3.1. IIC and CCCP: Principles and Differences
The IIC is a criterion of the predictive potential of models that combines observed conditions in two directions: (i) from the value of the coefficient of determination; and (ii) from the value of the dispersion of points in the coordinates of the experiment vs. model.
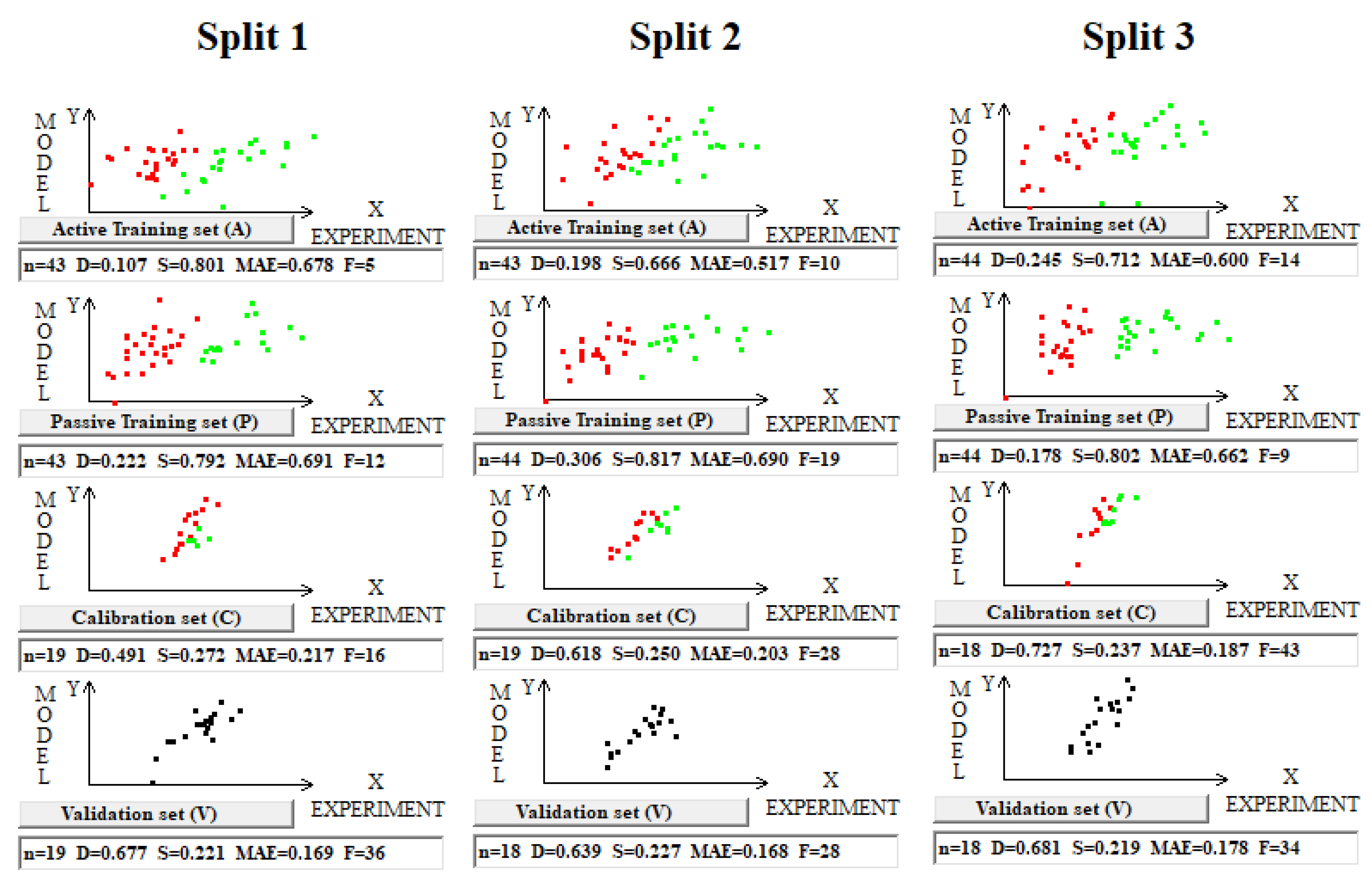
The CCCP criterion is a value developed based on the representation of points that are participants in correlations, similarly to social processes in terms of a “supporter” or “opponent” of the observed correlation. If removing a point results in an increase in the correlation coefficient for the remaining points, then that point is an “opponent” of the correlation. If removing a point results in a decrease in the correlation coefficient for the remaining points, then that point is a “supporter” of the correlation.
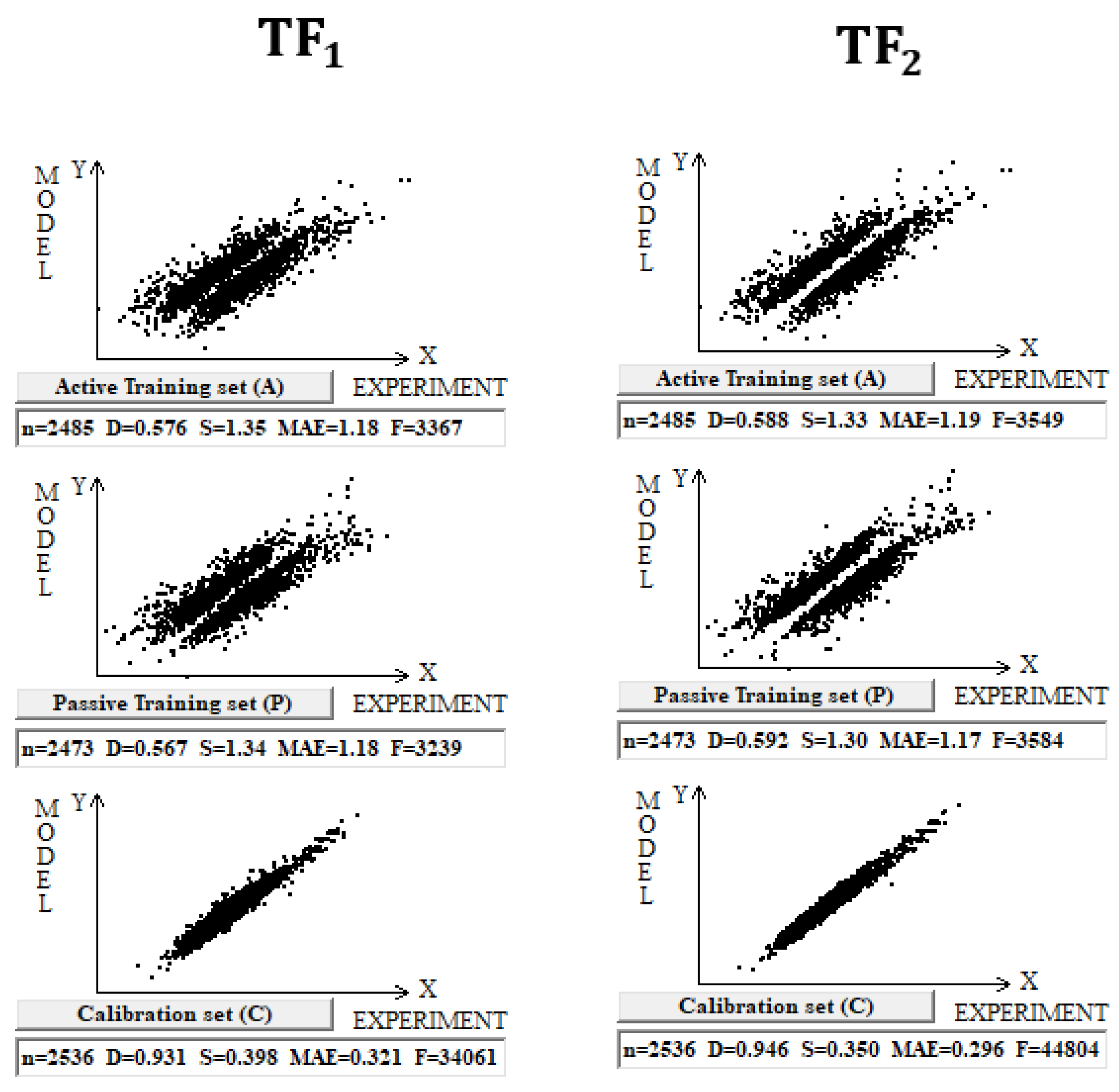
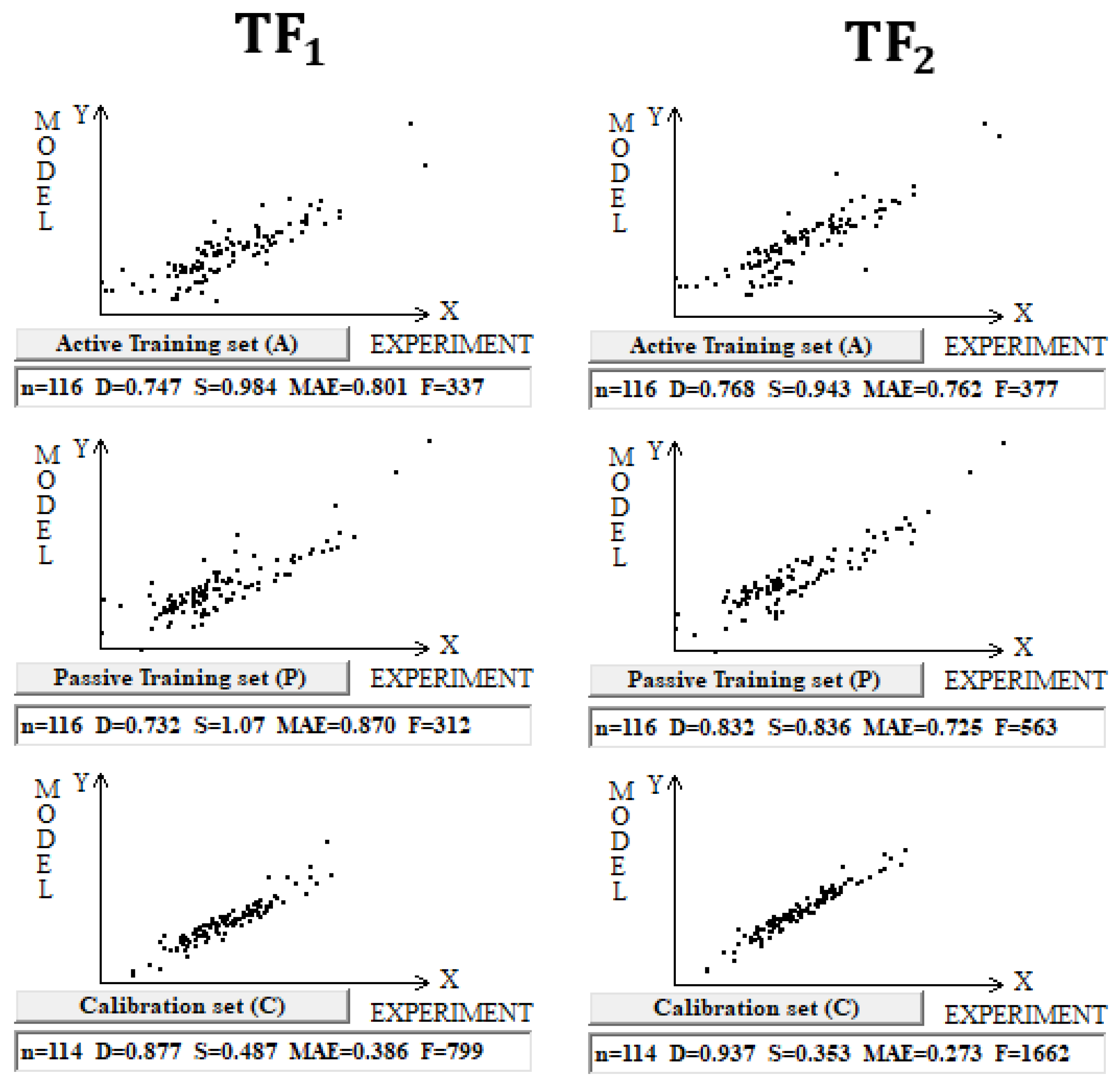
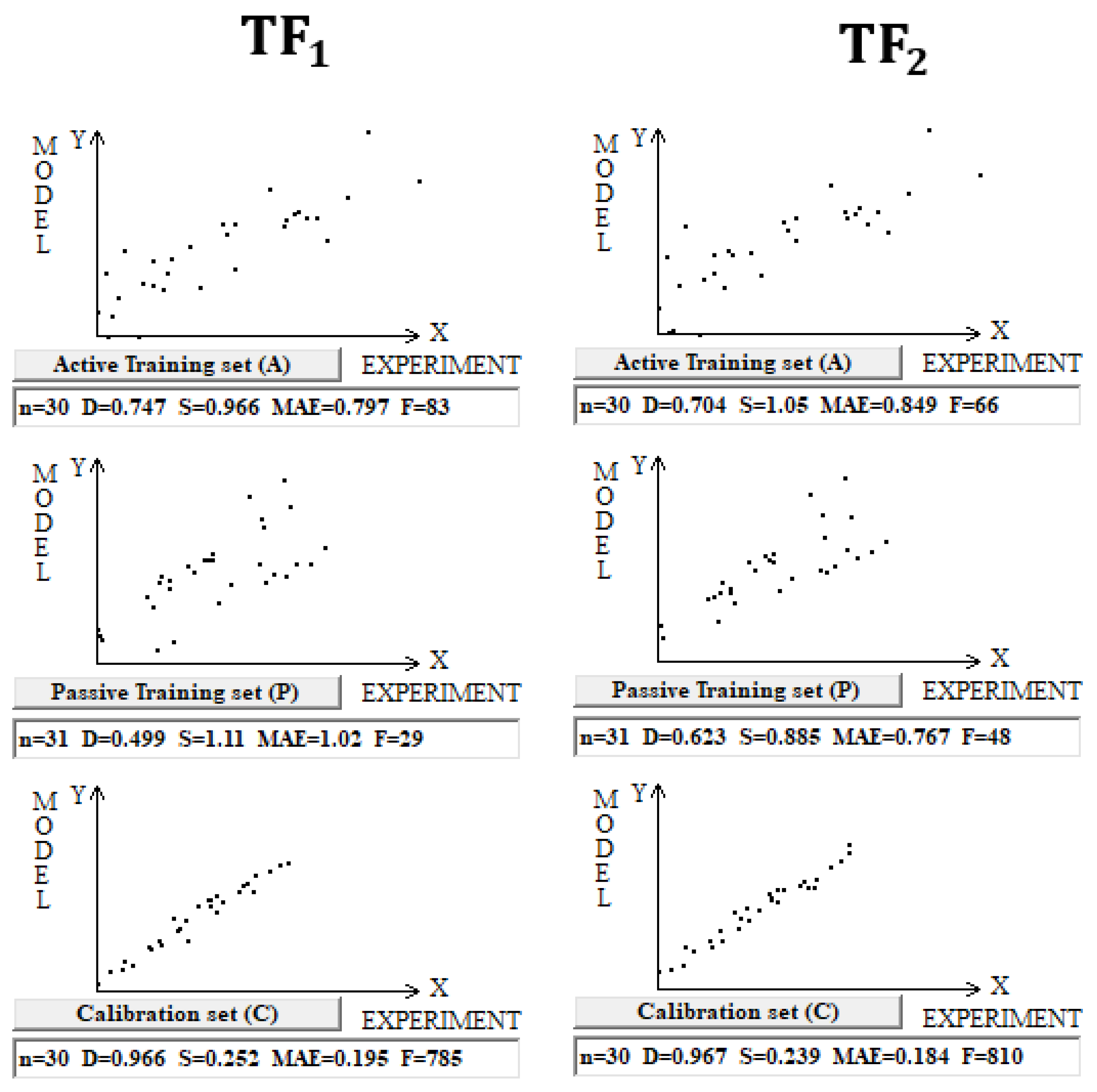
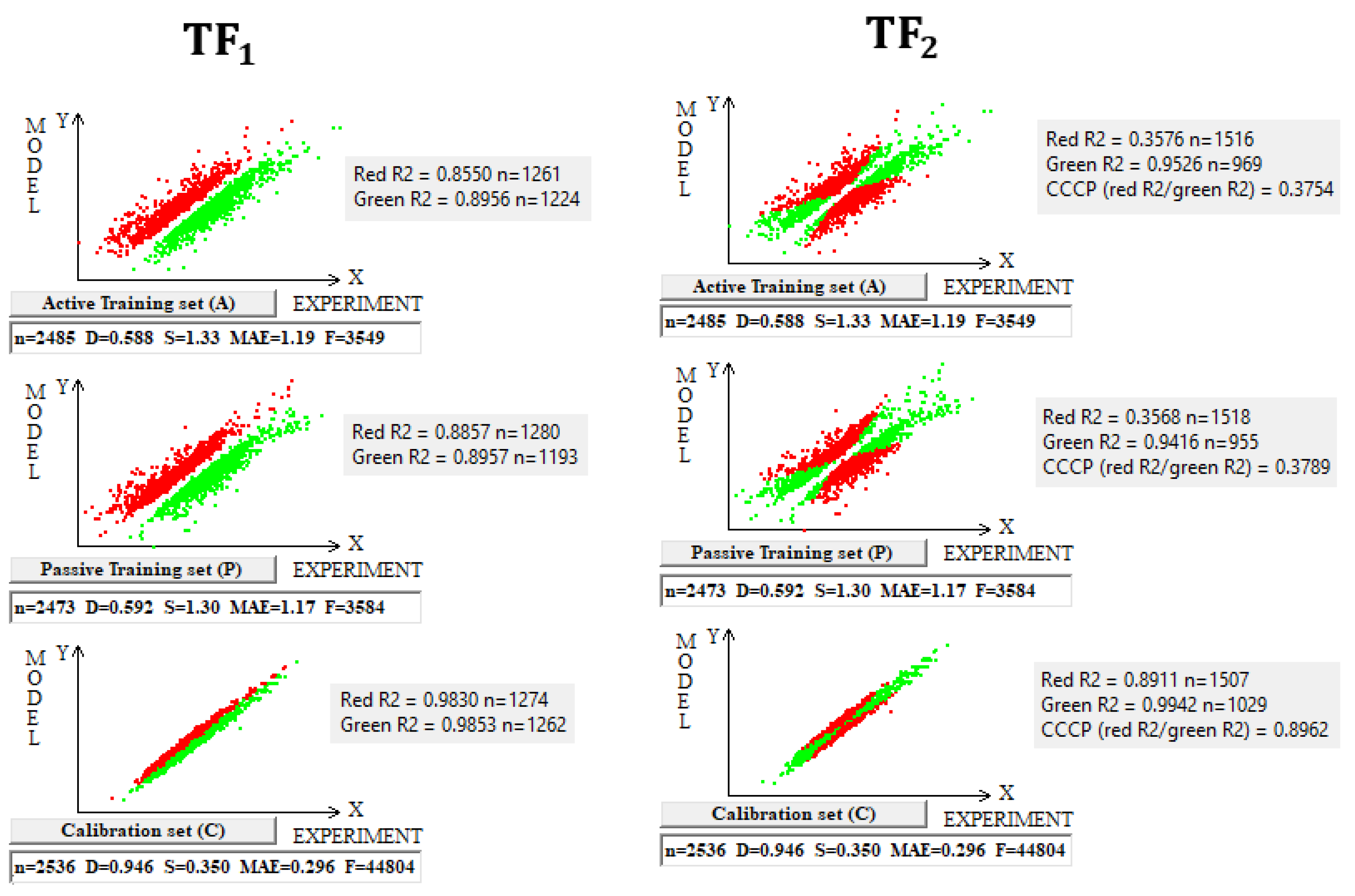
The use of the IIC leads to an improvement in the statistical quality of the model for the calibration set, but to the detriment of the statistical quality of the model for the active and passive training samples. Geometrically, this decrease in the statistical quality on the training samples looks like two parallel clusters (
Figure 6). The coefficient of determination calculated for both clusters (red and green) has a value lower than that for each of the mentioned clusters separately. Remarkably, such clusters for the calibration set turn out to be closer to each other than those on the active and passive training sets. There is a probability that these clusters can be close for the validation set too. If so, the model is successful.
The use of CCCP aims to obtain completely different correlation clusters to those obtained using the IIC (
Figure 6). CCCP aims to obtain close coefficients of determination for the clusters of “supporters” and “opponents” of the correlation. Again, the transformations have the maximum effect on the calibration set, giving the user of the method some hope that this useful effect will be observed in the external calibration set. The different datasets chosen in this study represent quite different properties, related to physicochemical and toxicological endpoints. The interactions of the molecules with the abiotic and biotic external situation are expected to play different roles. Thus, this study may help us explore the possible contributions of specific algorithms to the success of the models. The results of the computer experiments conducted indicate that the CCCP-based approach is effective for large datasets, i.e., for all considered models except for the toxicity of inorganic compounds to rats. Only for the last model was the application of the IIC better than that of CCCP. This may be due to the lower number of substances (a few hundred) in this case.
3.2. Genesis of Models for logP
Conducting a series of runs of the described optimization procedure allowed us to identify collections of molecular features extracted from the SMILES that influence the growth or, conversely, the decrease in the values of the octanol–water distribution coefficients on the datasets considered.
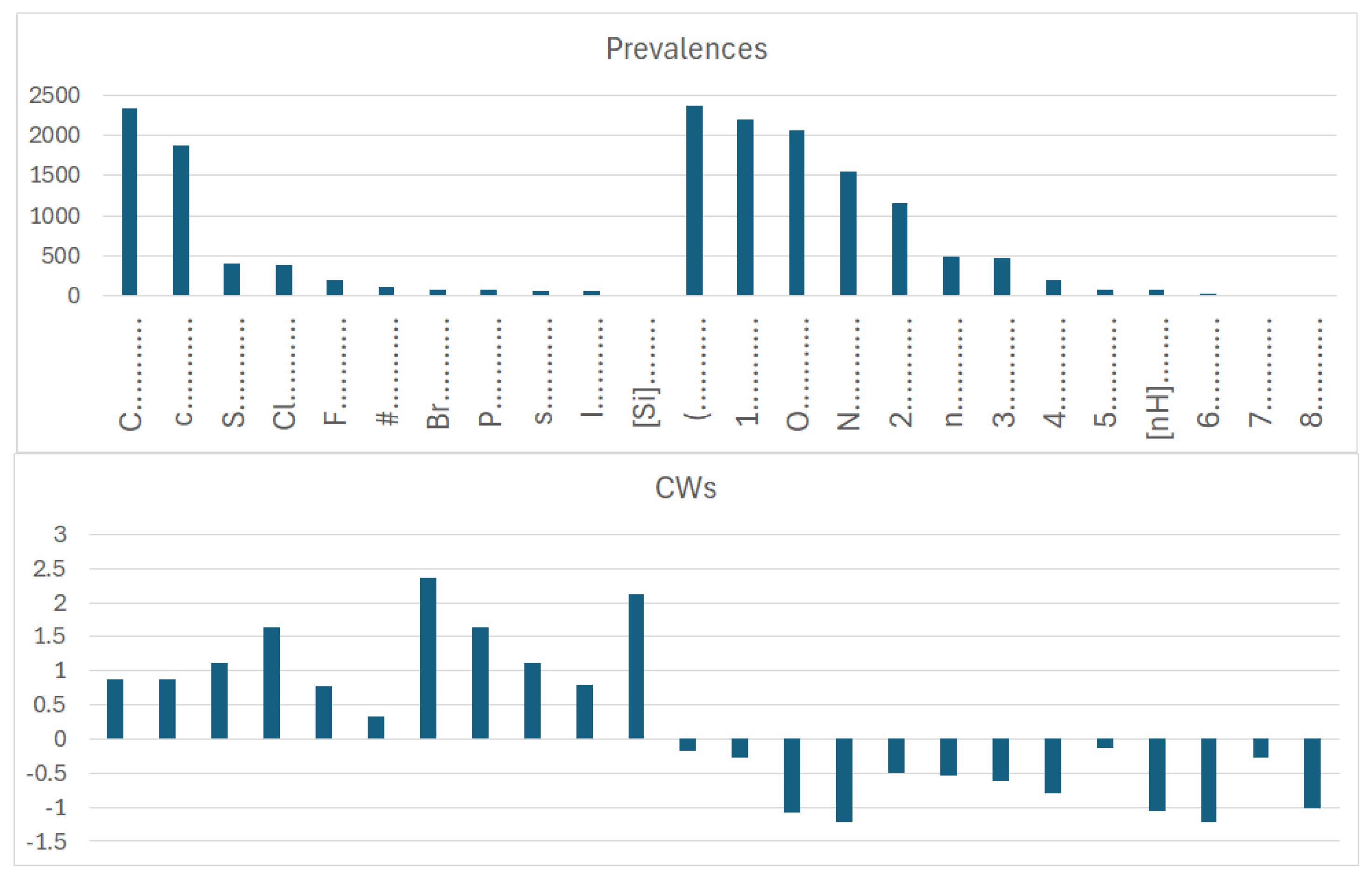
In dataset 1, involving organic and inorganic molecules, the number of promoters of the increase/decrease in logP (
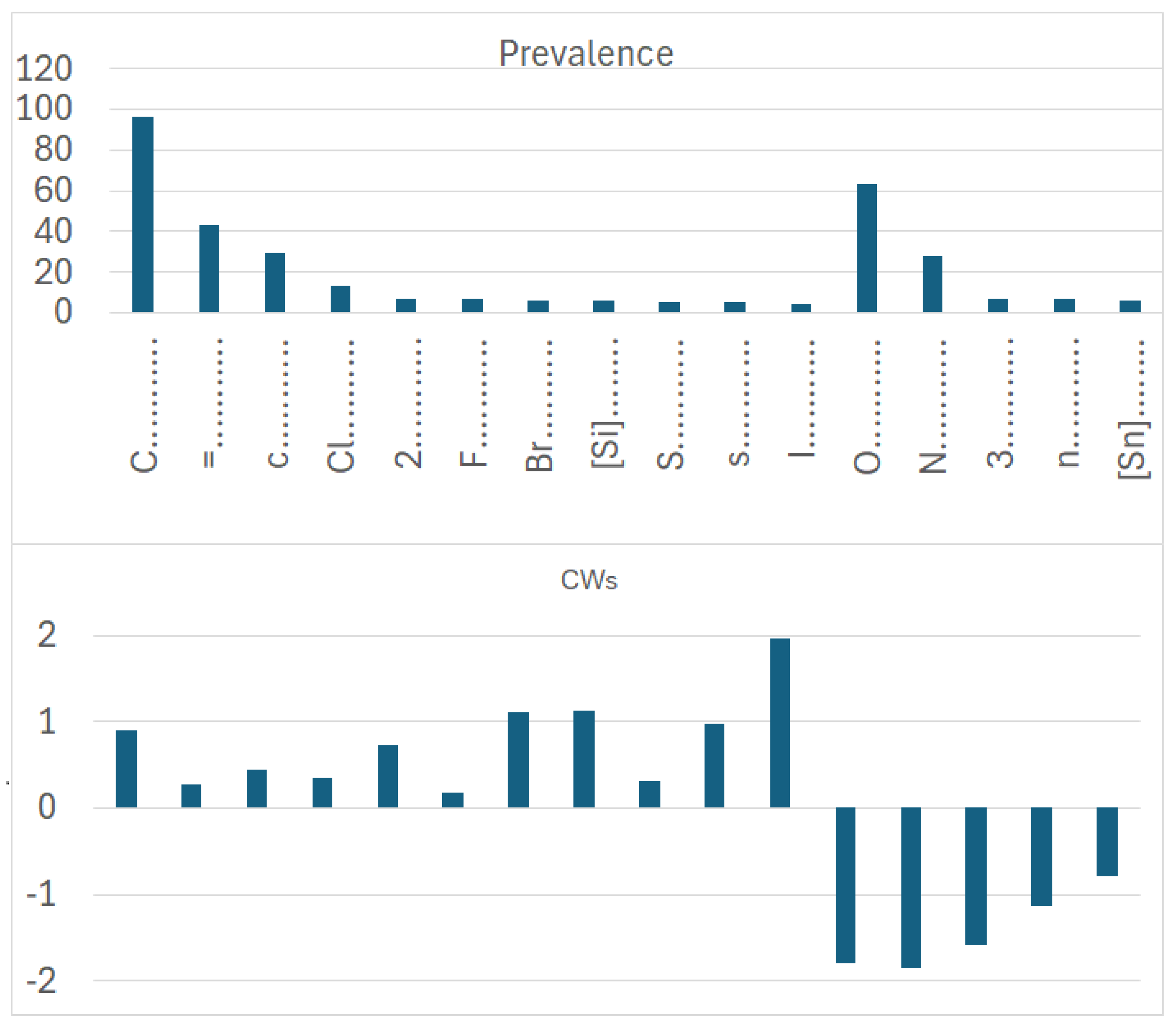
Figure 7) is larger than that of promoters of the increase/decrease in the logP of dataset 2 (
Figure 8). This is expected; indeed, our models, like most in silico models, have a statistical basis. Thus, if the dataset at the basis of the model is larger, the number of features extracted by the model is very likely larger than the number of features extracted from a smaller dataset. This kind of consideration underlines the relevance of efforts to build up models merging organic and inorganic substances, as we present here. Unfortunately, most in silico models eliminate information on salts due to the technical difficulty of coping with disconnected structures. Furthermore, several models deal only with “classical” atoms present in the organic substances, disregarding organometallic compounds. This aspect is partially due to technical reasons (it is easier to develop certain kinds of models) and partially related to the availability of experimental data for certain compounds (data on certain organometallic substances are more limited than data on classical substances). Thus, even from a technological point of view, it may be feasible to develop models including, for instance, substances like germanium; if the number of substances with germanium is low, we cannot expect to obtain good models based on a large population of substances with very few substances with a specific atom. Nevertheless, it is necessary to address the modeling from a broader perspective, and, in particular, to be aware of the limitations of the current models when dealing with properties where the neutralization of the structure produces a substance with very different properties. This is the case for logP, as studied here.
More specifically, for organic substances, the software correctly identifies the role of atoms, such as oxygen and nitrogen, which increase the polarity, and thus decrease logP; conversely, chlorine, bromine, fluorine, and carbon, for instance, increase logP. For inorganic compounds, the main features are present for the organic substances discussed above, but with very different relevance in some cases. In addition, some features are not present at all, such as in the cases with branching and a large number of rings. Obviously, these features are typical of organic substances.
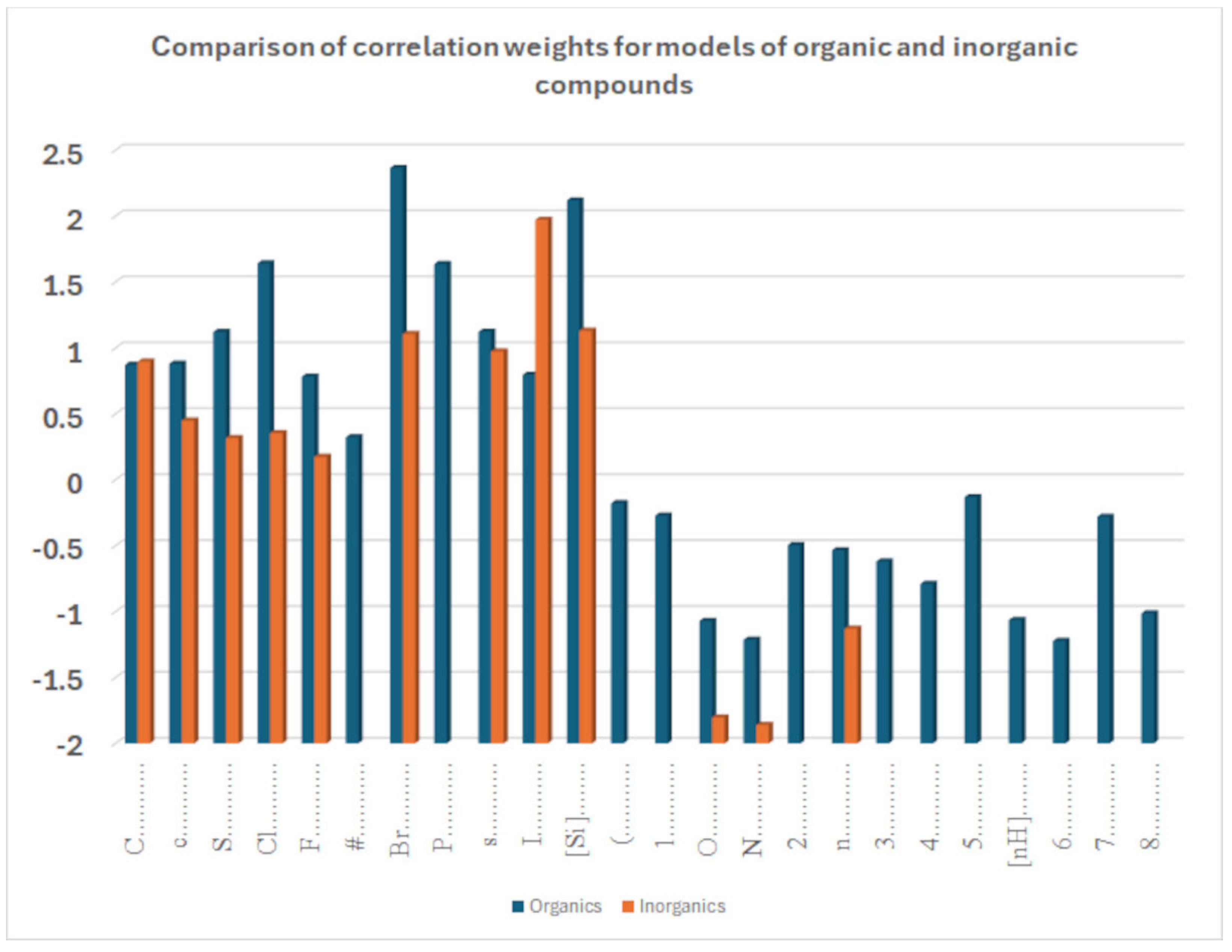
The coefficient serves to clarify the role of SMILES components in increasing or decreasing logP.
Figure 9 compares the influence of various correlation weights, indicating that there is some difference in the participation of different features in the organization of the models. Thus, for inorganic compounds, the same features present for organic compounds appear, but sometimes with a different relevance, as represented by the coefficients. This is the case with chlorine, for instance. It appears both in
Figure 7 and
Figure 8, indicating a role associated with organic and inorganic substances; however, in the case of organic substances, its coefficient is the third largest one, while in the case of inorganic compounds, its coefficient is quite small. This can be explained by the fact that many polychlorinated organic substances largely contribute to the model for logP in the case of organic substances. Bromine and fluorine have a similar behavior, as shown in
Figure 9.
The list of promoters for the increase and decrease in logP for dataset 3 (
Table 7) shows that the property is sensitive to the complexes possessing 3D features, as well as to presence of double bonds. This kind of information is present in dataset 3, which is a quite focused population of substances enabling the investigation of sophisticated features. Thus, one can identify an essential difference between the considered model for organic and inorganic compounds and the model for Pt (IV) complexes. Certain features are common to all datasets. Indeed, oxygen and nitrogen are responsible for an increased polarity, and thus a reduced logP, while branching and chlorine increase logP, as discussed above, for the previous datasets as well.
The features relevant for dataset 4 are shown in
Table 8. In the case of the models of enthalpy formation, some features increase the enthalpy, with different levels of effect, considering the sign and value of the coefficient. As in the other cases, it is necessary to have consistency between the three runs. Thus, mercury, tin, organic carbon, and a relatively large number of rings (three) increase enthalpy. Conversely, chlorine, boron, branching, and aliphatic carbon decrease the enthalpy. Chlorine is by far the most relevant feature playing a role in enthalpy, followed by boron and mercury.
Table 5 shows the promoters of an increase or decrease in the effect for dataset 5. This dataset concerns acute toxicity in rats. The endpoint is expressed as the negative decimal logarithm of the oral lethal dose. Thus, a higher value is associated with a higher toxicity. The SMILES for the toxicity case, studied with dataset 5, has a different configuration compared to the SMILESs used for the other models because, for these compounds, an important aspect of the structures is the charged particles, which are virtually absent for the other sets of molecules considered here. From
Table 9, we can see that nickel has the highest effect. Other salts have a role too. Regarding the organic components of the molecules, aliphatic carbon, double bonds, and branching have a negative coefficient, while the presence of rings has a positive coefficient.
Here, as in other datasets, it can be noted that the main differences between the models for organic and inorganic compounds are the sensitivity of organic compounds to branching and the presence of rings. This is quite obvious, since many organic substances are not linear and contain rings. The models correctly identified these peculiar features, and thus, this confirms the correctness of the modeling approach, from a practical point of view and based on the fact that the models are heuristic. The presence of metals, salts, etc., is another feature extracted by the models. This involves the fact that the modeling approach is versatile and can deal with organic and inorganic features simultaneously. Thus, the lack of models for inorganic compounds in the literature is explained only partially by the more limited number of inorganic compounds. A key point in covering the present gap of models on inorganic or combined substances is the use of adequate tools, as demonstrated here.