Reaction of Non-Symmetric Schiff Base Metallo-Ligand Complexes Possessing an Oxime Function with Ln Ions
Abstract
:1. Introduction
2. Results and Discussion
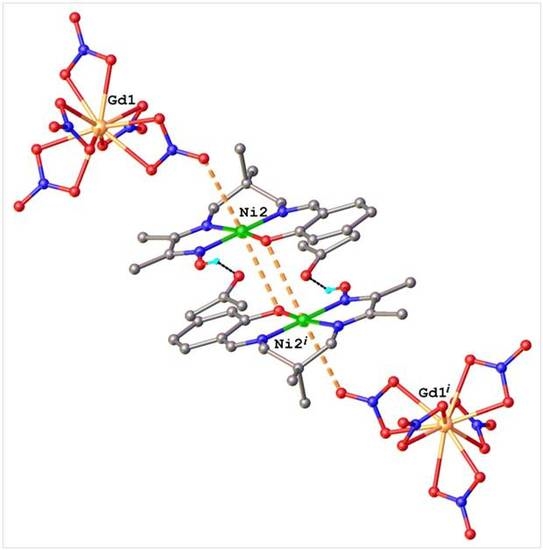
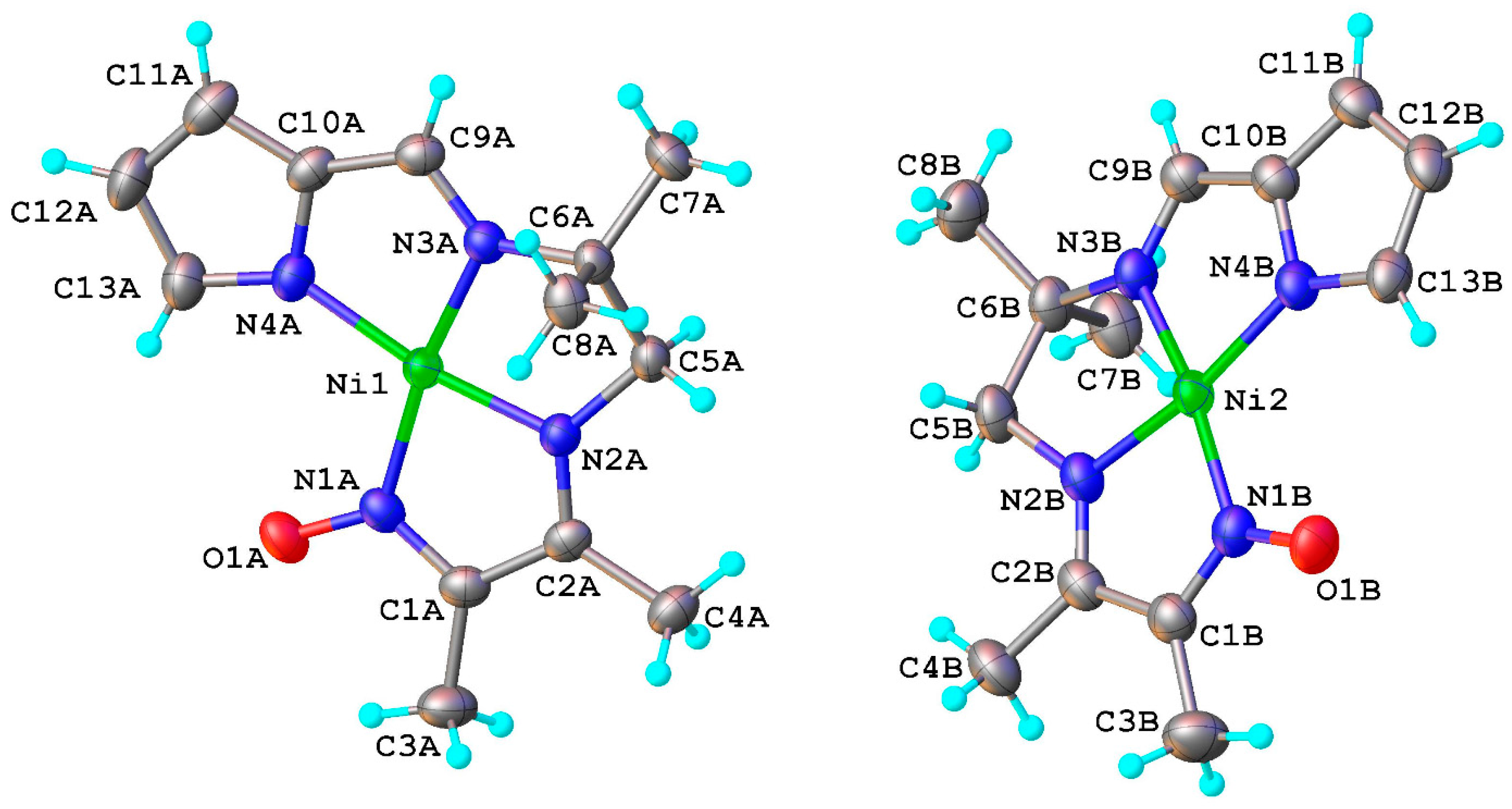
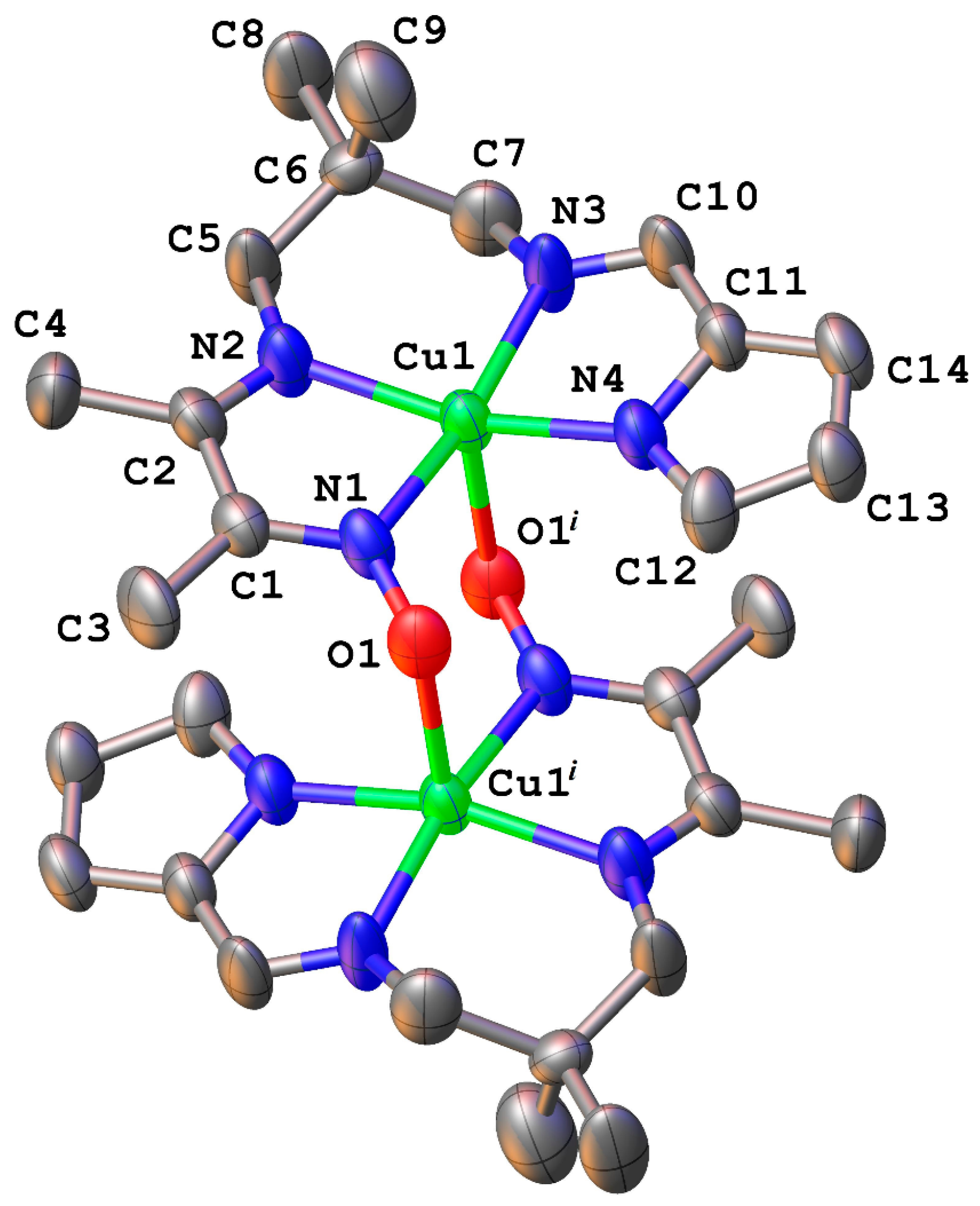
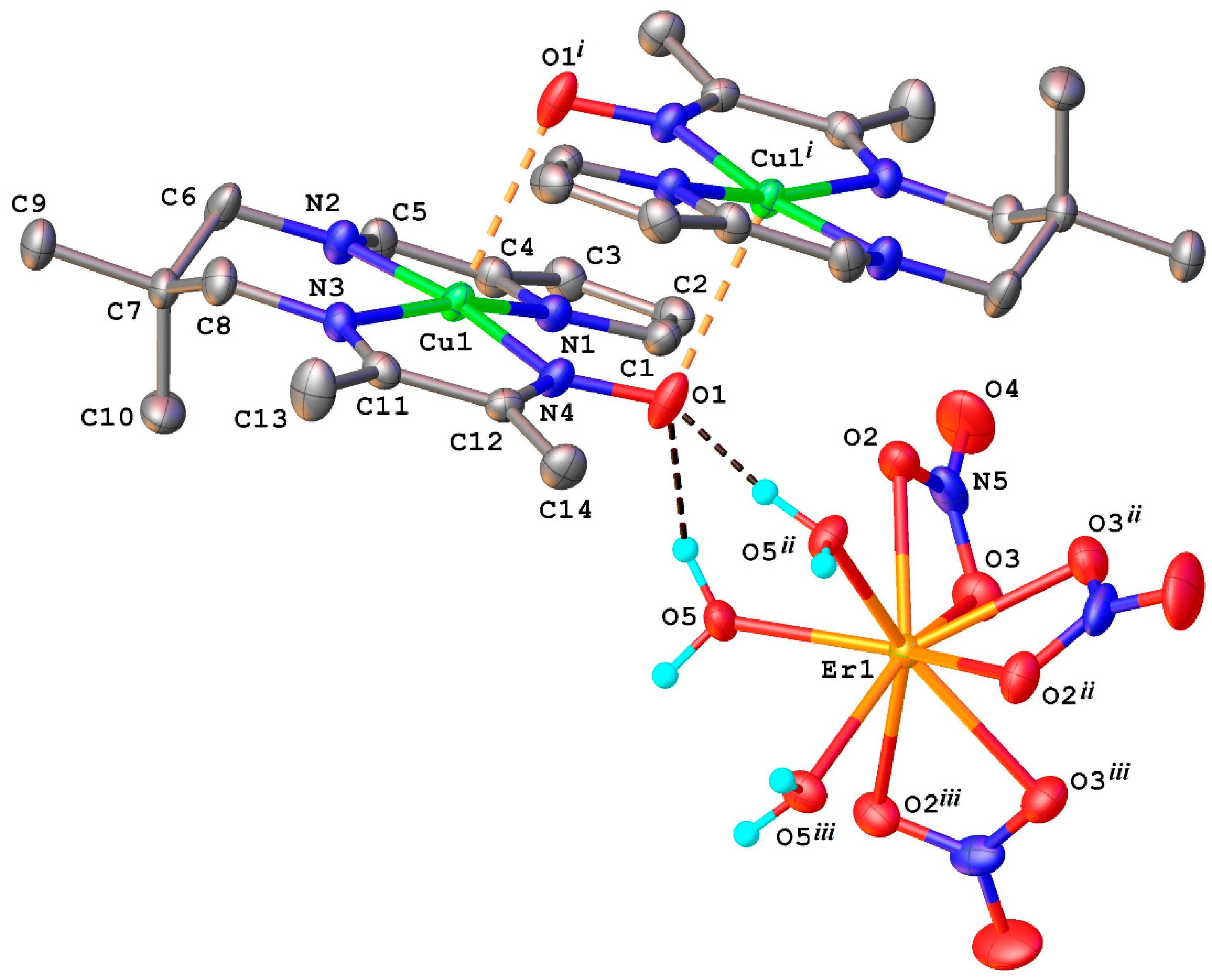
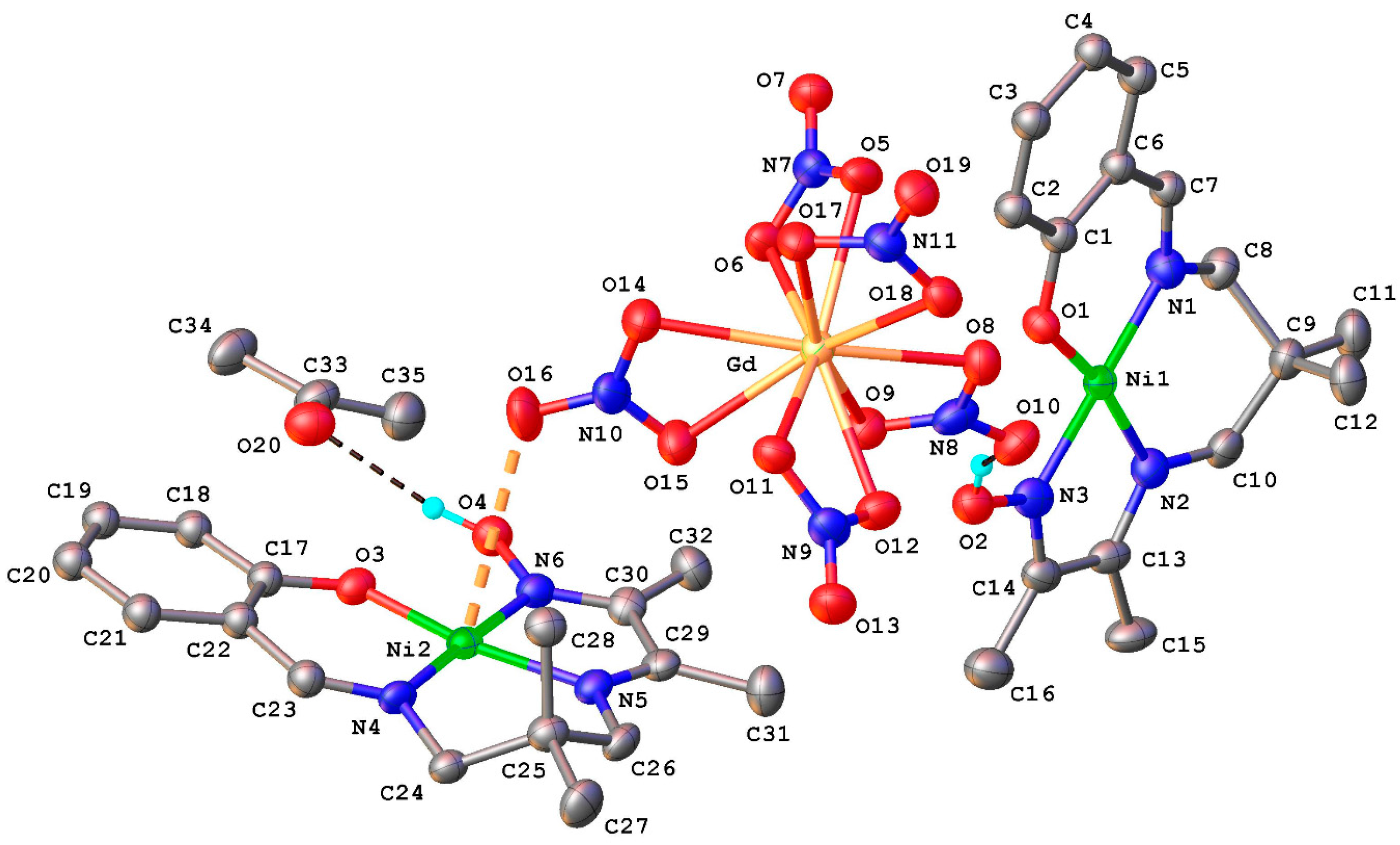
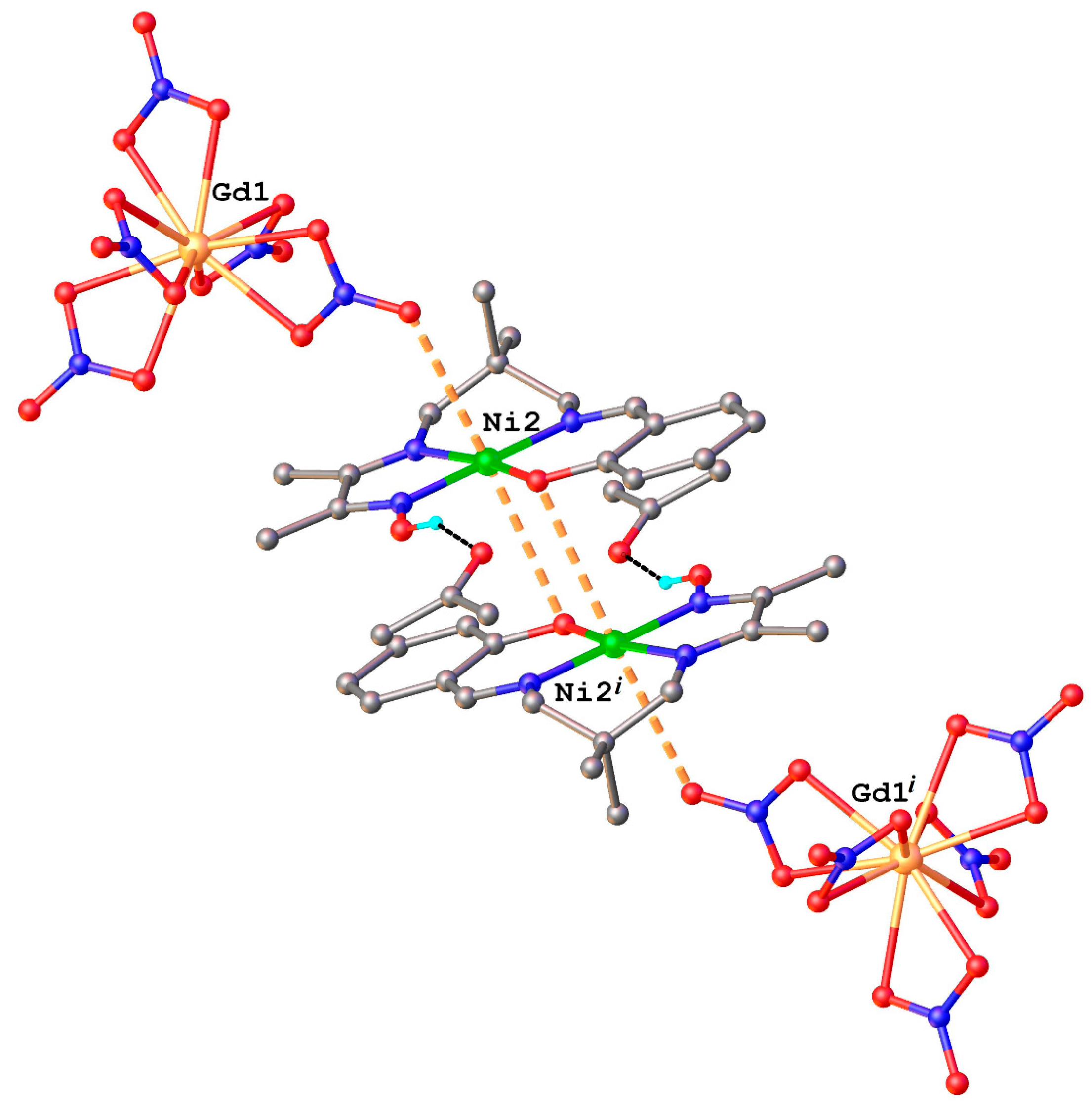
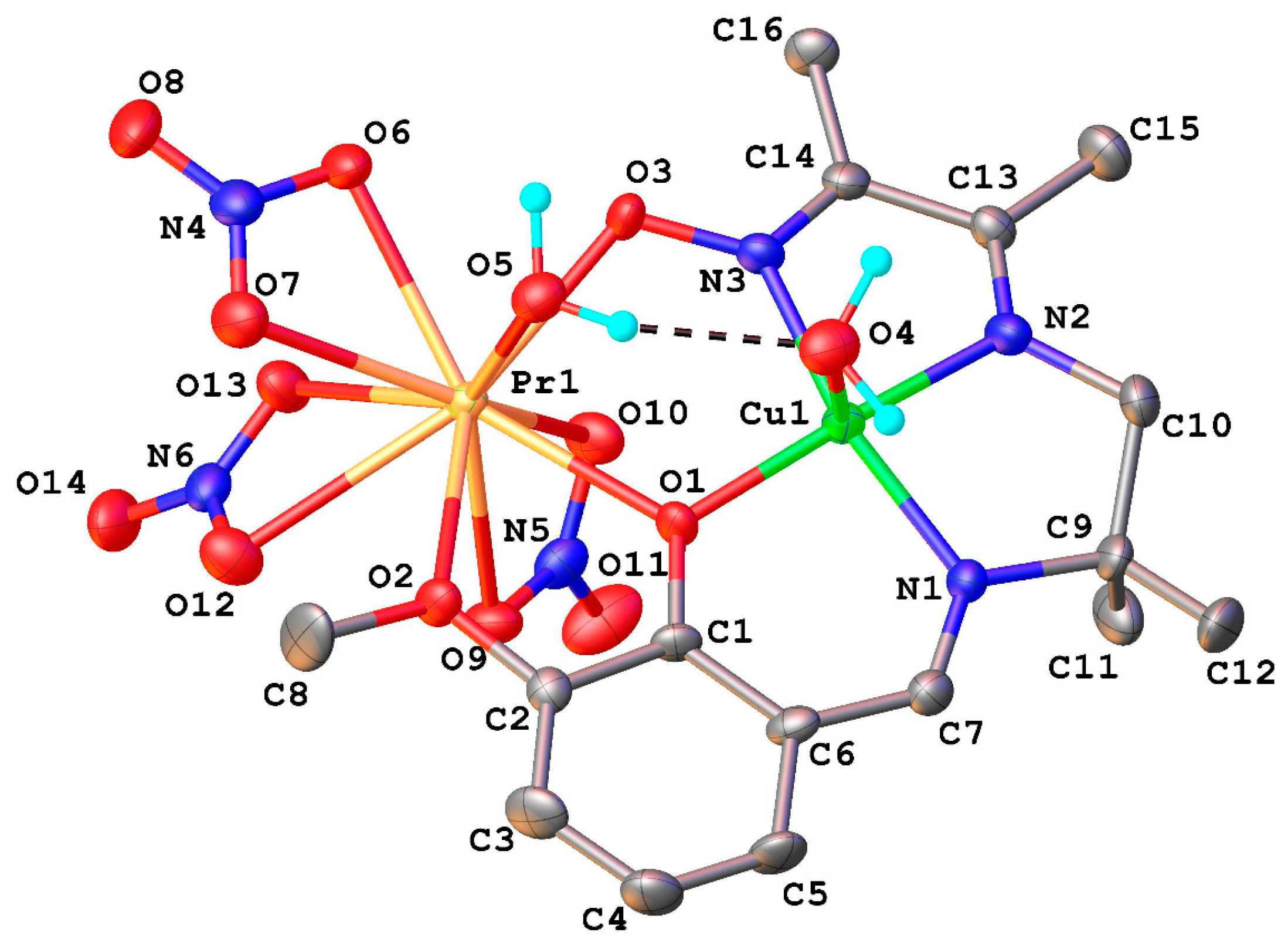
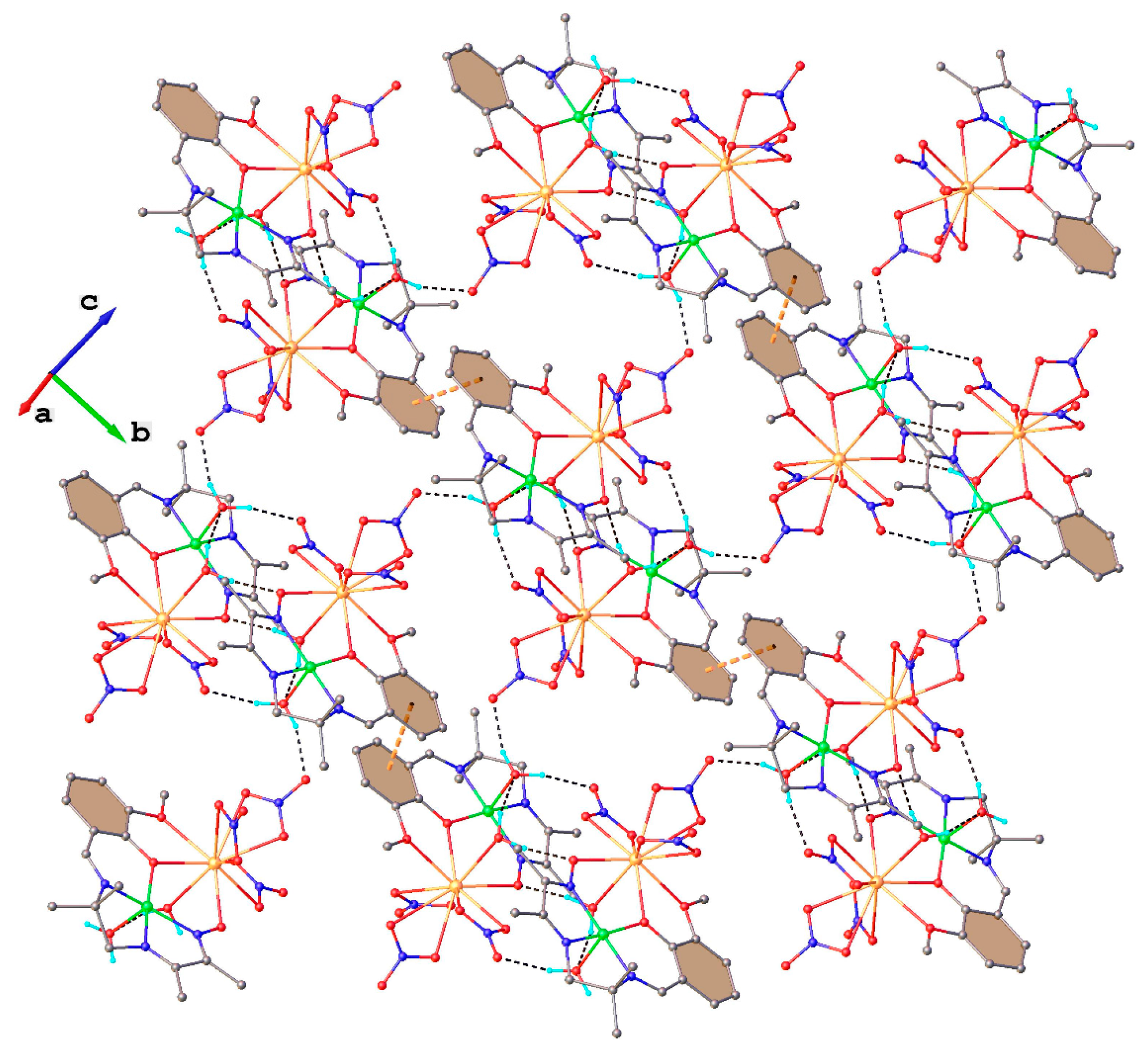
2.1. Structural Chracterisation
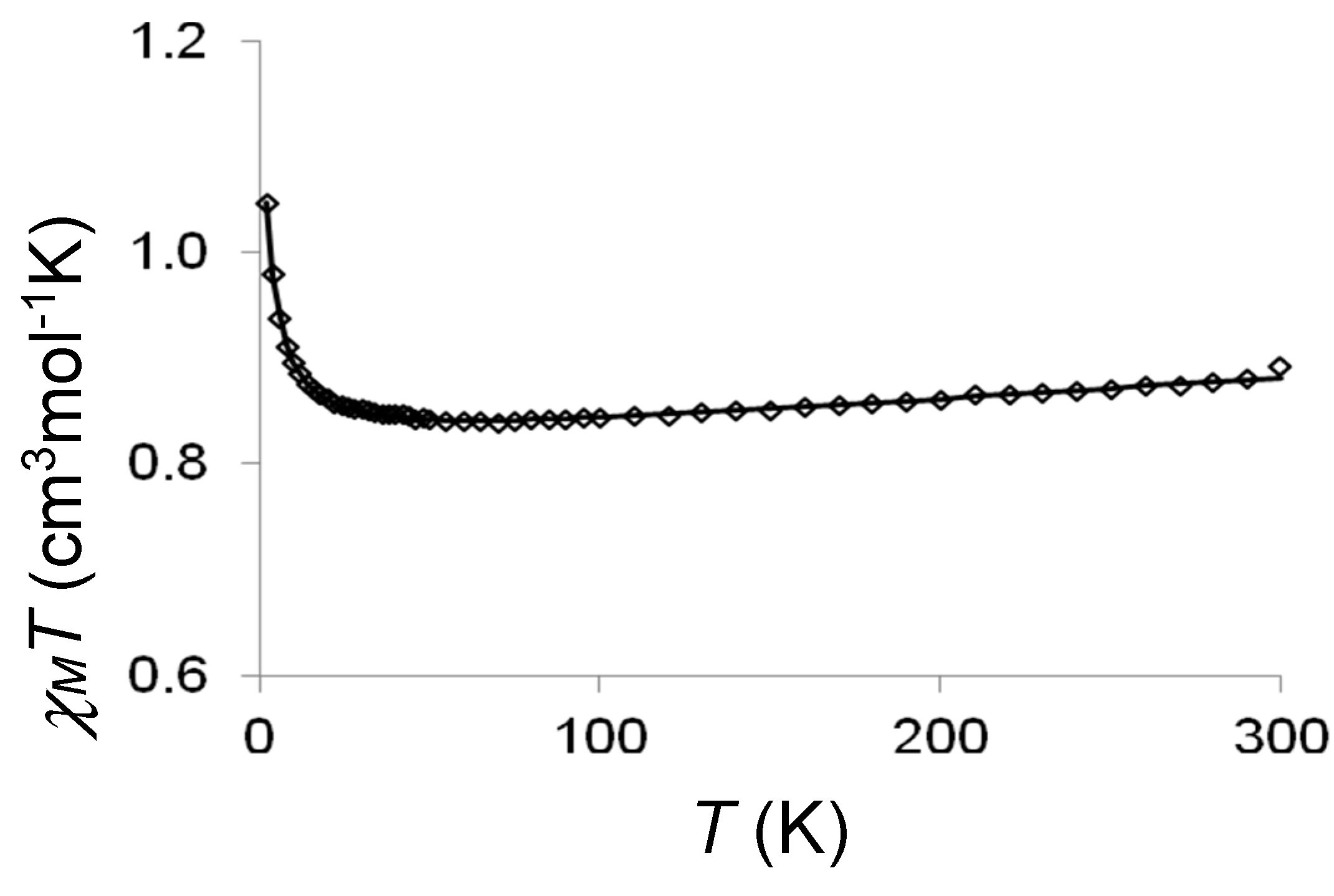
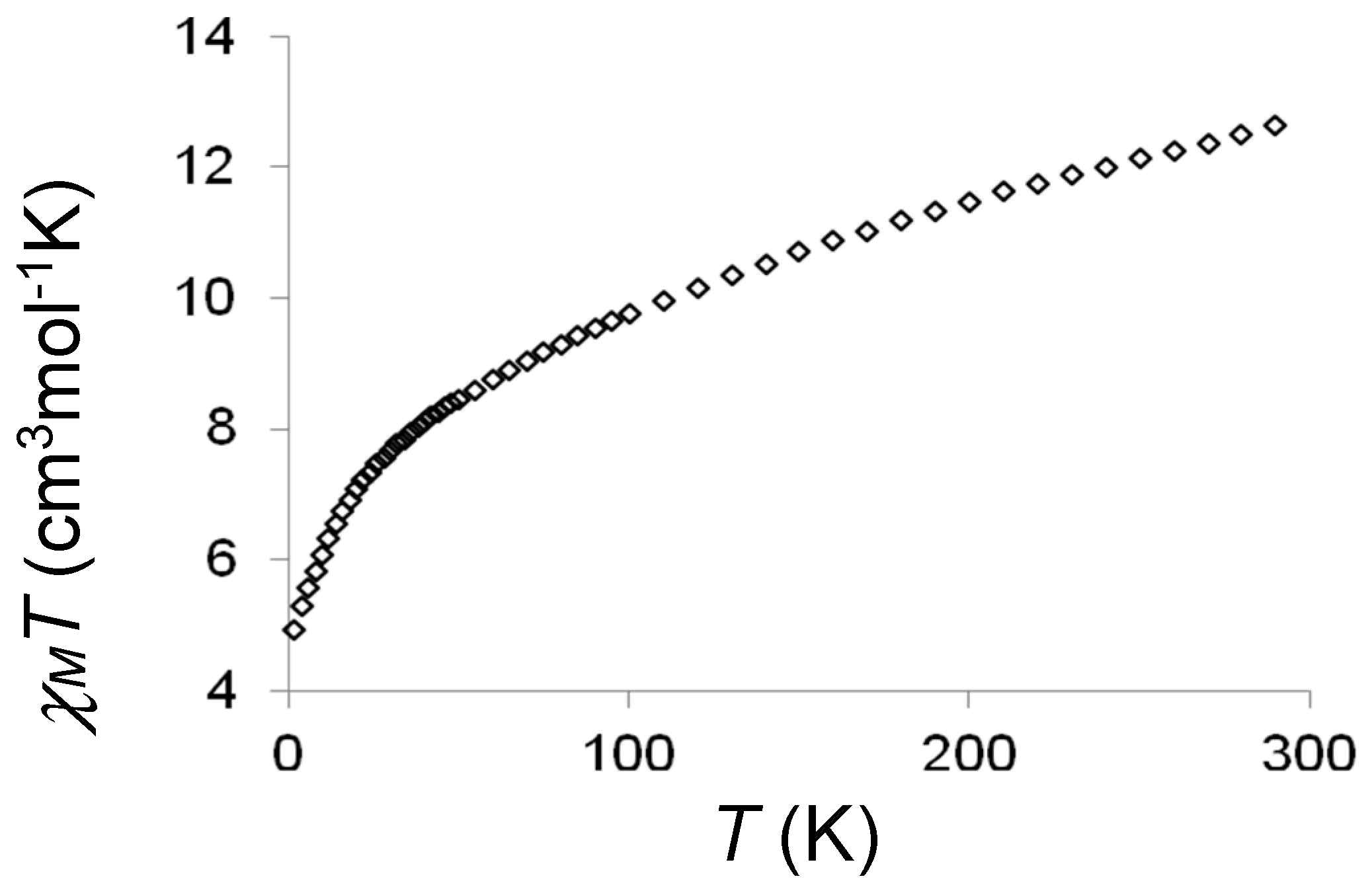
2.2. Magnetic Studies
2.3. Discussion
3. Experimental Section
3.1. Materials
3.2. Ligands
3.3. Complexes
3.4. Crystallographic Data Collections and Structure Determinations for 1, 2, 3, 5, 6, 7, and 8
4. Conclusions
Supplementary Materials
Author Contributions
Conflicts of Interest
References
- Kahn, O. Molecular Magnetism; VCH: New York, NY, USA, 1993. [Google Scholar]
- Benelli, C.; Gatteschi, D. Magnetism of Lanthanides in Molecular Materials with Transition-Metal Ions and Organic Radicals. Chem. Rev. 2002, 102, 2369–2387. [Google Scholar] [CrossRef] [PubMed]
- Lorion, M.; Maindan, K.; Kapdi, A.R.; Ackermann, L. Heteromultimetallic catalysis for sustainable organic syntheses. Chem. Soc. Rev. 2017, 46, 7399–7420. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Manzur, C.; Novoa, N.; Celedón, S.; Carrillo, D.; Hamon, J. Multidentate unsymmetrically-substituted Schiff bases and their metal complexes: Synthesis, functional materials properties, and applications to catalysis. Coord. Chem. Rev. 2018, 357, 144–172. [Google Scholar] [CrossRef]
- Pfeiffer, P.; Breith, E.; Lülle, E.; Tsumaki, T. Tricyclische orthokondensierte Nebenvalenzringe. Justus Liebigs Ann. Chem. 1933, 503, 84–130. [Google Scholar] [CrossRef]
- Vigato, P.A.; Tamburini, S. The challenge of cyclic and acyclic Schiff bases and related derivatives. Coord. Chem. Rev. 2004, 248, 1717–2128. [Google Scholar] [CrossRef]
- Vigato, P.A.; Tamburini, S.; Batolo, L. The development of compartmental macrocyclic Schiff bases and related polyamine derivatives. Coord. Chem. Rev. 2007, 251, 1311–1492. [Google Scholar] [CrossRef]
- Vigato, P.A.; Tamburini, S. Advances in acyclic compartmental ligands and related complexes. Coord. Chem. Rev. 2008, 252, 1871–1995. [Google Scholar] [CrossRef]
- Vigato, P.A.; Peruzzo, V.; Tamburini, S. Acyclic and cyclic compartmental ligands: Recent results and perspectives. Coord. Chem. Rev. 2012, 256, 953–1114. [Google Scholar] [CrossRef]
- Andruh, M. The exceptionally rich coordination chemistry generated by Schiff-base ligands derived from o-vanillin. Dalton Trans. 2015, 44, 16633–16653. [Google Scholar] [CrossRef] [PubMed]
- Kleij, A.W. Nonsymmetrical Salen Ligands and Their Complexes: Synthesis and Applications. Eur. J. Inorg. Chem. 2009, 2009, 193–205. [Google Scholar] [CrossRef]
- Costes, J.P.; Chiboub Fellah, F.Z.; Dahan, F.; Duhayon, C. Role of the kinetic template effect in the syntheses of non-symmetric Schiff base complexes. Polyhedron 2013, 52, 1065–1072. [Google Scholar] [CrossRef]
- Burke, P.J.; McMillin, D.R. Unsymmetrical Ligand Complexes of Cu, Ni, and Co derived from Salicylaldehyde and 1,3-Propanediamine with either Pyridine-2-carbaldehyde or Pyrrole-2-carbaldehyde. J. Chem. Soc. Dalton. 1980, 1794–1796. [Google Scholar] [CrossRef]
- Costes, J.P.; Cros, G.; Darbieu, M.H.; Laurent, J.P. The non-template synthesis of novel non-symmetrical, tetradentate Schiff bases. Their nickel(II) and cobalt(III) complexes. Inorg. Chim. Acta 1982, 60, 111–114. [Google Scholar] [CrossRef]
- Costes, J.P.; Dahan, F.; Laurent, J.P. New route to bimetallic imidazolate-bridged complexes. 6. Structural and magnetic consequences of steric effects in mono and dinuclear nickel(II) and copper(II) complexes involving 4-methylimidazole as ligand. Inorg. Chem. 1991, 30, 1887–1892. [Google Scholar] [CrossRef]
- Béreau, V.; Dhers, S.; Costes, J.P.; Duhayon, C.; Sutter, J.P. Syntheses, Structures, and Magnetic Properties of Symmetric and Dissymmetric Ester-Functionalized 3d-4f Schiff Base Complexes. Eur. J. Inorg. Chem. 2018, 2018, 66–73. [Google Scholar] [CrossRef]
- Costes, J.P.; Dahan, F.; Dupuis, A. Laurent, Solid state and solution studies of the compounds resulting from the equimolecular condensation of a diamine with a dione monoxime. New J. Chem. 1997, 21, 1211–1217. [Google Scholar]
- Lah, M.S.; Pecoraro, V.L. Isolation and Characterization of {MnII[MnIII(salicylhydroximate)]4 (acetate)2(DMF)6}·2DMF: An Inorganic Analogue of M2+(12-crown-4). J. Am. Chem. Soc. 1989, 111, 7258–7259. [Google Scholar] [CrossRef]
- Mezei, G.; Zaleski, C.M.; Pecoraro, V.L. Structural and Functional Evolution of Metallacrowns. Chem. Rev. 2007, 1074, 4933–5003. [Google Scholar] [CrossRef] [PubMed]
- Bain, G.A.; Berry, J.F. Diamagnetic Corrections and Pascal’s Constants. J. Chem. Educ. 2008, 85, 532–536. [Google Scholar] [CrossRef]
- Costes, J.P.; Vendier, L. Cu–Ln complexes with a single μ-oximato bridge. C. R. Chimie 2010, 13, 661–667. [Google Scholar] [CrossRef]
- Bencini, A.; Benelli, C.; Caneschi, A.; Carlin, R.L.; Dei, A.; Gatteschi, D. Crystal and Molecular Structure of and Magnetic Coupling in Two Complexes Containing Gadolinium(III) and Copper(II) Ions. J. Am. Chem. Soc. 1985, 107, 8128–8136. [Google Scholar] [CrossRef]
- Costes, J.P.; Dahan, F.; Dupuis, A.; Laurent, J.P. Is ferromagnetism an intrinsic property of the CuII/GdIII couple? Part 1. Structures and magnetic properties of two novel dinuclear complexes with a μ-phenolato-μ-oximato (Cu,Gd) core. Inorg. Chem. 2000, 39, 169–173. [Google Scholar] [CrossRef] [PubMed]
- Costes, J.P.; Dahan, F.; Dupuis, A. Is ferromagnetism an intrinsic property of the CuII/GdIII couple? Part 2. Structures and magnetic properties of novel trinuclear complexes with a μ-phenolato-μ-oximato (Cu,Gd) core. Inorg. Chem. 2000, 39, 5994–6000. [Google Scholar] [CrossRef] [PubMed]
- Boudalis, A.K.; Clemente-Juan, J.M.; Dahan, F.; Tuchagues, J.P. New Poly-Iron(II) Complexes of N4O Dinucleating Schiff Bases and Pseudohalides: Syntheses, Structures, and Magnetic and Mossbauer Properties. Inorg. Chem. 2004, 43, 1574–1586. [Google Scholar] [CrossRef] [PubMed]
- James, F.; Roos, M. MINUIT Program, a System for Function Minimization and Analysis of the Parameters Errors and Correlations. Comput. Phys. Commun. 1975, 10, 343–367. [Google Scholar] [CrossRef]
- Fair, C.K. MolEN. An Interactive Structure Solution Procedure; Enraf-Nonius: Delft, The Netherlands, 1990. [Google Scholar]
- North, A.C.T.; Phillips, D.C.; Mathews, F.S. A semi-empirical method of absorption correction. Acta Crystallogr. Sect. A 1968, A24, 351–359. [Google Scholar] [CrossRef]
- Sheldrick, G.M. SHELX97 [Includes SHELXS97, SHELXL97, CIFTAB]; Programs for Crystal Structure Analysis (Release 97-2); Institüt für Anorganische Chemie der Universität: Tammanstrasse 4, Göttingen, Germany, 1998. [Google Scholar]
- Farrugia, L.J. WinGX suite for small-molecule single-crystal crystallography. J. Appl. Crystallogr. 1999, 32, 837–838. [Google Scholar] [CrossRef]
- Ibers, J.A.; Hamilton, W.C. International Tables for X-ray Crystallography. Vol. IV; Kynoch Press: Birmingham, UK, 1974. [Google Scholar]
- Dolomanov, O.V.; Bourhis, L.J.; Gildea, R.J.; Howard, J.A.K.; Puschmann, H. OLEX2: A complete structure solution, refinement and analysis program. J. Appl. Cryst. 2009, 42, 339–341. [Google Scholar] [CrossRef]


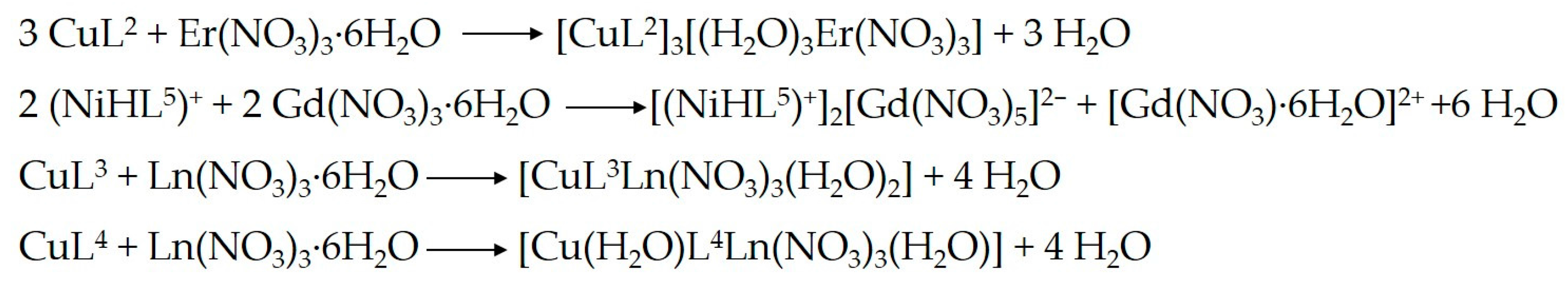

© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Costes, J.-P.; Dahan, F.; Dupuis, A.; Shova, S.; Tojal, J.G. Reaction of Non-Symmetric Schiff Base Metallo-Ligand Complexes Possessing an Oxime Function with Ln Ions. Inorganics 2018, 6, 33. https://doi.org/10.3390/inorganics6010033
Costes J-P, Dahan F, Dupuis A, Shova S, Tojal JG. Reaction of Non-Symmetric Schiff Base Metallo-Ligand Complexes Possessing an Oxime Function with Ln Ions. Inorganics. 2018; 6(1):33. https://doi.org/10.3390/inorganics6010033
Chicago/Turabian StyleCostes, Jean-Pierre, Françoise Dahan, Arnaud Dupuis, Sergiu Shova, and Javier Garcia Tojal. 2018. "Reaction of Non-Symmetric Schiff Base Metallo-Ligand Complexes Possessing an Oxime Function with Ln Ions" Inorganics 6, no. 1: 33. https://doi.org/10.3390/inorganics6010033
APA StyleCostes, J.-P., Dahan, F., Dupuis, A., Shova, S., & Tojal, J. G. (2018). Reaction of Non-Symmetric Schiff Base Metallo-Ligand Complexes Possessing an Oxime Function with Ln Ions. Inorganics, 6(1), 33. https://doi.org/10.3390/inorganics6010033