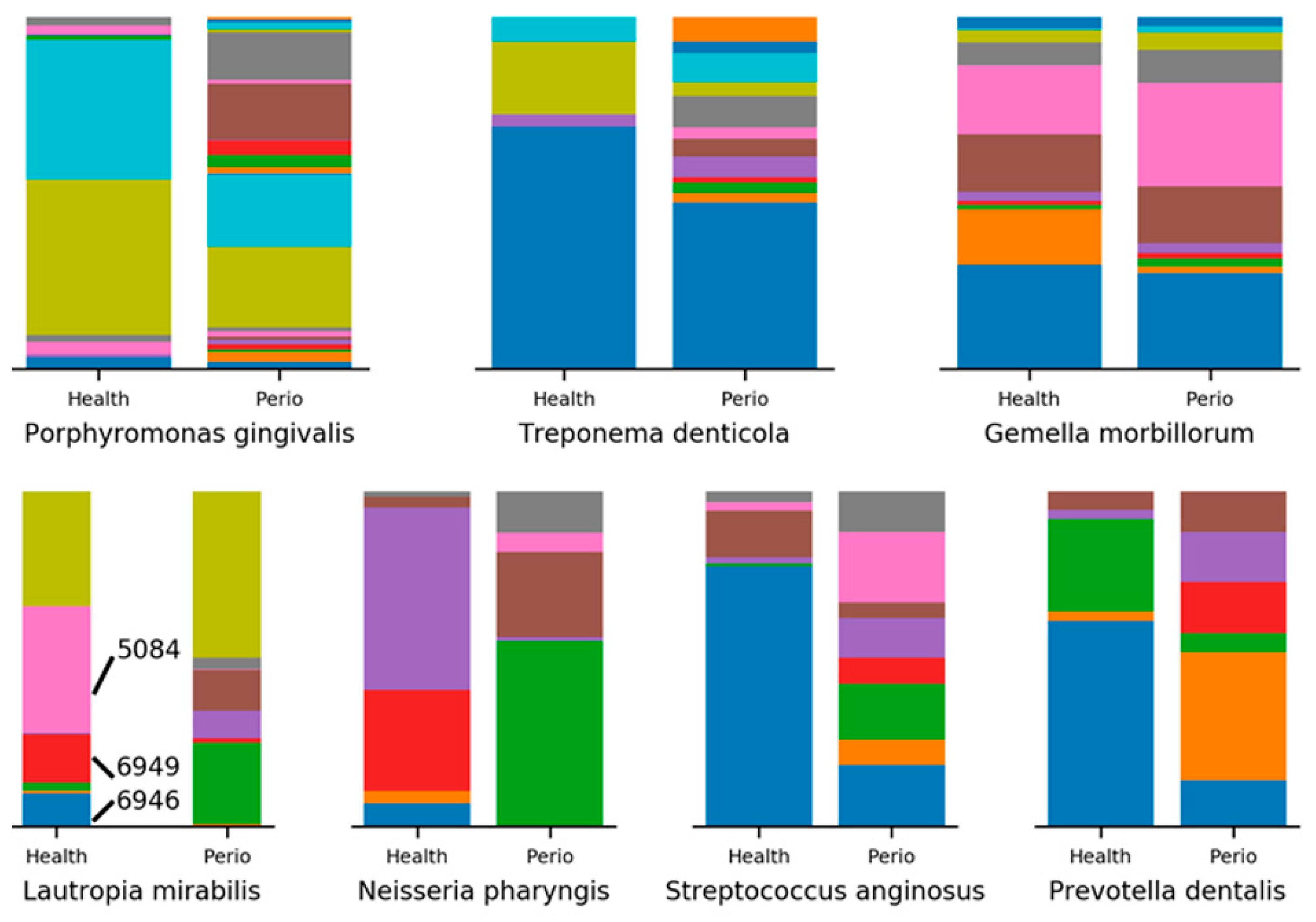
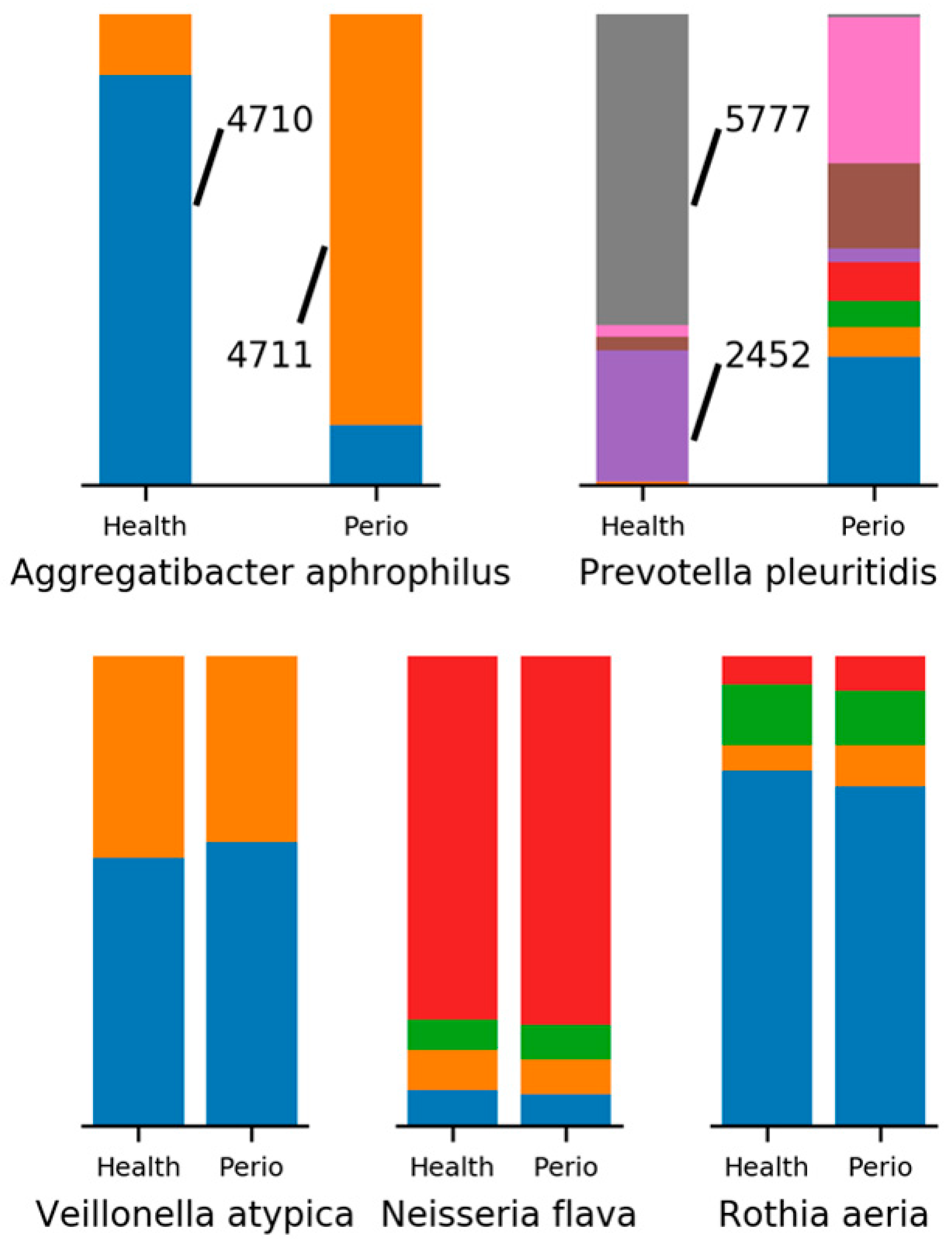
2.3. Minimum Entropy Decomposition Analysis
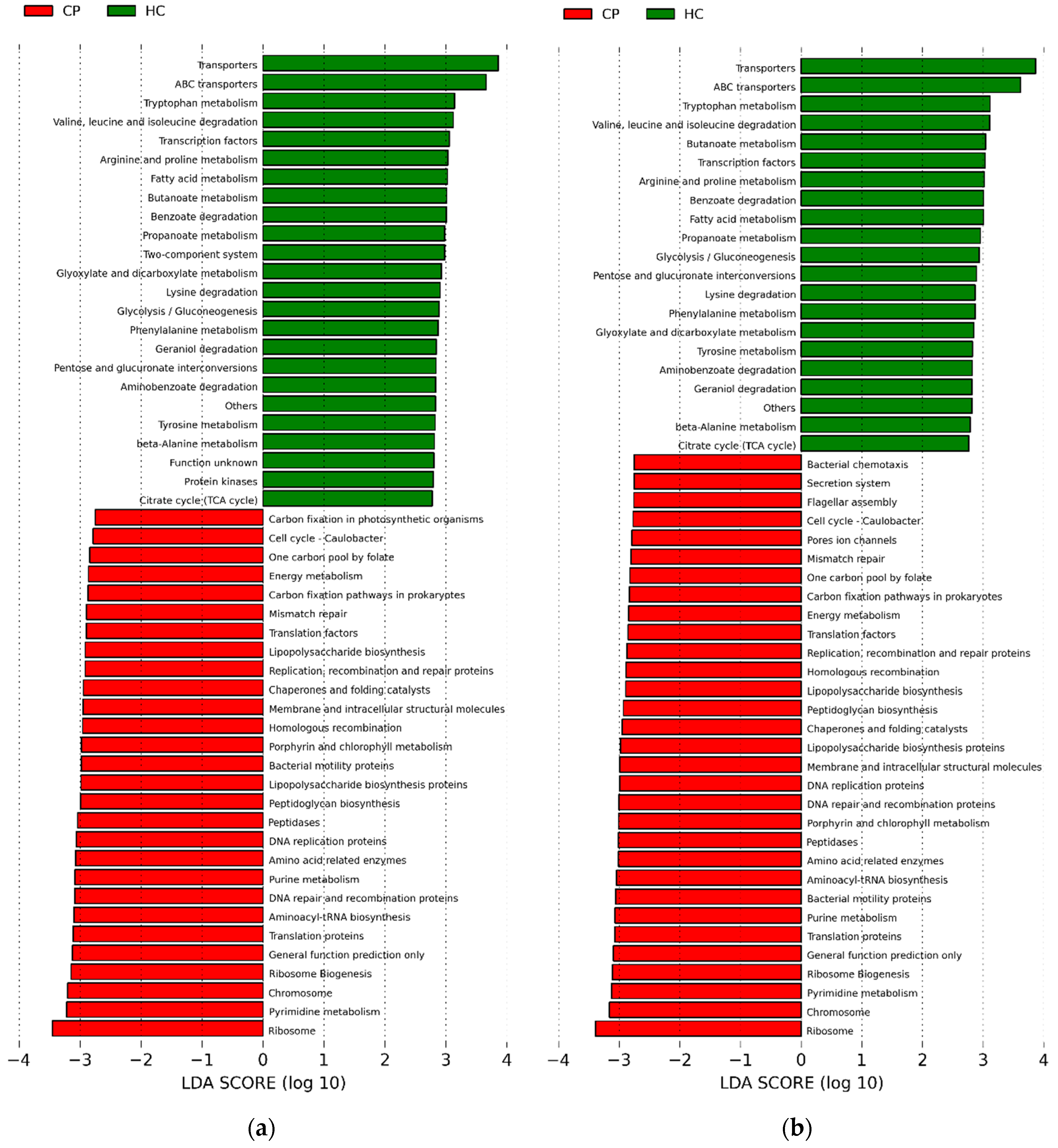
2.4. Potential Functions of Subgingival Bacteria
3. Discussion
4. Materials and Methods
4.1. Participants
4.2. Subgingival Plaque Samples
4.3. 16S rRNA Sequencing, Processing, and Data Analysis
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Availability of Data and Materials:
Abbreviations
| CAL | clinical attachment level |
| DNA | deoxyribonucleic acid |
| HOMD | human oral microbiome database |
| LDA | linear discriminant analysis |
| MED | minimum entropy decomposition analysis |
| NGS | next generation sequencing |
| OS | observational study |
| OsteoPerio | osteoporosis and periodontal disease study |
| OTU | operational taxonomic unit |
| PD | pocket depth |
| rRNA | ribosomal ribonucleic acid |
| WHI | women’s health initiative |
References
- Weinert, B.T.; Timiras, P.S. Invited review: Theories of aging. J. Appl. Physiol. 2003, 95, 1706–1716. [Google Scholar] [CrossRef] [PubMed]
- Dewhirst, F.E.; Chen, T.; Izard, J.; Paster, B.J.; Tanner, A.; Yu, W.H.; Lakshamanan, A.; Wade, W.G. The Human Oral Microbiome. J. Bacteriol. 2010, 192, 5002–5017. [Google Scholar] [CrossRef] [PubMed]
- Feres, M.; Teles, F.; Teles, R.; Figueiredo, L.C.; Faveri, M. The subgingival periodontal microbiota of the aging mouth. Periodontology 2016, 72, 30–53. [Google Scholar] [CrossRef] [PubMed]
- Langille, M.G.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Vega Thurber, R.L.; Knight, R.; et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef] [PubMed]
- Griffen, A.L.; Beall, C.; Campbell, J.H.; Firestone, N.; Kumar, P.; Yang, Z.; Podar, M.; Leys, E.J. Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. ISME J. 2012, 6, 1176–1185. [Google Scholar] [CrossRef] [PubMed]
- Paulson, J.N.; Stine, O.C.; Bravo, H.C.; Pop, M. Differential abundance analysis for microbial marker-gene surveys. Nat. Methods 2013, 10, 1200–1205. [Google Scholar] [CrossRef] [PubMed]
- Paster, B.J.; Olsen, I.; Aas, J.A.; Dewhirst, F.E. The breadth of bacterial diversity in the human periodontal pocket and other oral sites. Periodontology 2006, 42, 80–87. [Google Scholar] [CrossRef] [PubMed]
- Keijser, B.J.F.; Huse, S.M.; van der Vossen, J.; Schuren, F.; Martijin, R.; Cate, J.; Crelaard, W. Pyrosequencing analysis of the oral microflora of health adults. J. Dent. Res. 2008, 87, 1016–1020. [Google Scholar] [CrossRef] [PubMed]
- Kirst, M.E.; Li, E.C.; Alfant, B.; Chi, Y.-Y.; Walker, C.; Magnusson, I.; Wang, G.P. Dysbiosis and alterations in predicted functions of the subgingival microbiome in chronic periodontitis. Appl. Environ. Microbiol. 2015, 81, 783–793. [Google Scholar] [CrossRef] [PubMed]
- Socransky, S.S.; Haffajee, A.D. Periodontal microbial ecology. Periodontology 2005, 38, 135–187. [Google Scholar] [CrossRef] [PubMed]
- Teles, R.; Teles, F.; Frias-Lopez, J.; Paster, B.; Haffajee, A. Lessons learned and unlearned in periodontal microbiology. Periodontology 2013, 62, 95–162. [Google Scholar] [CrossRef] [PubMed]
- Maraki, S.; Papadakis, I.; Chronakis, E.; Panagopolos, D.; Vakis, A. Aggregatibacter aphrophilus brain abscess secondary to primary tooth extraction: Case report and literature review. J. Microbiol. Immunol. Infect. 2016, 49, 119–122. [Google Scholar] [CrossRef] [PubMed]
- Akondi, H.; Rahimi, A. Haemophilus aphrophilus endocarditis after tongue piercing. Emerg. Infect. Dis. 2002, 8, 850. [Google Scholar] [CrossRef]
- Sakamoto, M.; Ohkusu, K.; Masaki, T.; Kako, H.; Ezaki, T.; Benno, Y. Prevotella pluritidis sp. nov.; isolated from pleural fluid. Int. J. Syst. Evolut. Microbiol. 2007, 57, 1725–1728. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.Y.; Antonopoulos, A.; Kalra, A.; Tonelli, A.D.; Khalife, W.T.; Schmidt, T.M.; Young, V.B. Decreased diversity of the fecal microbiome in recurrent Clostridium difficile-associated diarrhea. J. Infect. Dis. 2008, 197, 434–438. [Google Scholar] [CrossRef] [PubMed]
- Jorth, P.; Turner, K.H.; Gumus, P.; Nizam, N.; Buduneli, N.; Whiteley, M. Metatranscriptomics of the human oral microbiome during health and disease. mBio 2014, 5, e01012–14. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Qi, J.; Zhao, H.; He, S.; Zhang, Y.; Wei, S.; Zhao, F. Metagenomic sequencing reveals microbiota and its functional potential associated with periodontal disease. Sci. Rep. 2013, 3, 1843. [Google Scholar] [CrossRef] [PubMed]
- Yost, S.; Duran-Pinedo, A.E.; Teles, R.; Krishnan, K.; Frias-Lopez, J. Functional signatures of oral dysbiosis during periodontitis progression revealed by microbial metatranscriptome analysis. Genom. Med. 2015, 7, 27. [Google Scholar] [CrossRef] [PubMed]
- Duran-Pinedo, A.E.; Yost, S.; Frias-Lopez, J. Small RNA transcriptome of the oral microbiome during periodontitis progression. Appl. Environ. Microbiol. 2015, 81, 6688–6699. [Google Scholar] [CrossRef] [PubMed]
- Dabdoub, S.M.; Ganesan, S.M.; Kumar, P.S. Comparative metagenomics reveals taxonomically idiosyncratic yet functionally congruent communities in periodontitis. Sci. Rep. 2016, 6, 38993. [Google Scholar] [CrossRef] [PubMed]
- Duran-Pinedo, A.E.; Chen, T.; Teles, R.; Starr, J.R.; Wang, X.; Krishnan, K.; Frias-Lopez, J. Community-wide transcriptome of the oral microbiome in subjects with and without periodontitis. ISME J. 2014, 8, 1659–1672. [Google Scholar] [CrossRef] [PubMed]
- Griffin, S.O.; Jones, J.A.; Brunson, D.; Friffin, P.M.; Bailey, W.D. Burden of oral disease among older adults and implications for public health priorities. Am. J. Public Health 2012, 102, 411–418. [Google Scholar] [CrossRef] [PubMed]
- Scannapieco, F.A.; Cantos, A. Oral inflammation and infection, and chronic medical diseases: Implications for the elderly. Periodontology 2016, 72, 153–175. [Google Scholar] [CrossRef] [PubMed]
- World Medical Association. Declaration of Helsinki. Ethical principles for medical research involving human subject. JAMA 2013, 310, 2191–2194. [Google Scholar] [CrossRef] [PubMed]
- Langer, R.D.; White, E.; Lewis, C.E.; Kotchen, J.M.; Hendrix, S.L.; Trevisan, M. The Women’s Health Initiative Observational Study: Baseline characteristics of participants and reliability of baseline measures. Ann. Epidemiol. 2003, 13 (Suppl. 9), S107–S121. [Google Scholar] [CrossRef]
- Wactawski-Wende, J.; Hausmann, E.; Hovey, K.; Trevisan, M.; Grossi, S.; Genco, R.J. The association between osteoporosis and alveolar crestal height in postmenopausal women. J. Periodontol. 2005, 76 (Suppl. 11), 2116–2124. [Google Scholar] [CrossRef] [PubMed]
- LaMonte, M.J.; Hovey, K.M.; Genco, R.J.; Millen, A.E.; Trevisan, M.; Wactawski-Wende, J. Five year changes in periodontal disease measures among postmenopausal females: The Buffalo OsteoPerio Study. J. Periodontol. 2013, 84, 572–584. [Google Scholar] [CrossRef] [PubMed]
- Eke, P.I.; Page, R.C.; Wei, L.; Thornton-Evans, G.; Genco, R.J. Update of the case definitions for population-based surveillance of periodontitis. J. Periodontol. 2012, 83, 1449–1454. [Google Scholar] [CrossRef] [PubMed]
- Brennan, R.M.; Genco, R.J.; Wilding, G.E.; Hovey, K.M.; Trevisan, M.; Wactawski-Wende, J. Bacterial species in subgingival plaque and oral bone loss in postmenopausal women. J. Periodontol. 2007, 78, 1051–1061. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pena, A.G.; Goodrich, J.K.; Gordon, J.I. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 75, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genom. Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. 1995, 57, 289–300. [Google Scholar]

| Characteristic | Overall | Periodontal Disease Status | |
|---|---|---|---|
| - | - | None | Severe |
| N | 30 | 15 | 15 |
| Age (years) | 70.5 ± 7.6 | 67.7 ± 7.3 | 73.2 ± 7.1 |
| BMI * (kg/m2) | 25.6 ± 4.8 | 27.3 ± 5.4 | 23.9 ± 3.6 |
| Caucasian | 30 (100.0) | 15 (100.0) | 15 (100.0) |
| Education | - | - | - |
| High school | 8 (26.7) | 3 (20.0) | 5 (33.3) |
| College | 12 (40.0) | 6 (40.0) | 4 (26.7) |
| Postgraduate | 10 (33.3) | 6 (40.0) | 4 (26.7) |
| Smoking | - | - | - |
| Never | 14 (46.7) | 8 (53.3) | 6 (40.0) |
| Former | 16 (53.3) | 7 (46.7) | 9 (60.0) |
| Current | 0 | 0 | 0 |
| Hormone therapy use | - | - | - |
| Never | 11 (36.7) | 5 (33.3) | 6 (40.0) |
| Former | 12 (40.0) | 6 (40.0) | 6 (40.0) |
| Current | 7 (23.3) | 4 (26.7) | 3 (20.0) |
| History of diagnosed treated diabetes | 1 (3.3) | 0 | 1 (6.7) |
| No. teeth present | 23.9 ± 3.3 | 25.7 ± 2.4 | 22.1 ± 3.1 |
| Whole-mouth PD ** (mm) | 2.2 ± 0.6 | 1.9 ± 0.2 | 2.5 ± 0.7 |
| Whole-mouth CAL *** (mm) | 2.6 ± 0.9 | 1.8 ± 0.2 | 3.3 ± 0.8 |
| Sites bleeding on probing (%) | 17.0 ± 19.0 | 9.0 ± 6.0 | 24.0 ± 25.0 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
LaMonte, M.J.; Genco, R.J.; Zheng, W.; McSkimming, D.I.; Andrews, C.A.; Hovey, K.M.; Li, L.; Sun, Y.; Buck, M.J.; Millen, A.E.; et al. Substantial Differences in the Subgingival Microbiome Measured by 16S Metagenomics According to Periodontitis Status in Older Women. Dent. J. 2018, 6, 58. https://doi.org/10.3390/dj6040058
LaMonte MJ, Genco RJ, Zheng W, McSkimming DI, Andrews CA, Hovey KM, Li L, Sun Y, Buck MJ, Millen AE, et al. Substantial Differences in the Subgingival Microbiome Measured by 16S Metagenomics According to Periodontitis Status in Older Women. Dentistry Journal. 2018; 6(4):58. https://doi.org/10.3390/dj6040058
Chicago/Turabian StyleLaMonte, Michael J., Robert J. Genco, Wei Zheng, Daniel I. McSkimming, Christopher A. Andrews, Kathleen M. Hovey, Lu Li, Yijun Sun, Michael J. Buck, Amy E. Millen, and et al. 2018. "Substantial Differences in the Subgingival Microbiome Measured by 16S Metagenomics According to Periodontitis Status in Older Women" Dentistry Journal 6, no. 4: 58. https://doi.org/10.3390/dj6040058
APA StyleLaMonte, M. J., Genco, R. J., Zheng, W., McSkimming, D. I., Andrews, C. A., Hovey, K. M., Li, L., Sun, Y., Buck, M. J., Millen, A. E., Falkner, K. L., & Wactawski-Wende, J. (2018). Substantial Differences in the Subgingival Microbiome Measured by 16S Metagenomics According to Periodontitis Status in Older Women. Dentistry Journal, 6(4), 58. https://doi.org/10.3390/dj6040058





