Synthesis and Structural Characterization of Half-Sandwich Arene–Ruthenium(II) Complexes with Bis(imidazol-1-yl)methane, Imidazole and Benzimidazole
Abstract
:1. Introduction
2. Results and Discussion
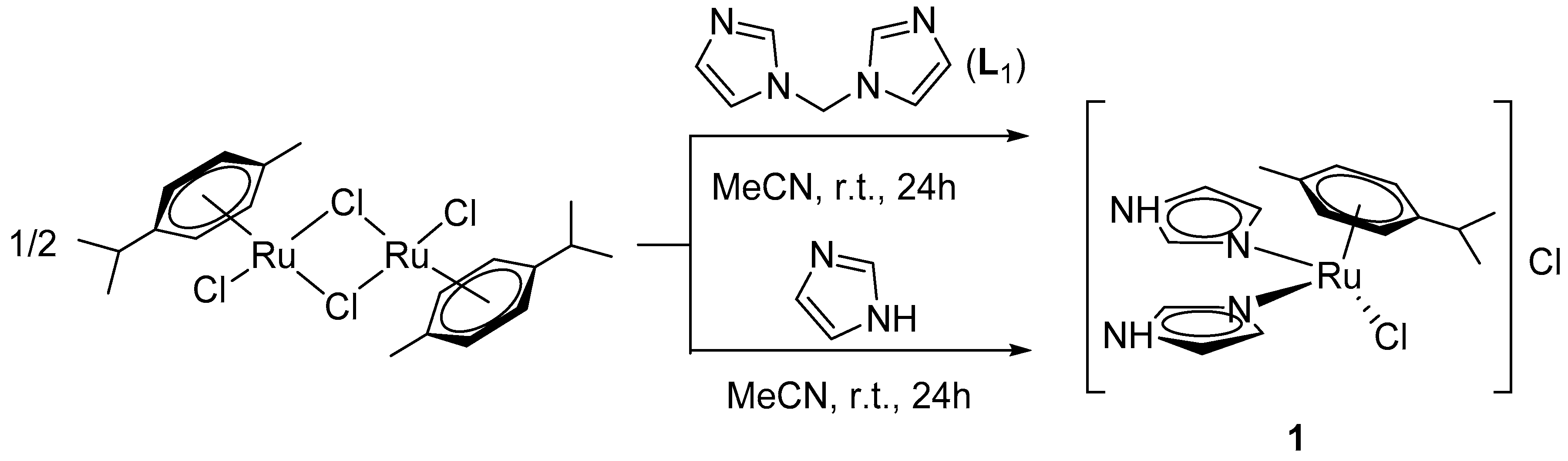
2.1. Synthesis of Coordination Compounds
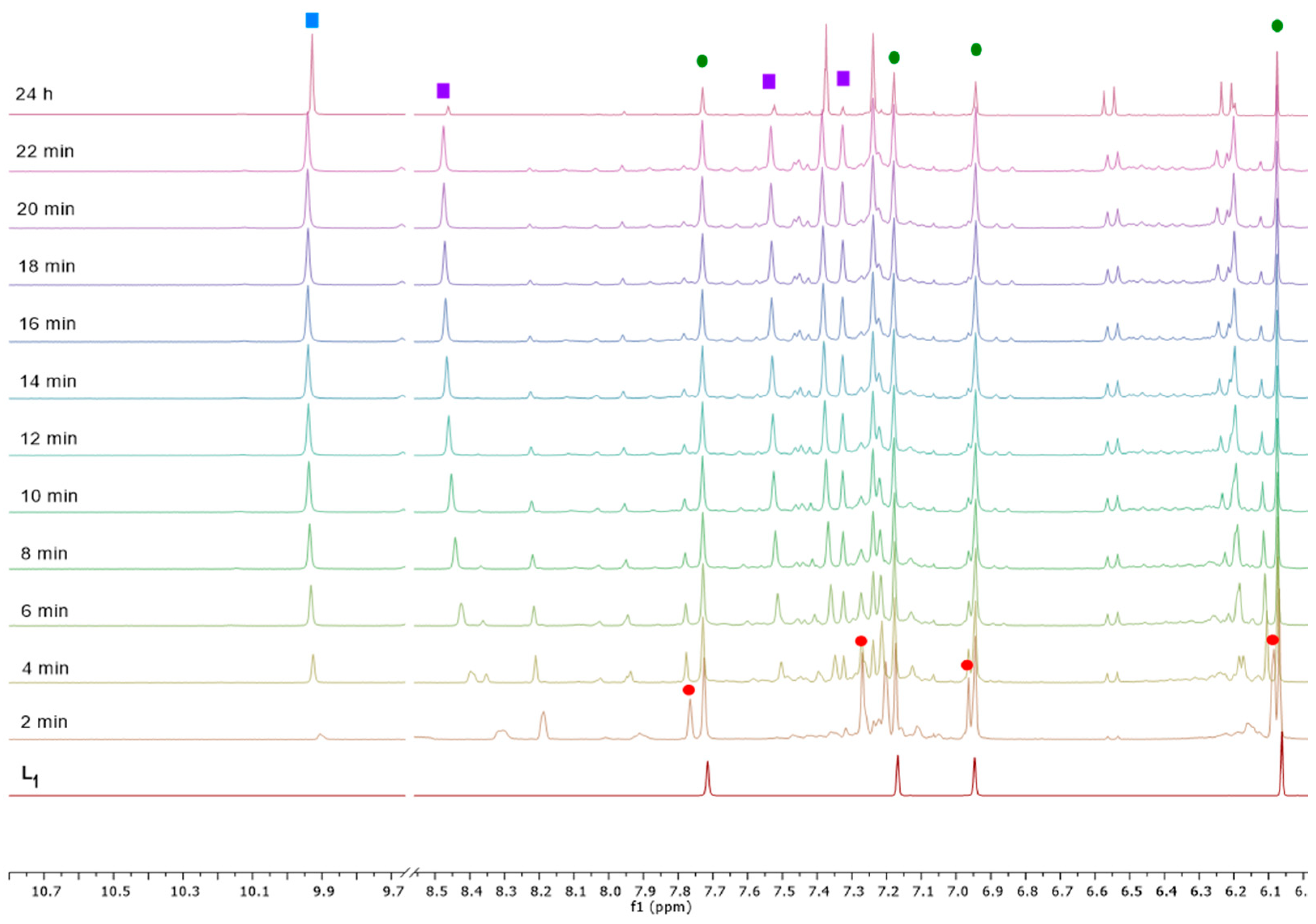
2.2. Spectroscopic Characterization
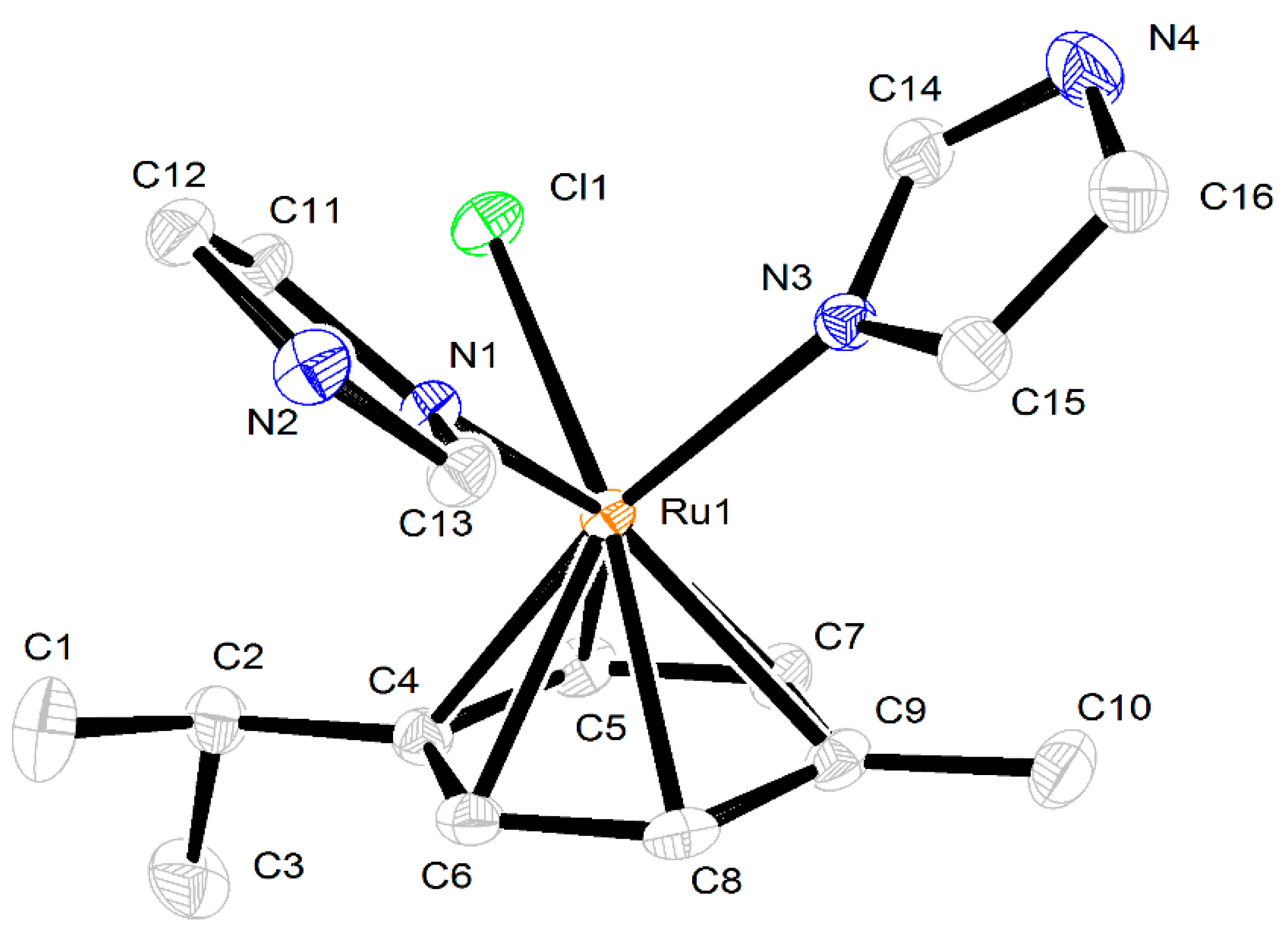
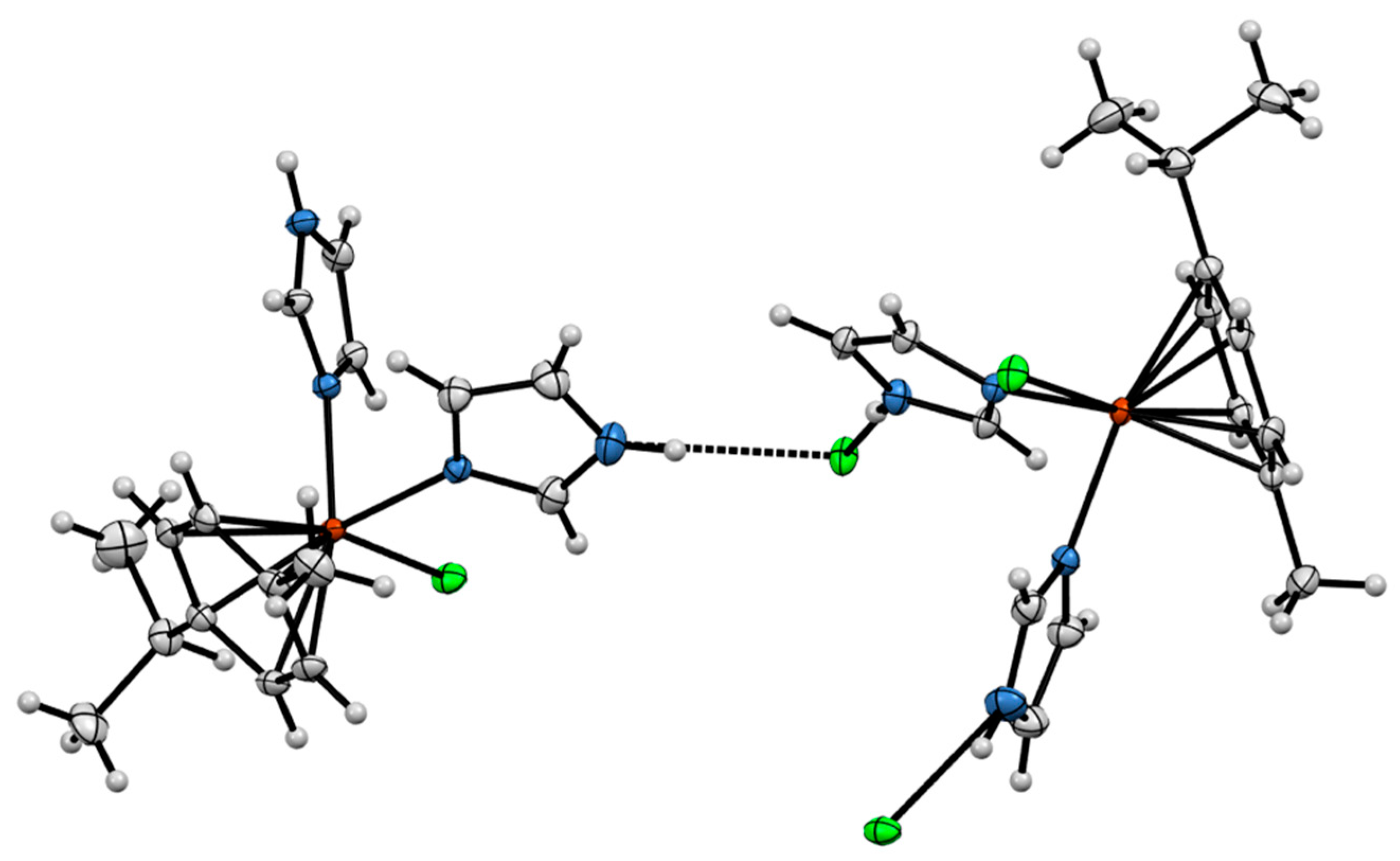
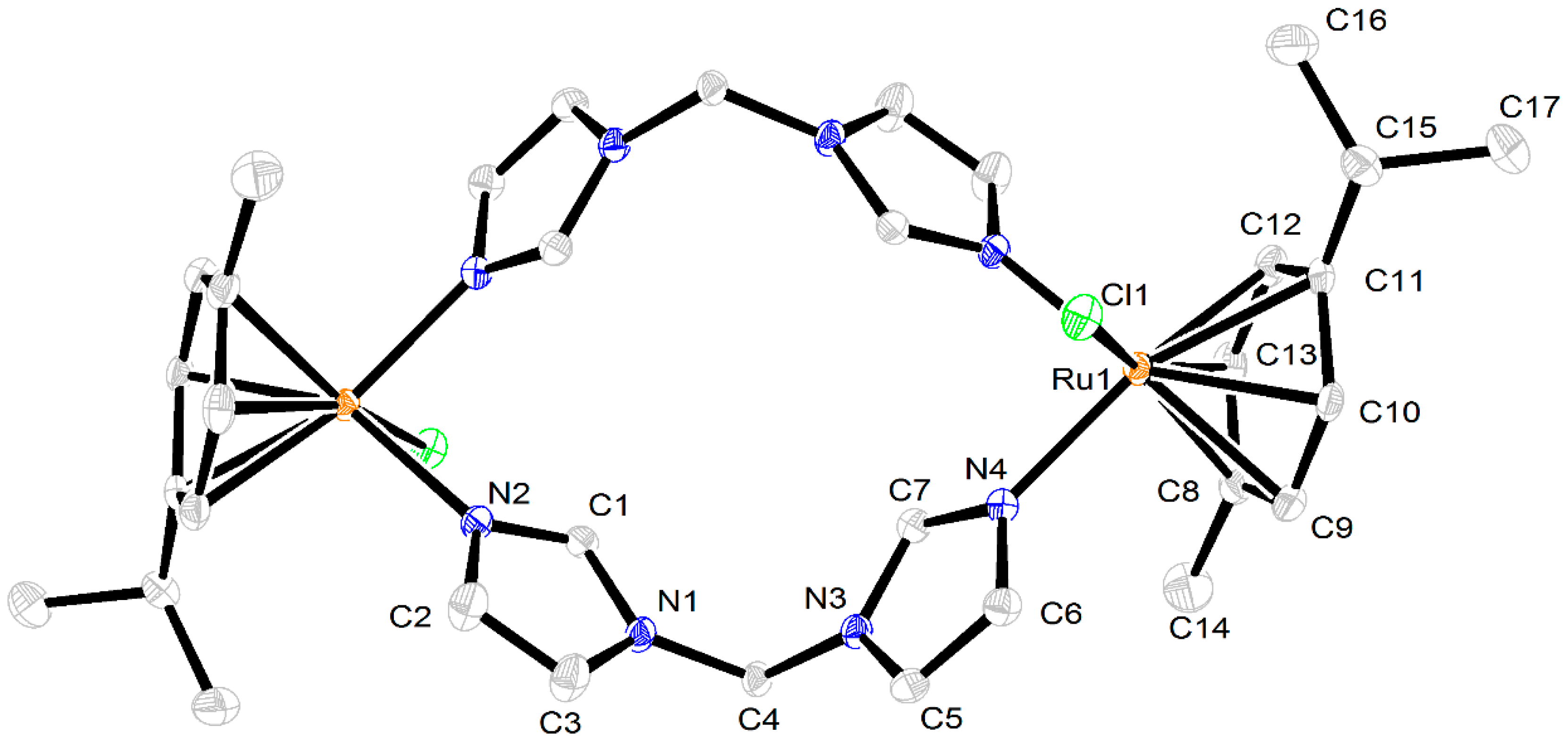
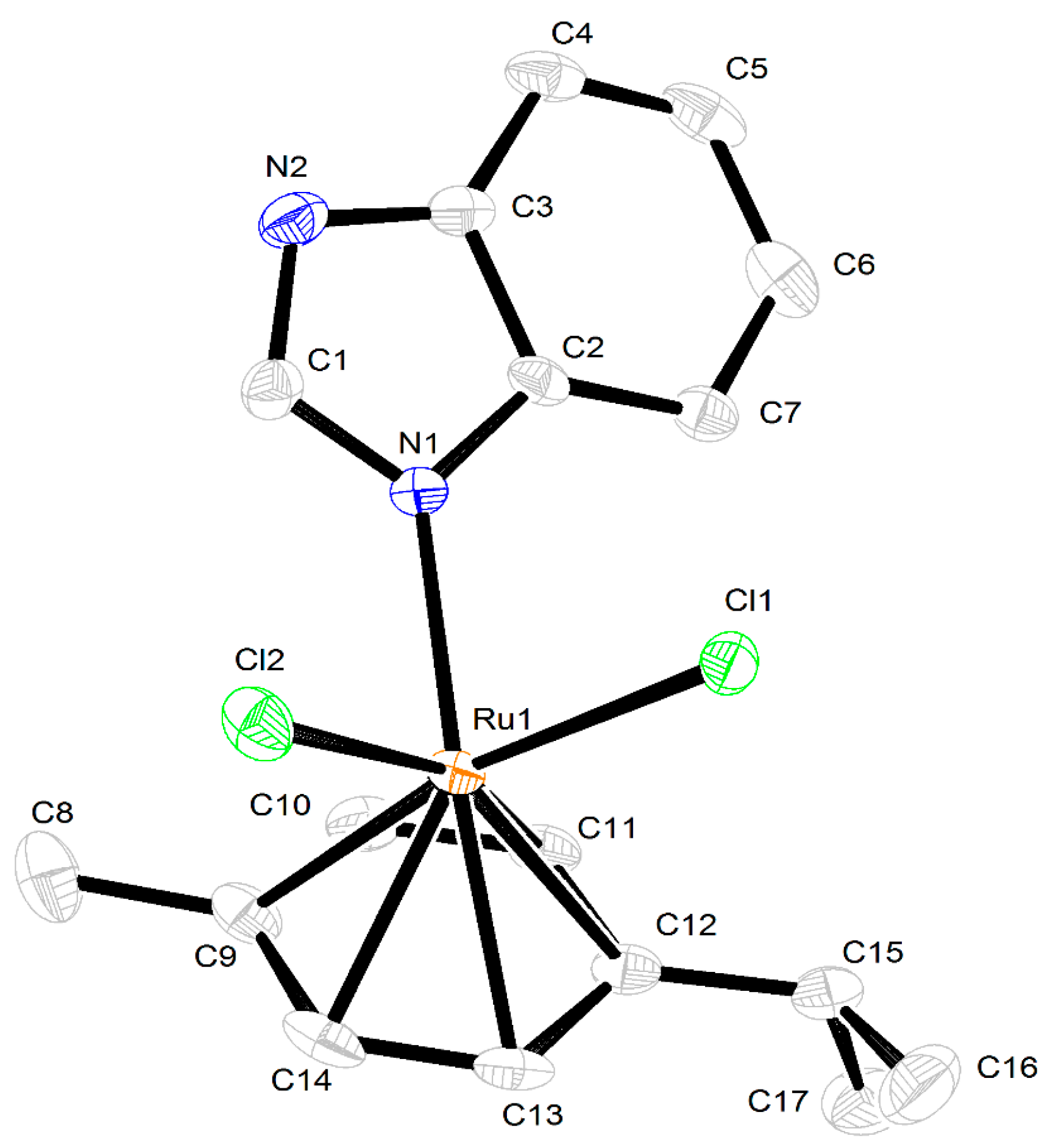
2.3. Crystal Structures of the Complexes
2.4. Cytoxicity Evaluation
2.5. Catalytic Activity of Complexes 1–3 in Transfer Hydrogenation
3. Materials and Methods
3.1. Synthesis of the Complexes
3.2. Spectral Methods and Elemental Analysis
3.3. X-ray Crystal Structure Determination
3.4. Cytotoxicity Study
3.5. General Procedure for Catalytic Transfer Hydrogenation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Štarha, P. Multinuclear biologically active Ru, Rh, Os and Ir arene complexes. Coord. Chem. Rev. 2021, 431, 213690. [Google Scholar] [CrossRef]
- Su, W.; Li, Y.; Li, P. Design of Ru-arene Complexes for Antitumor Drugs. Mini-Rev. Med. Chem. 2018, 18, 184–193. [Google Scholar] [CrossRef]
- Rilak Simović, A.; Masnikosa, R.; Bratsos, I.; Alessio, E. Chemistry and reactivity of ruthenium(II) complexes: DNA/protein binding mode and anticancer activity are related to the complex structure. Coord. Chem. Rev. 2019, 398, 113011. [Google Scholar] [CrossRef]
- Lee, S.Y.; Kim, C.Y.; Nam, T.-G. Ruthenium Complexes as Anticancer Agents: A Brief History and Perspectives. Drug Des. Devel. Ther. 2020, 14, 5375–5392. [Google Scholar] [CrossRef] [PubMed]
- Kar, B.; Roy, N.; Pete, S.; Moharana, P.; Paira, P. Ruthenium and iridium based mononuclear and multinuclear complexes: A Breakthrough of Next-Generation anticancer metallopharmaceuticals. Inorg. Chim. Acta 2020, 512, 119858. [Google Scholar] [CrossRef]
- Moharana, P.; Ghosh, D.; Paira, P. Drive to organoruthenium and organoiridium complexes from organoplatinum: Next-generation anticancer metallotherapeutics. Inorg. Chem. Commun. 2021, 124, 108364. [Google Scholar] [CrossRef]
- Rademaker-Lakhai, J.M.; Van Den Bongard, D.; Pluim, D.; Beijnen, J.H.; Schellens, J.H.M. A phase I and pharmacological study with imidazolium-trans-DMSO-imidazole-tetrachlororuthenate, a novel ruthenium anticancer agent. Clin. Cancer Res. 2004, 10, 3717–3727. [Google Scholar] [CrossRef] [Green Version]
- Hartinger, C.G.; Jakupec, M.A.; Zorbas-Seifried, S.; Groessl, M.; Egger, A.; Berger, W.; Zorbas, H.; Dyson, P.J.; Keppler, B.K. KP1019, A New Redox-Active Anticancer Agent—Preclinical Development and Results of a Clinical Phase I Study in Tumor Patients. Chem. Biodivers. 2008, 5, 2140–2155. [Google Scholar] [CrossRef]
- Monro, S.; Colón, K.L.; Yin, H.; Roque, J.; Konda, P.; Gujar, S.; Thummel, R.P.; Lilge, L.; Cameron, C.G.; McFarland, S.A. Transition Metal Complexes and Photodynamic Therapy from a Tumor-Centered Approach: Challenges, Opportunities, and Highlights from the Development of TLD1433. Chem. Rev. 2019, 119, 797–828. [Google Scholar] [CrossRef] [PubMed]
- Zeng, L.; Gupta, P.; Chen, Y.; Wang, E.; Ji, L.; Chao, H.; Chen, Z.-S. The development of anticancer ruthenium(II) complexes: From single molecule compounds to nanomaterials. Chem. Soc. Rev. 2017, 46, 5771–5804. [Google Scholar] [CrossRef]
- Meier-Menches, S.M.; Gerner, C.; Berger, W.; Hartinger, C.G.; Keppler, B.K. Structure-activity relationships for ruthenium and osmium anticancer agents-towards clinical development. Chem. Soc. Rev. 2018, 47, 909–928. [Google Scholar] [CrossRef]
- Bhambri, S.; Tocher, D.A. Synthesis and characterisation of ruthenium(II) arene complexes containing κ3- and κ2-poly(pyrazolyl)borates and methanes. J. Chem. Soc. Dalt. Trans. 1997, 18, 3367–3372. [Google Scholar] [CrossRef]
- Marchetti, F.; Pettinari, C.; Pettinari, R.; Cerquetella, A.; Di Nicola, C.; Macchioni, A.; Zuccaccia, D.; Monari, M.; Piccinelli, F. Synthesis and Intramolecular and Interionic Structural Characterization of Half-Sandwich (Arene)Ruthenium(II) Derivatives of Bis(Pyrazolyl)Alkanes. Inorg. Chem. 2008, 47, 11593–11603. [Google Scholar] [CrossRef] [PubMed]
- Montani, M.; Pazmay, G.V.B.; Hysi, A.; Lupidi, G.; Pettinari, R.; Gambini, V.; Tilio, M.; Marchetti, F.; Pettinari, C.; Ferraro, S.; et al. The water soluble ruthenium(II) organometallic compound [Ru(p-cymene)(bis(3,5-dimethylpyrazol-1-yl)methane)Cl]Cl suppresses triple negative breast cancer growth by inhibiting tumor infiltration of regulatory T cells. Pharmacol. Res. 2016, 107, 202–290. [Google Scholar] [CrossRef]
- Nielsen, T.O.; Hsu, F.D.; Jensen, K.; Cheang, M.; Karaca, G.; Hu, Z.; Hernandez-Boussard, T.; Livasy, C.; Cowan, D.; Dressler, L.; et al. Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clin. Cancer Res. 2004, 10, 5367–5374. [Google Scholar] [CrossRef] [Green Version]
- Bellachioma, G.; Cardaci, G.; Gramlich, V.; Macchioni, A.; Pieroni, F.; Venanzi, L.M. Synthesis and characterisation of bis- and tris-(pyrazol-1-yl)borate acetyl complexes of FeII and RuII and isolation of an intermediate of B–N bond hydrolysis. J. Chem. Soc. Dalt. Trans. 1998, 947–951. [Google Scholar] [CrossRef]
- Herrera-Castro, F.; Torres, L.A. Understanding the solvation process and solute-solvent interactions of imidazole compounds in three different solvents through solution calorimetry and 1H NMR. J. Mol. Liq. 2019, 284, 232–240. [Google Scholar] [CrossRef]
- Shapiro, B.L.; Kopchik, R.M.; Ebersole, S.J. Proton NMR Studies of CHDO and CH2O. J. Chem. Phys. 1963, 39, 3154–3155. [Google Scholar] [CrossRef]
- Chen, L.; Malollari, K.G.; Uliana, A.; Hartwig, J.F. Ruthenium-Catalyzed, Chemoselective and Regioselective Oxidation of Polyisobutene. J. Am. Chem. Soc. 2021, 143, 4531–4535. [Google Scholar] [CrossRef] [PubMed]
- Vock, C.A.; Scolaro, C.; Phillips, A.D.; Scopelliti, R.; Sava, G.; Dyson, P.J. Synthesis, Characterization, and in Vitro Evaluation of Novel Ruthenium(II) η6-Arene Imidazole Complexes. J. Med. Chem. 2006, 49, 5552–5561. [Google Scholar] [CrossRef] [PubMed]
- Albertin, G.; Antoniutti, S.; Castro, J.; García-Fontán, S. Preparation of Pyrazole–Pyrazolate Half-Sandwich Complexes of Ruthenium and Osmium. Eur. J. Inorg. Chem. 2011, 2011, 510–520. [Google Scholar] [CrossRef]
- Fernández, R.; Melchart, M.; Habtemariam, A.; Parsons, S.; Sadler, P.J. Use of Chelating Ligands to Tune the Reactive Site of Half-Sandwich Ruthenium(II)–Arene Anticancer Complexes. Chem. A Eur. J. 2004, 10, 5173–5179. [Google Scholar] [CrossRef] [PubMed]
- Peacock, A.F.A.; Melchart, M.; Deeth, R.J.; Habtemariam, A.; Parsons, S.; Sadler, P.J. Osmium(II) and Ruthenium(II) Arene Maltolato Complexes: Rapid Hydrolysis and Nucleobase Binding. Chem. A Eur. J. 2007, 13, 2601–2613. [Google Scholar] [CrossRef] [PubMed]
- Małecki, J.G.; Dziȩgielewski, J.O.; Jaworska, M.; Kruszynski, R.; Bartczak, T.J. Synthesis, molecular, crystal and electronic structures of [(C6H6)RuCl(HPz)2]Cl and [(C6H6)RuCl2(Me2HPz)]. Polyhedron 2004, 23, 885–894. [Google Scholar] [CrossRef]
- Field, L.D.; Messerle, B.A.; Soler, L.; Buys, I.E.; Hambley, T.W. Polypyrazolylmethane complexes of ruthenium. J. Chem. Soc. Dalt. Trans. 2001, 1959–1965. [Google Scholar] [CrossRef]
- Huynh, M.H.V.; Lasker, J.M.; Wetzler, M.; Mort, B.; Szczepura, L.F.; Witham, L.M.; Cintron, J.M.; Marschilok, A.C.; Ackerman, L.J.; Castellano, R.K.; et al. Remarkable spectator ligand effect on the rate constant of ligand substitution of (Aqua)ruthenium(II) complexes. J. Am. Chem. Soc. 2001, 123, 8780–8784. [Google Scholar] [CrossRef]
- Eremina, J.A.; Lider, E.V.; Kuratieva, N.V.; Samsonenko, D.G.; Klyushova, L.S.; Sheven’, D.G.; Trifonov, R.E.; Ostrovskii, V.A. Synthesis and crystal structures of cytotoxic mixed-ligand copper(II) complexes with alkyl tetrazole and polypyridine derivatives. Inorganica Chim. Acta 2021, 516, 120169. [Google Scholar] [CrossRef]
- Semitut, E.; Sukhikh, T.; Filatov, E.; Ryadun, A.; Potapov, A. Synthesis, Crystal Structure and Luminescent Properties of 2D Zinc Coordination Polymers Based on Bis(1,2,4-triazol-1-yl)methane and 1,3-Bis(1,2,4-triazol-1-yl)propane. Crystals 2017, 7, 354. [Google Scholar] [CrossRef] [Green Version]
- APEX2 (Version 2.0), SAINT (Version 8.18c), and SADABS (Version 2.11), Bruker Advanced X-ray Solutions; Bruker AXS Inc.: Madison, WI, USA, 2012.
- Sheldrick, G.M. SHELXT-Integrated space-group and crystal-structure determination. Acta Crystallogr. Sect. A 2015, 71, 3–8. [Google Scholar] [CrossRef] [Green Version]
- Sheldrick, G.M. IUCr Crystal structure refinement with SHELXL. Acta Crystallogr. Sect. C Struct. Chem. 2015, 71, 3–8. [Google Scholar] [CrossRef]
- Dolomanov, O.V.; Bourhis, L.J.; Gildea, R.J.; Howard, J.A.K.; Puschmann, H. OLEX2: A complete structure solution, refinement and analysis program. J. Appl. Crystallogr. 2009, 42, 339–341. [Google Scholar] [CrossRef]



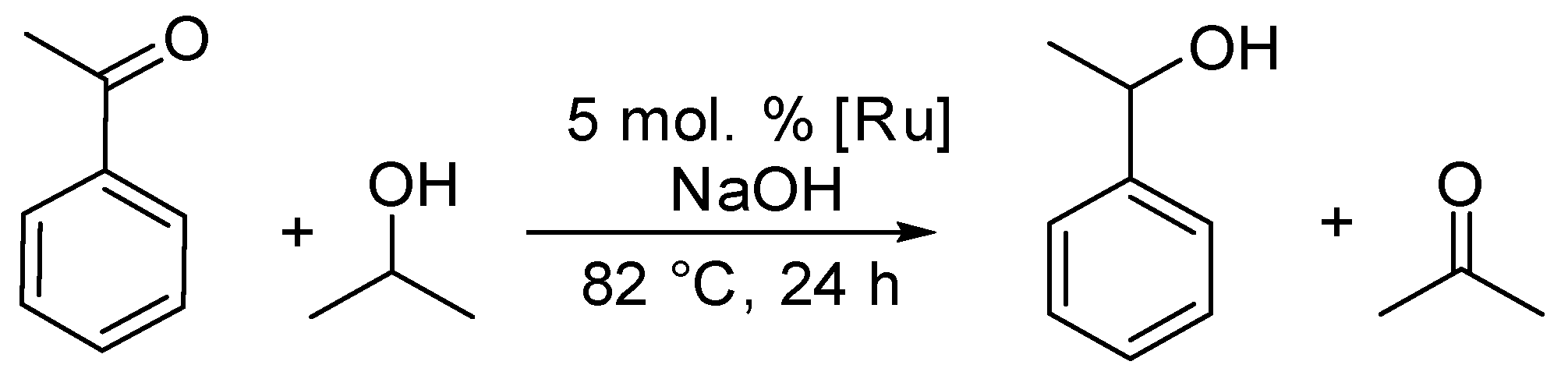
| Compound | 1 | 2 | 3 |
|---|---|---|---|
| Empirical formula | C16H22Cl2N4Ru | C34H44Cl4N8Ru2 | C17H20Cl2N2Ru |
| Formula weight | 442.34 | 972.91 | 424.32 |
| Temperature, K | 300(5) | 140(2) | 140(2) |
| Crystal system | monoclinic | monoclinic | orthorhombic |
| Space group | P21/n | P21/c | Pna21 |
| a, Å | 9.3972(2) | 11.7136(4) | 6.8320(3) |
| b, Å | 17.3629(3) | 10.3400(3) | 18.2051(8) |
| c, Å | 12.9511(3) | 17.0882(6) | 13.6361(5) |
| α, ° | 90 | 90 | 90 |
| β, ° | 97.019(2) | 101.060(4) | 90 |
| γ, ° | 90 | 90 | 90 |
| Volume, Å3 | 2097.30(8) | 2031.26(12) | 1696.02(12) |
| Z | 4 | 2 | 4 |
| ρcalc, g/cm3 | 1.401 | 1.591 | 1.662 |
| μ, mm−1 | 1.006 | 1.050 | 1.237 |
| F(000) | 896 | 992 | 856 |
| Crystal size, mm3 | 0.23 × 0.08 × 0.05 | 0.21 × 0.19 × 0.05 | 0.15 × 0.10 × 0.07 |
| 2Θ range for data collection, ° | 5.662 to 64.854 | 5.04 to 58.78 | 4.474 to 57.594 |
| Index ranges | −14 ≤ h ≤ 13 −25 ≤ k ≤ 24 −19 ≤ l ≤ 12 | −14 ≤ h ≤ 15, −12 ≤ k ≤ 13, −16 ≤ l ≤ 23 | −8 ≤ h ≤ 7 −18 ≤ k ≤ 24 −17 ≤ l ≤ 13 |
| Reflections collected | 12806 | 10166 | 8203 |
| Independent reflections | 6659 (Rint = 0.0242, Rsigma = 0.0372) | 4548 (Rint = 0.0204, Rsigma = 0.0305) | 3157 (Rint = 0.0210, Rsigma = 0.0258) |
| Restraints/Parameters | 0/211 | 0/241 | 1/202 |
| Goodness-of-fit on F2 | 1.032 | 1.045 | 1.073 |
| Final R indexes (I ≥ 2σ (I)) | R1 = 0.0274, wR2 = 0.0600 | R1 = 0.0267, wR2 = 0.0591 | R1 = 0.0193, wR2 = 0.0401 |
| Final R indexes (all data) | R1 = 0.0340, wR2 = 0.0624 | R1 = 0.0348, wR2 = 0.0616 | R1 = 0.0207, wR2 = 0.0406 |
| Largest diff. peak/hole, e·Å−3 | 0.768/−0.440 | 1.081/−0.589 | 0.322/−0.302 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Matveevskaya, V.V.; Pavlov, D.I.; Samsonenko, D.G.; Ermakova, E.A.; Klyushova, L.S.; Baykov, S.V.; Boyarskiy, V.P.; Potapov, A.S. Synthesis and Structural Characterization of Half-Sandwich Arene–Ruthenium(II) Complexes with Bis(imidazol-1-yl)methane, Imidazole and Benzimidazole. Inorganics 2021, 9, 34. https://doi.org/10.3390/inorganics9050034
Matveevskaya VV, Pavlov DI, Samsonenko DG, Ermakova EA, Klyushova LS, Baykov SV, Boyarskiy VP, Potapov AS. Synthesis and Structural Characterization of Half-Sandwich Arene–Ruthenium(II) Complexes with Bis(imidazol-1-yl)methane, Imidazole and Benzimidazole. Inorganics. 2021; 9(5):34. https://doi.org/10.3390/inorganics9050034
Chicago/Turabian StyleMatveevskaya, Vladislava V., Dmitry I. Pavlov, Denis G. Samsonenko, Ekaterina A. Ermakova, Lyubov S. Klyushova, Sergey V. Baykov, Vadim P. Boyarskiy, and Andrei S. Potapov. 2021. "Synthesis and Structural Characterization of Half-Sandwich Arene–Ruthenium(II) Complexes with Bis(imidazol-1-yl)methane, Imidazole and Benzimidazole" Inorganics 9, no. 5: 34. https://doi.org/10.3390/inorganics9050034
APA StyleMatveevskaya, V. V., Pavlov, D. I., Samsonenko, D. G., Ermakova, E. A., Klyushova, L. S., Baykov, S. V., Boyarskiy, V. P., & Potapov, A. S. (2021). Synthesis and Structural Characterization of Half-Sandwich Arene–Ruthenium(II) Complexes with Bis(imidazol-1-yl)methane, Imidazole and Benzimidazole. Inorganics, 9(5), 34. https://doi.org/10.3390/inorganics9050034









