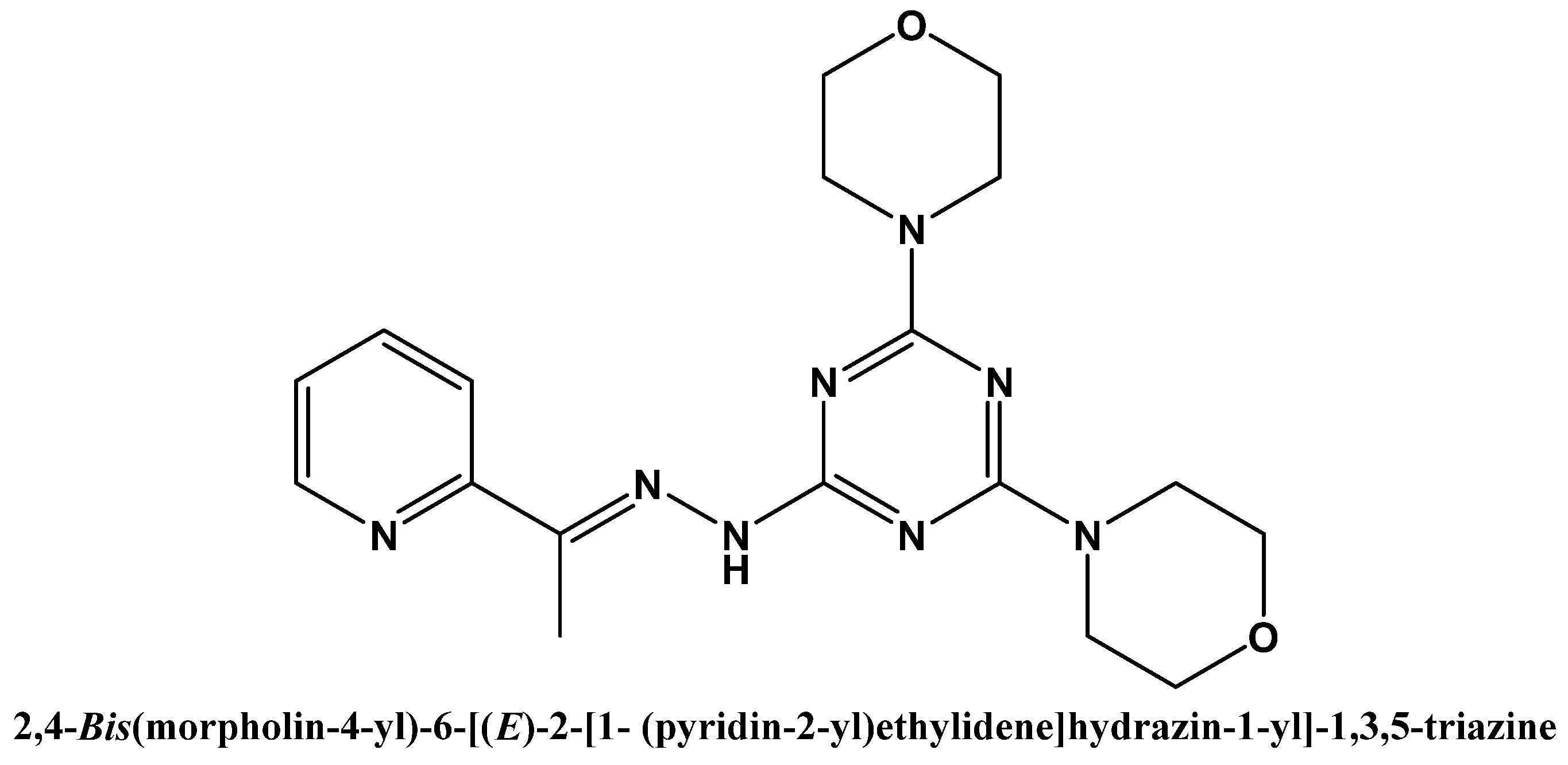
Synthesis, X-ray Structure, Cytotoxic, and Anti-Microbial Activities of Zn(II) Complexes with a Hydrazono s-Triazine Bearing Pyridyl Arm
Abstract
1. Introduction
2. Results and Discussion
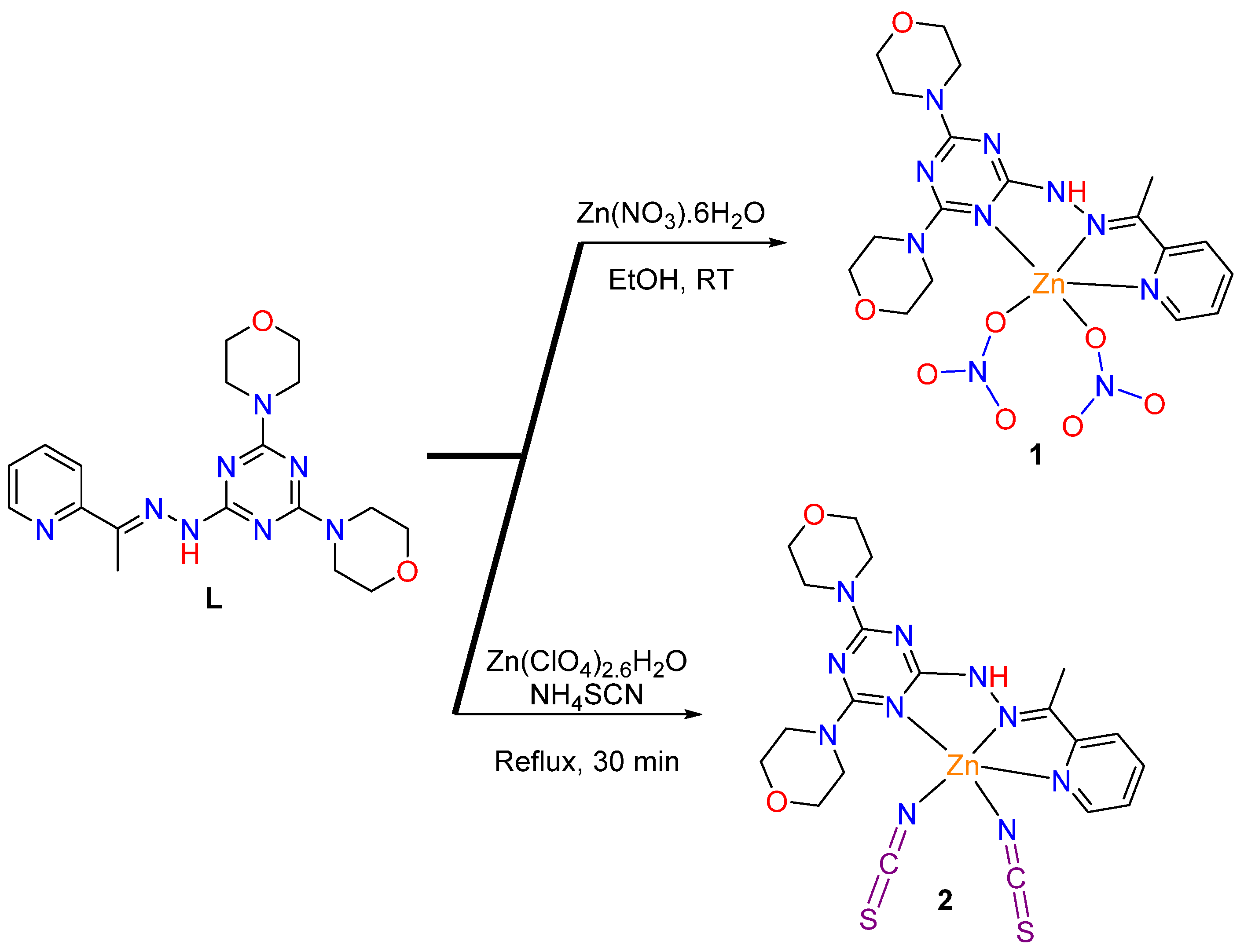
2.1. Synthesis and Characterizations
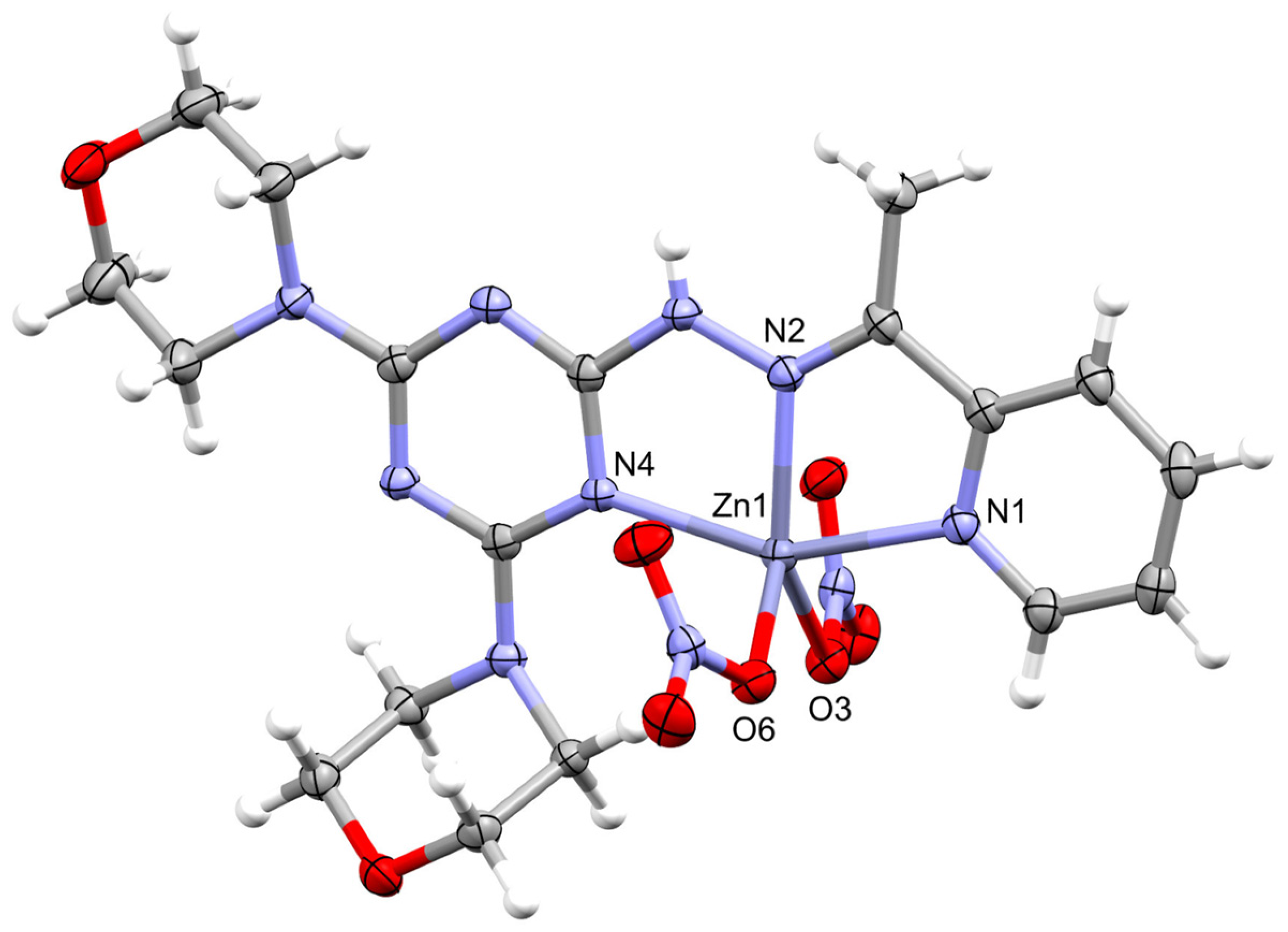
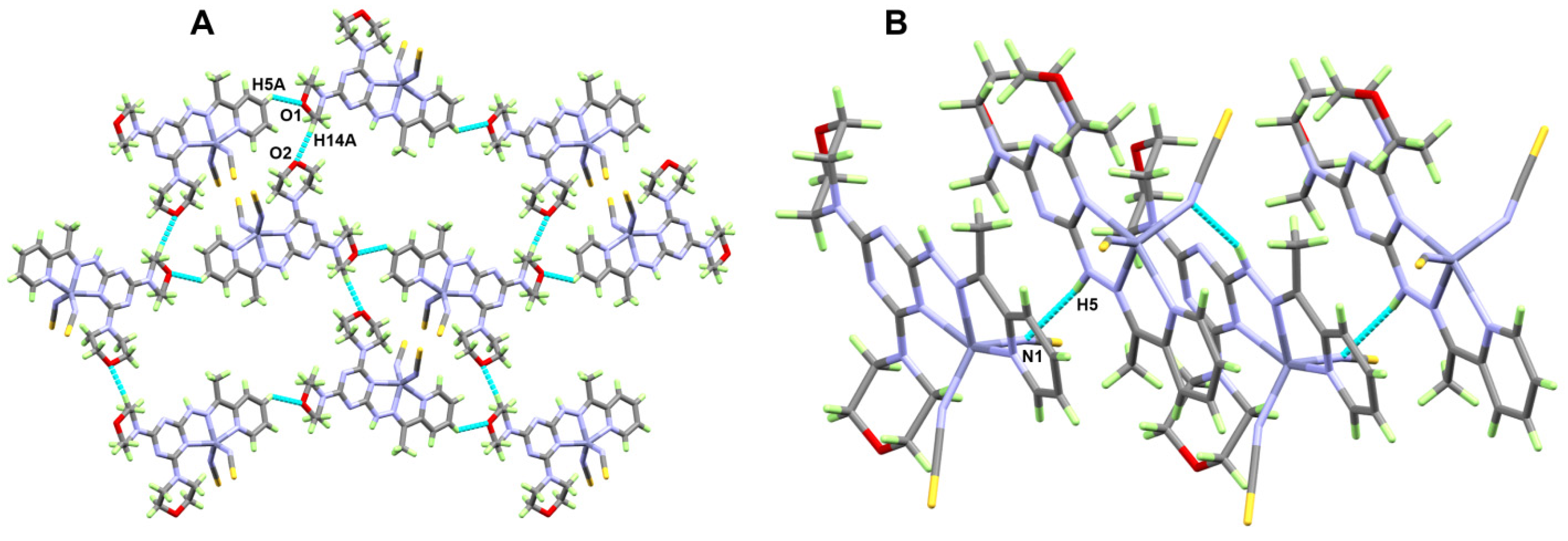
2.2. X-ray Structure Description for [ZnL(ONO2)2]; 1
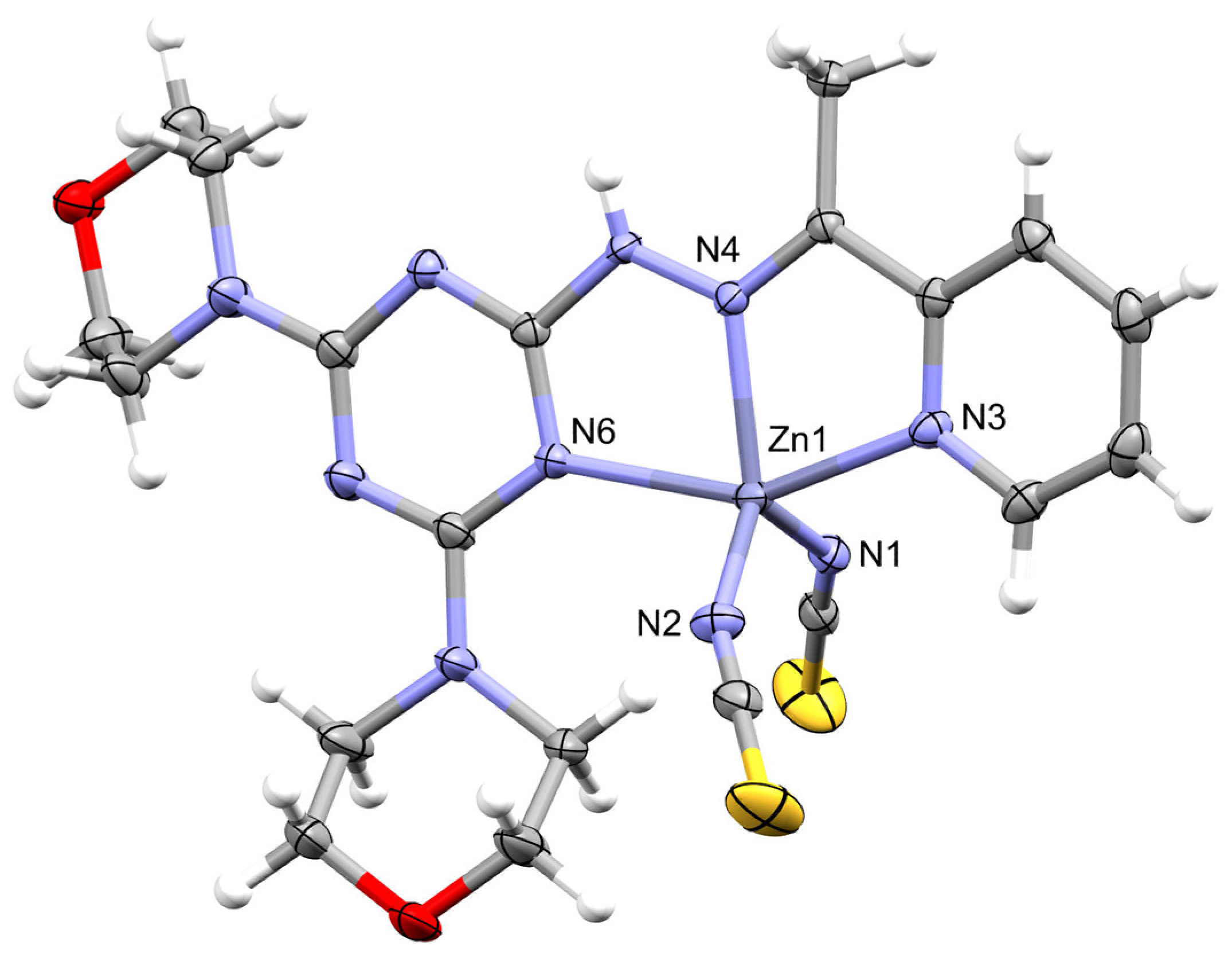
2.3. X-ray Structure Description for [ZnL(NCS)2]; 2
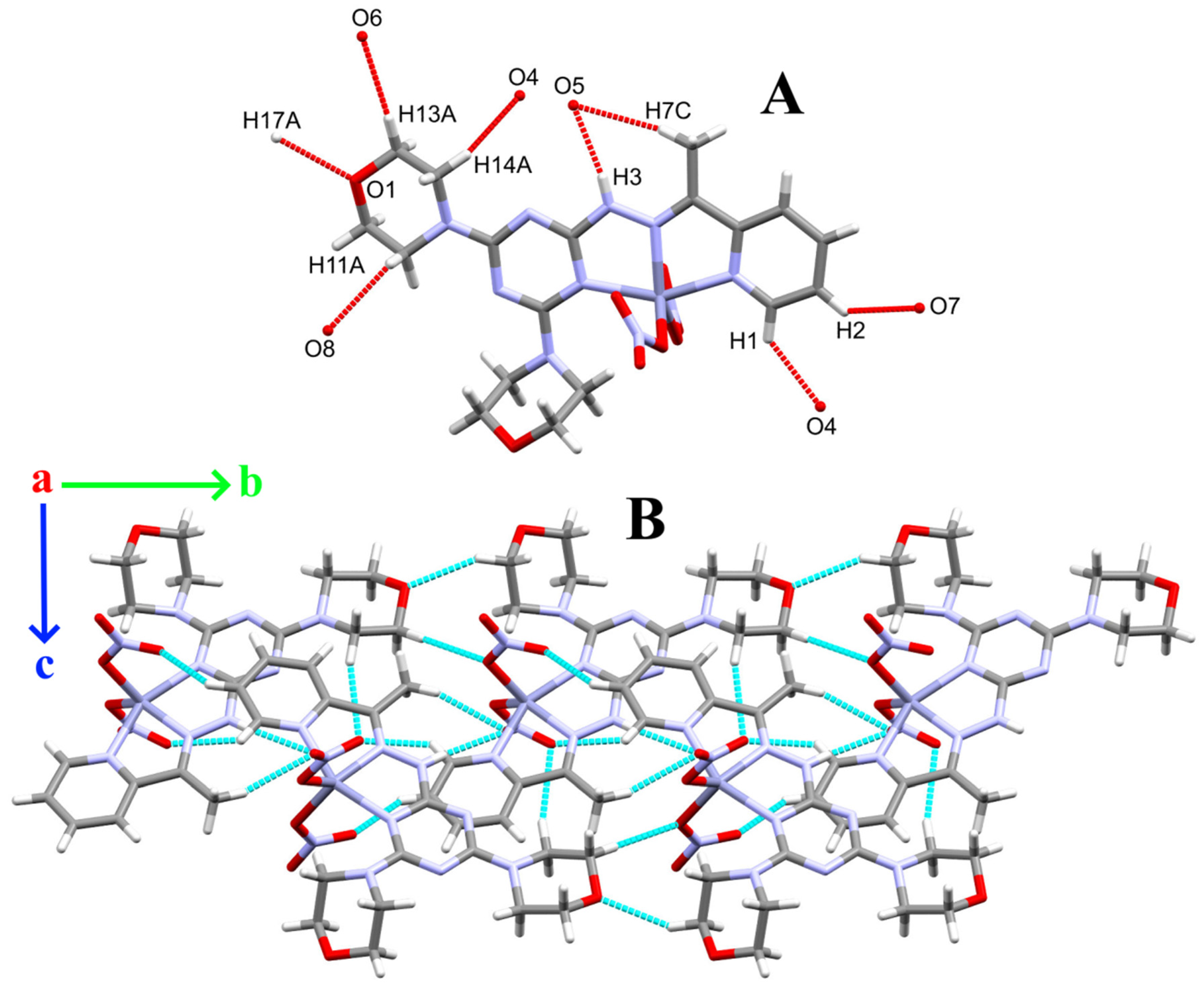
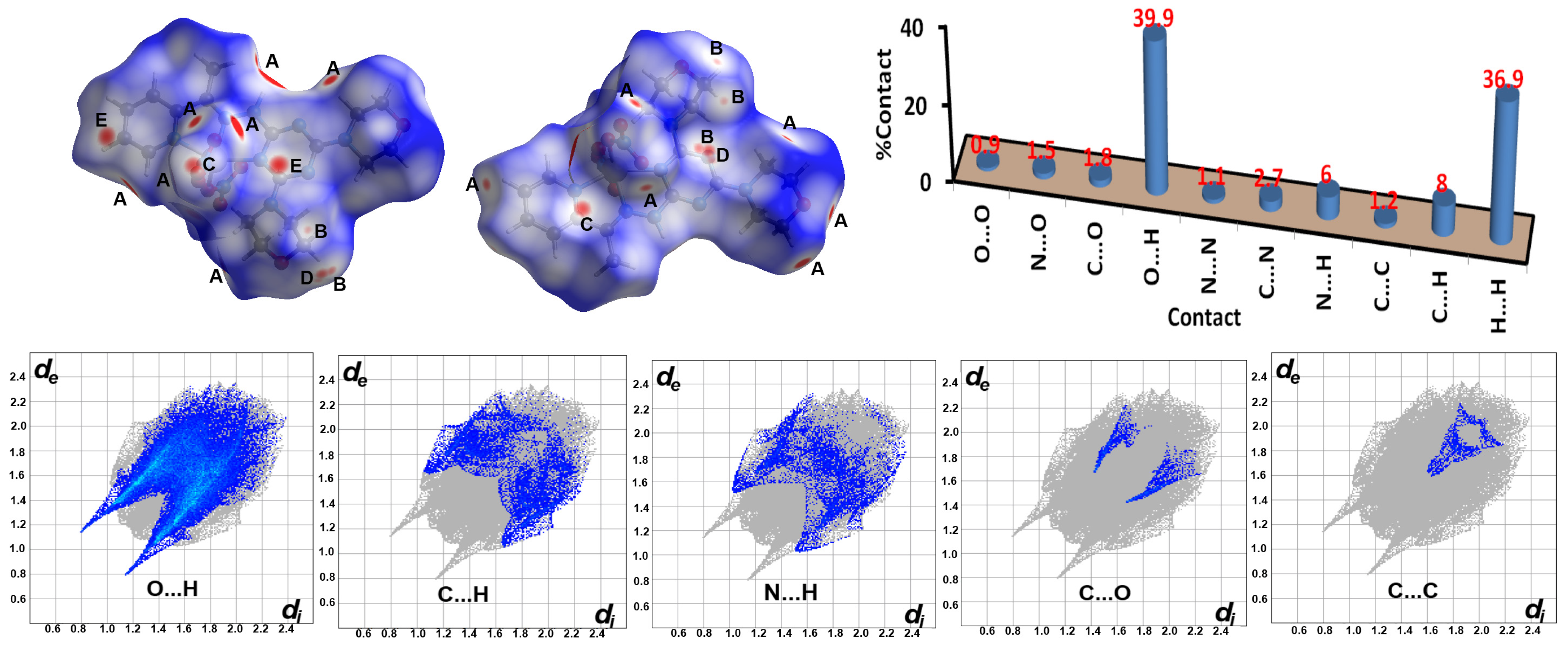
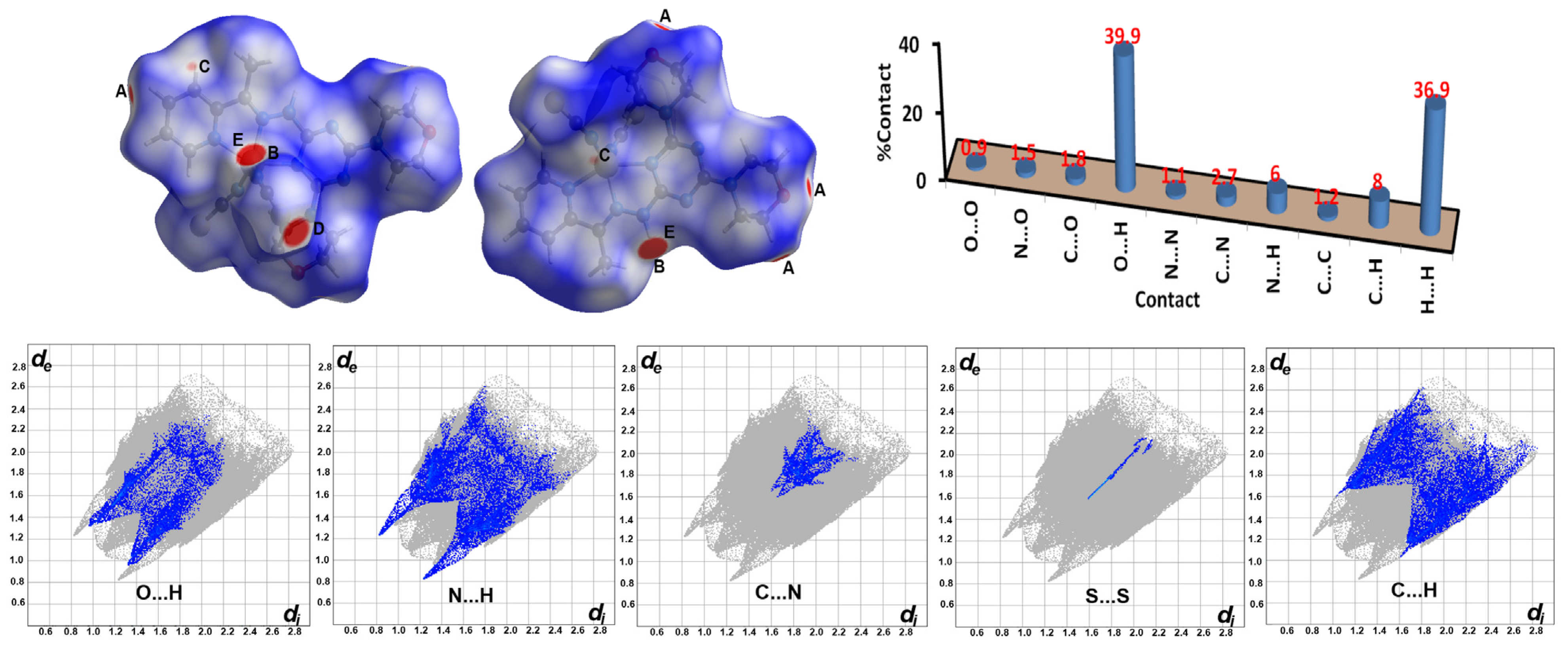
2.4. Hirshfeld Analysis
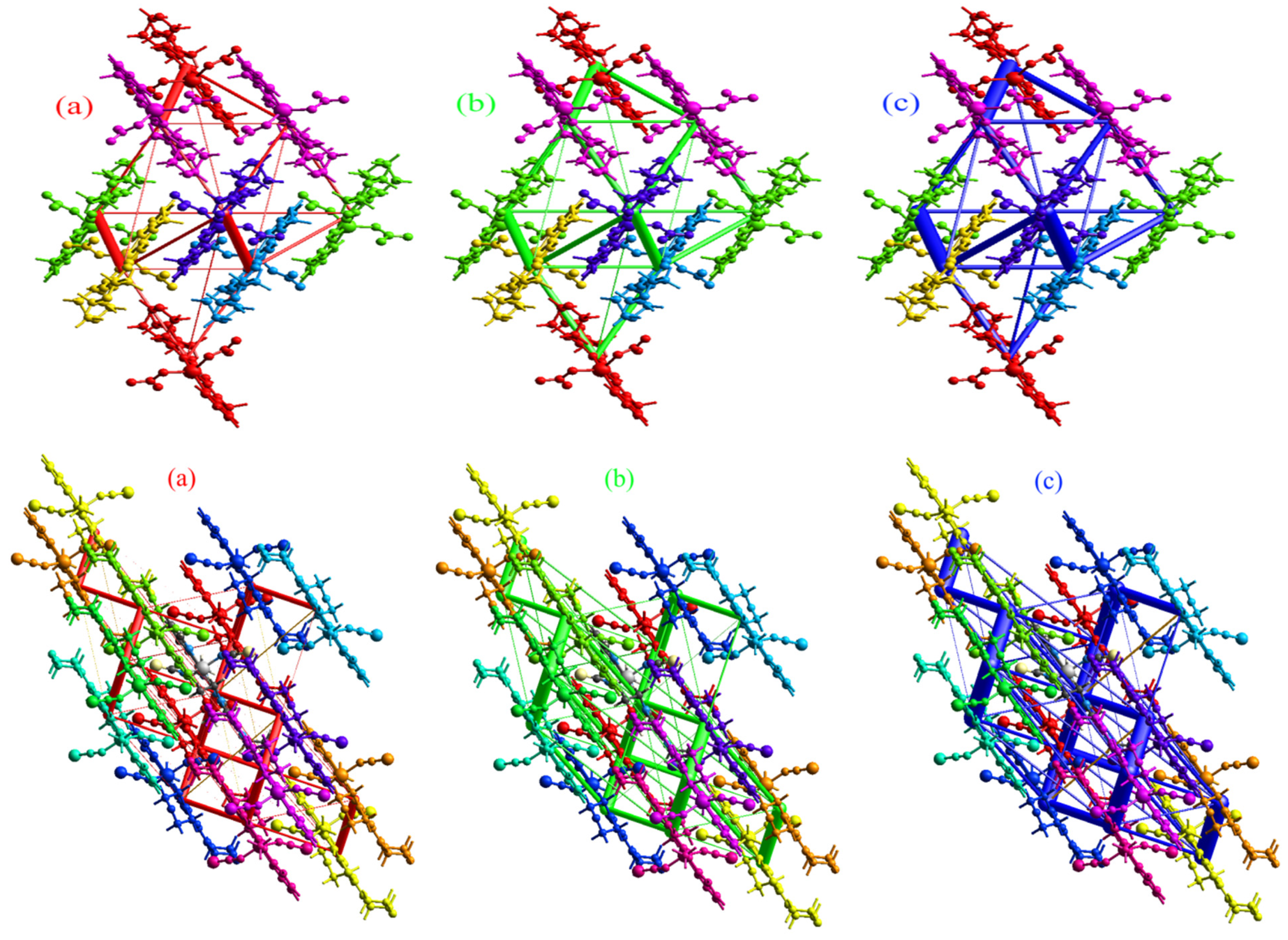
2.5. Energy Framework Analysis
2.6. NBO Charge Analysis
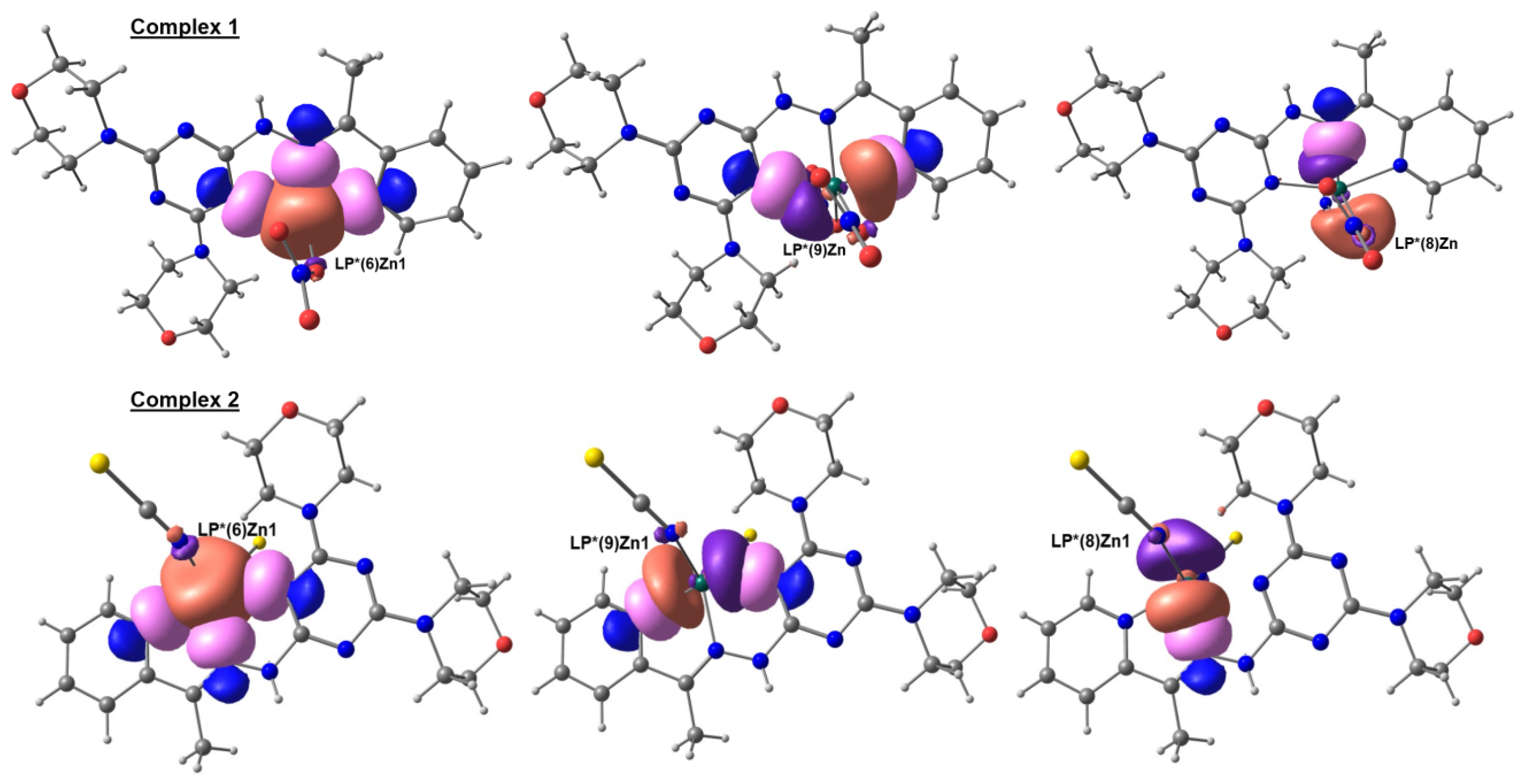
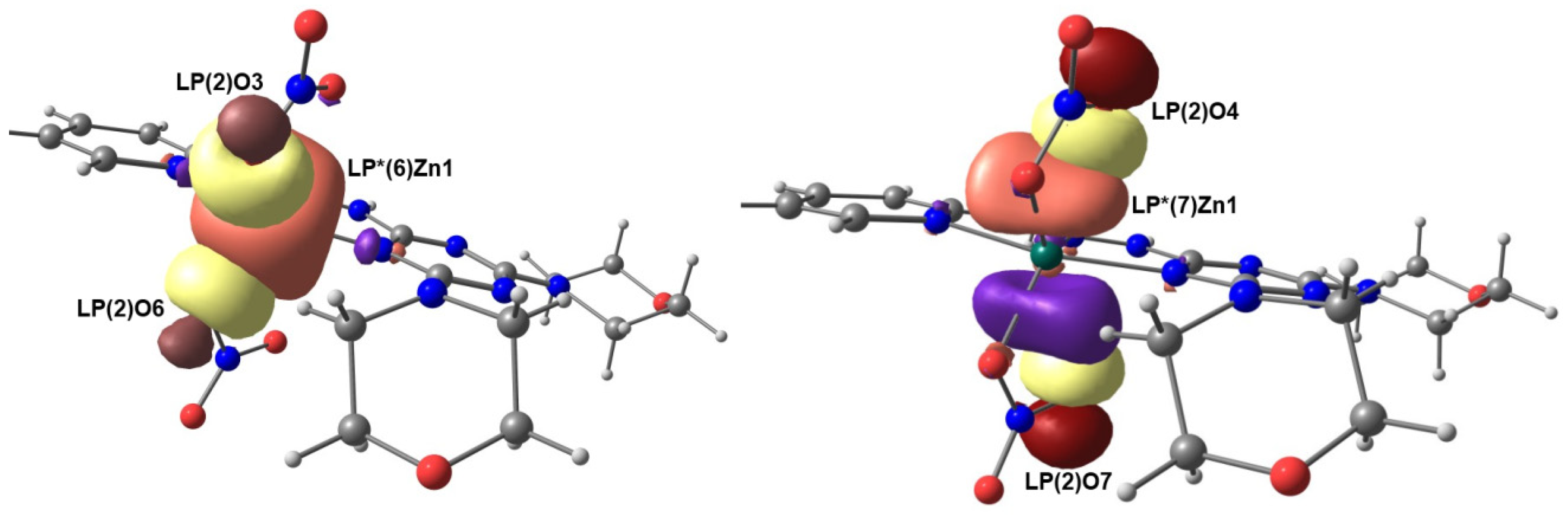
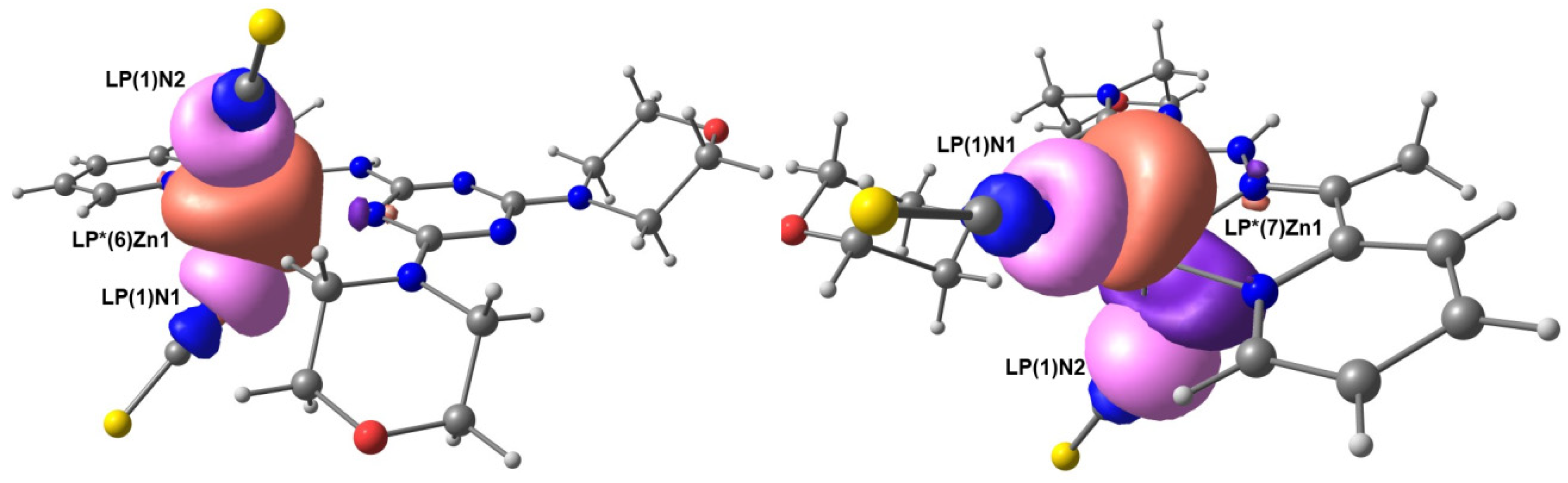
2.7. NBO Analysis
2.8. Antimicrobial Activity
2.9. Cytotoxic Activity
3. Materials and Methods
3.1. Chemicals and Instruments
3.2. Syntheses
3.2.1. Synthesis of [ZnL(ONO2)2]; 1
3.2.2. Synthesis of [ZnL(NCS)2]; 2
3.3. X-ray Structure Determinations
3.4. Hirshfeld Analysis
3.5. Computational Details
3.6. Biological Studies
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pellei, M.; Del Bello, F.; Porchia, M.; Santini, C. Zinc coordination complexes as anticancer agents. Coord. Chem. Rev. 2021, 445, 214088. [Google Scholar] [CrossRef]
- Vallee, B.L.; Auld, D.S. Zinc coordination, function, and structure of zinc enzymes and other proteins. Biochemistry 1990, 29, 5647–5659. [Google Scholar] [CrossRef]
- Vallee, B.L.; Auld, D.S. Active-site zinc ligands and activated H2O of zinc enzymes. Proc. Nat. Acad. Sci. USA 1990, 87, 220–224. [Google Scholar] [CrossRef] [PubMed]
- Lipscomb, W.N.; Sträter, N. Recent advances in zinc enzymology. Chem. Rev. 1996, 96, 2375–2434. [Google Scholar] [CrossRef] [PubMed]
- Andreini, C.; Bertini, I. A bioinformatics view of zinc enzymes. J. Inorg. Biochem. 2012, 111, 150–156. [Google Scholar] [CrossRef]
- Costello, L.C.; Fenselau, C.C.; Franklin, R.B. Evidence for operation of the direct zinc ligand exchange mechanism for trafficking, transport, and reactivity of zinc in mammalian cells. J. Inorg. Biochem. 2011, 105, 589–599. [Google Scholar] [CrossRef] [PubMed]
- Federico, A.; Iodice, P.; Federico, P.; Del Rio, A.; Mellone, M.; Catalano, G. Effects of selenium and zinc supplementation on nutritional status in patients with cancer of digestive tract. Eur. J. Clin. Nutr. 2001, 55, 293–297. [Google Scholar] [CrossRef]
- Prasad, A.S.; Beck, F.W.; Doerr, T.D.; Shamsa, F.H.; Penny, H.S.; Marks, S.C.; Kaplan, J.; Kucuk, O.; Mathog, R.H. Nutritional and zinc status of head and neck cancer patients: An interpretive review. J. Am. Coll. Nutr. 1998, 17, 409–418. [Google Scholar] [CrossRef] [PubMed]
- d’Angelo, J.; Morgant, G.; Ghermani, N.E.; Desmaële, D.; Fraisse, B.; Bonhomme, F.; Dichi, E.; Sghaier, M.; Li, Y.; Journaux, Y. Crystal structures and physico-chemical properties of Zn(II) and Co(II) tetraaqua (3-nitro-4-hydroxybenzoato) complexes: Their anticonvulsant activities as well as related (5-nitrosalicylato)–metal complexes. Polyhedron 2008, 27, 537–546. [Google Scholar] [CrossRef]
- Sakurai, H.; Kojima, Y.; Yoshikawa, Y.; Kawabe, K.; Yasui, H. Antidiabetic vanadium(IV) and zinc(II) complexes. Coord. Chem. Rev. 2002, 226, 187–198. [Google Scholar] [CrossRef]
- Zhou, Q.; Hambley, T.W.; Kennedy, B.J.; Lay, P.A.; Turner, P.; Warwick, B.; Biffin, J.R.; Regtop, H.L. Syntheses and characterization of anti-inflammatory dinuclear and mononuclear zinc indomethacin complexes. Crystal structures of [Zn2(indomethacin)4(L)2](L = N,N-dimethylacetamide,pyridine,1-methyl-2-pyrrolidinone) and [Zn(indomethacin)2(L1)2](L1 = ethanol, methanol). Inorg. Chem. 2000, 39, 3742–3748. [Google Scholar] [PubMed]
- Kasuga, N.C.; Sekino, K.; Ishikawa, M.; Honda, A.; Yokoyama, M.; Nakano, S.; Shimada, N.; Koumo, C.; Nomiya, K. Synthesis, structural characterization and antimicrobial activities of 12 zinc(II) complexes with four thiosemicarbazone and two semicarbazone ligands. J. Inorg. Biochem. 2003, 96, 298–310. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.Q.; Wu, F.J.; Gong, Y.; Hu, C.W.; Zhang, Y.H.; Gan, M.Y. Synthesis, characterization and activity against staphylococcus of metal(II)-gatifloxacin complexes. Chin. J. Chem. 2007, 25, 1809–1814. [Google Scholar] [CrossRef]
- Chen, Z.-F.; Xiong, R.-G.; Zhang, J.; Chen, X.-T.; Xue, Z.-L.; You, X.-Z. 2D molecular square grid with strong blue fluorescent emission: A complex of norfloxacin with zinc(II). Inorg. Chem. 2001, 40, 4075–4077. [Google Scholar] [CrossRef]
- Lopez-Gresa, M.; Ortiz, R.; Perelló, L.; Latorre, J.; Liu-Gonzalez, M.; Garcıa-Granda, S.; Perez-Priede, M.; Canton, E. Interactions of metal ions with two quinolone antimicrobial agents (cinoxacin and ciprofloxacin): Spectroscopic and X-ray structural characterization. Antibacterial studies. J. Inorg. Biochem. 2002, 92, 65–74. [Google Scholar] [CrossRef]
- Xiao, D.R.; Wang, E.B.; An, H.Y.; Su, Z.M.; Li, Y.G.; Gao, L.; Sun, C.Y.; Xu, L. Rationally designed, polymeric, extended metal–ciprofloxacin complexes. Chem. A Eur. J. 2005, 11, 6673–6686. [Google Scholar] [CrossRef]
- Tarushi, A.; Lafazanis, K.; Kljun, J.; Turel, I.; Pantazaki, A.A.; Psomas, G.; Kessissoglou, D.P. First-and second-generation quinolone antibacterial drugs interacting with zinc(II): Structure and biological perspectives. J. Inorg. Biochem. 2013, 121, 53–65. [Google Scholar] [CrossRef] [PubMed]
- Tarushi, A.; Karaflou, Z.; Kljun, J.; Turel, I.; Psomas, G.; Papadopoulos, A.N.; Kessissoglou, D.P. Antioxidant capacity and DNA-interaction studies of zinc complexes with a non-steroidal anti-inflammatory drug, mefenamic acid. J. Inorg. Biochem. 2014, 128, 85–96. [Google Scholar] [CrossRef]
- Tarushi, A.; Totta, X.; Papadopoulos, A.; Kljun, J.; Turel, I.; Kessissoglou, D.P.; Psomas, G. Antioxidant activity and interaction with DNA and albumins of zinc–tolfenamato complexes. Crystal structure of [Zn(tolfenamato)2(2,2′-dipyridylketoneoxime)2]. Eur. J. Med. Chem. 2014, 74, 187–198. [Google Scholar] [CrossRef]
- Kovala-Demertzi, D.; Yadav, P.N.; Wiecek, J.; Skoulika, S.; Varadinova, T.; Demertzis, M.A. Zinc(II) complexes derived from pyridine-2-carbaldehyde thiosemicarbazone and (1E)-1-pyridin-2-ylethan-1-one thiosemicarbazone. Synthesis, crystal structures and antiproliferative activity of zinc(II) complexes. J. Inorg. Biochem. 2006, 100, 1558–1567. [Google Scholar] [CrossRef]
- Belicchi Ferrari, M.; Bisceglie, F.; Pelosi, G.; Tarasconi, P.; Albertini, R.; Pinelli, S. New methyl pyruvate thiosemicarbazones and their copper and zinc complexes: Synthesis, characterization, X-ray structures and biological activity. J. Inorg. Biochem. 2001, 87, 137–147. [Google Scholar] [CrossRef]
- Trávníček, Z.; Kryštof, V.; Šipl, M. Zinc(II) complexes with potent cyclin-dependent kinase inhibitors derived from 6-benzylaminopurine: Synthesis, characterization, X-ray structures and biological activity. J. Inorg. Biochem. 2006, 100, 214–225. [Google Scholar] [CrossRef] [PubMed]
- Di Vaira, M.; Bazzicalupi, C.; Orioli, P.; Messori, L.; Bruni, B.; Zatta, P. Clioquinol, a Drug for Alzheimer’s disease specifically interfering with brain metal metabolism: structural characterization of its zinc(II) and copper(II) complexes. Inorg. Chem. 2004, 43, 3795–3797. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.M.; Frei, E.; Straub, J.; Breuer, A.; Wiessler, M. Induction of metallothionein by zinc protects from daunorubicin toxicity in rats. Toxicology 2002, 179, 85–93. [Google Scholar] [CrossRef] [PubMed]
- Klenke, B.; Barrett, M.P.; Brun, R.; Gilbert, I.H. Antiplasmodial activity of a series of 1,3,5-triazine-substituted polyamines. J. Antimicrob. Chemother. 2003, 52, 290–293. [Google Scholar] [CrossRef] [PubMed]
- Srinivas, K.; Srinivas, U.; Bhanuprakash, K.; Harakishore, K.; Murthy, U.; Rao, V.J. Synthesis and antibacterial activity of various substituted s-triazines. Eur. J. Med. Chem. 2006, 41, 1240–1246. [Google Scholar] [CrossRef] [PubMed]
- Mandal, S.; Bérubé, G.; Asselin, É.; Mohammad, I.; Richardson, V.J.; Gupta, A.; Pramanik, S.K.; Williams, A.L.; Mandal, S.K. A novel series of potent cytotoxic agents targeting G2/M phase of the cell cycle and demonstrating cell killing by apoptosis in human breast cancer cells. Bioorg. Med. Chem. Lett. 2007, 17, 4955–4960. [Google Scholar] [CrossRef] [PubMed]
- Garaj, V.; Puccetti, L.; Fasolis, G.; Winum, J.-Y.; Montero, J.-L.; Scozzafava, A.; Vullo, D.; Innocenti, A.; Supuran, C.T. Carbonic anhydrase inhibitors: Novel sulfonamides incorporating 1,3,5-triazine moieties as inhibitors of the cytosolic and tumour-associated carbonic anhydrase isozymes I, II and IX. Bioorg. Med. Chem. Lett. 2005, 15, 3102–3108. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Zhou, H.-C. A metal− organic framework with entatic metal centers exhibiting high gas adsorption affinity. J. Am. Chem. Soc. 2006, 128, 11734–11735. [Google Scholar] [CrossRef]
- Chae, H.K.; Kim, J.; Friedrichs, O.D.; O’Keeffe, M.; Yaghi, O.M. Design of frameworks with mixed triangular and octahedral building blocks exemplified by the structure of [Zn4O(TCA)2] having the pyrite topology. Angew. Chem. Int. Ed. 2003, 42, 3907–3909. [Google Scholar] [CrossRef]
- Kepert, C.J.; Rosseinsky, M.J. Zeolite-like crystal structure of an empty microporous molecular framework. Chem. Commun. 1999, 4, 375–376. [Google Scholar] [CrossRef]
- Wang, S.-N.; Xing, H.; Li, Y.-Z.; Bai, J.; Scheer, M.; Pan, Y.; You, X.-Z. Unprecedented interweaving of single-helical and unequal double-helical chains into chiral metal–organic open frameworks with multiwalled tubular structures. Chem. Commun. 2007, 22, 2293–2295. [Google Scholar] [CrossRef]
- Soliman, S.M.; Elsilk, S.E.; El-Faham, A. Synthesis, structure and biological activity of zinc(II) pincer complexes with 2,4-bis(3,5-dimethyl-1H-pyrazol-1-yl)-6-methoxy-1,3,5-triazine. Inorg. Chim. Acta 2020, 508, 119627. [Google Scholar] [CrossRef]
- Fathalla, E.M.; Abu-Youssef, M.A.; Sharaf, M.M.; El-Faham, A.; Barakat, A.; Badr, A.M.; Soliman, S.M.; Slawin, A.M.; Woollins, J.D. Synthesis, characterizations, antitumor and antimicrobial evaluations of novel Mn(II) and Cu(II) complexes with NNN-tridentate s-triazine-Schiff base ligand. Inorg. Chim. Acta 2023, 555, 121586. [Google Scholar] [CrossRef]
- Fathalla, E.M.; Abu-Youssef, M.A.; Sharaf, M.M.; El-Faham, A.; Barakat, A.; Haukka, M.; Soliman, S.M. Synthesis, X-ray structure of two hexa-coordinated Ni(II) complexes with s-triazine hydrazine Schiff base ligand. Inorganics 2023, 11, 222. [Google Scholar] [CrossRef]
- Addison, A.W.; Rao, T.N.; Reedijk, J.; van Rijn, J.; Verschoor, G.C. Synthesis, structure, and spectroscopic properties of copper(II) compounds containing nitrogen–sulphur donor ligands; the crystal and molecular structure of aqua [1,7-bis(N-methylbenzimidazol-2′-yl)-2,6-dithiaheptane]copper(II) perchlorate. J. Chem. Soc. Dalton Trans. 1984, 7, 1349–1356. [Google Scholar] [CrossRef]
- Morozov, I.; Serezhkin, V.; Troyanov, S. Modes of coordination and stereochemistry of the NO3− anions in inorganic nitrates. Russ. Chem. Bull. 2008, 57, 439–450. [Google Scholar] [CrossRef]
- Kleywegt, G.J.; Wiesmeijer, W.G.; Van Driel, G.J.; Driessen, W.L.; Reedijk, J.; Noordik, J.H. Unidentate versus symmetrically and unsymmetrically bidentate nitrate co-ordination in pyrazole-containing chelates. The crystal and molecular structures of (nitrato-O)[tris(3,5-dimethylpyrazol-1-ylmethyl)amine]copper(II) nitrate,(nitrato-O,O′)[tris(3,5-dimethylpyrazol-1-ylmethyl)amine]nickel(II) nitrate, and (nitrato-O)(nitrato-O,O′)[tris(3,5-dimethylpyrazol-1-ylmethyl) amine]cadmium(II). J. Chem. Soc. Dalton Trans. 1985, 10, 2177–2184. [Google Scholar]
- Janiak, C. A critical account on π–π stacking in metal complexes with aromatic nitrogen-containing ligands. J. Chem. Soc. Dalton Trans. 2000, 21, 3885–3896. [Google Scholar] [CrossRef]
- Turner, M.J.; Thomas, S.P.; Shi, M.W.; Jayatilaka, D.; Spackman, M.A. Energy frameworks: Insights into interaction anisotropy and the mechanical properties of molecular crystals. Chem. Commun. 2015, 51, 3735–3738. [Google Scholar]
- Bakheit, A.H.; Attwa, M.W.; Kadi, A.A.; Alkahtani, H.M. Structural Analysis and Reactivity Insights of (E)-Bromo-4-((4-((1-(4-chlorophenyl)ethylidene)amino)-5-phenyl-4H-1,2,4-triazol-3-yl)thio)-5-((2-isopropylcyclohexyl)oxy) Furan-2(5H)-one: A combined approach using single-crystal X-ray diffraction, Hirshfeld surface analysis, and conceptual density functional theory. Crystals 2023, 13, 1313. [Google Scholar] [CrossRef]
- Turner, M.J.; Grabowsky, S.; Jayatilaka, D.; Spackman, M.A. Accurate and efficient model energies for exploring intermolecular interactions in molecular crystals. J. Phys. Chem. Lett. 2014, 5, 4249–4255. [Google Scholar] [CrossRef]
- Hajji, M.; Mtiraoui, H.; Amiri, N.; Msaddek, M.; Guerfel, T. Crystallographic and first-principles density functional theory study on the structure, noncovalent interactions, and chemical reactivity of 1,5-benzodiazepin-2-ones derivatives. Int. J. Quantum Chem. 2019, 119, e26000. [Google Scholar] [CrossRef]
- Abad, N.; Sallam, H.H.; Al-Ostoot, F.H.; Khamees, H.A.; Al-horaibi, S.A.; Khanum, S.A.; Madegowda, M.; El Hafi, M.; Mague, J.T.; Essassi, E.M. Synthesis, crystal structure, DFT calculations, Hirshfeld surface analysis, energy frameworks, molecular dynamics and docking studies of novel isoxazolequinoxaline derivative (IZQ) as anti-cancer drug. J. Mol. Struct. 2021, 1232, 130004. [Google Scholar] [CrossRef]
- Edwards, A.J.; Mackenzie, C.F.; Spackman, P.R.; Jayatilaka, D.; Spackman, M.A. Intermolecular interactions in molecular crystals: What’s in a name? Faraday Discuss. 2017, 203, 93–112. [Google Scholar] [CrossRef] [PubMed]
- Mackenzie, C.F.; Spackman, P.R.; Jayatilaka, D.; Spackman, M.A. CrystalExplorer model energies and energy frameworks: Extension to metal coordination compounds, organic salts, solvates and open-shell systems. IUCrJ 2017, 4, 575–587. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; He, X.; Rong, C.; Zhong, A.; Liu, S.; Zhao, D. On the origin and nature of internal methyl rotation barriers: An information-theoretic approach study. Theor. Chem. Acc. 2022, 141, 68. [Google Scholar] [CrossRef]
- Zhong, A.; Chen, D.; Li, R. Revisiting the beryllium bonding interactions from energetic and wavefunction perspectives. Chem. Phys. Lett. 2015, 633, 265–272. [Google Scholar] [CrossRef]
- Tan, S.L.; Jotani, M.M.; Tiekink, E.R. Utilizing Hirshfeld surface calculations, non-covalent interaction (NCI) plots and the calculation of interaction energies in the analysis of molecular packing. Acta Crystallogr. E Crystallogr. Commun. 2019, 75, 308–318. [Google Scholar] [CrossRef]
- Sreenatha, N.; Chakravarthy, A.J.; Suchithra, B.; Lakshminarayana, B.; Hariprasad, S.; Ganesha, D. Crystal, spectral characterization, molecular docking, Hirshfeld computational studies and 3D-energy framework analysis of a novel puckered compound (C14H15ClO): 2-Chloro-3-phenyl-5,5-dimethylcyclohex-2-en-1-one. J. Mol. Struct. 2020, 1210, 127979. [Google Scholar] [CrossRef]
- Guo, Z.A.; Xian, J.Y.; Rong, L.R.; Qin, H.; Jie, Z. Theoretical study of metal ion impact on geometric and electronic properties of terbutaline compounds. Monatsh. Chem. 2019, 150, 1355–1364. [Google Scholar] [CrossRef]
- Kheiralla, Z.M.; Abo-Ghalia, H.H.; Elaasser, M.M.; Hemeda, M.F.; Ibrahim, S.Y. Endophyte Chaetomium, Potential Bioactivity: Pharmaceutical and Phytochemical Analyses. Res. Sq. 2023. [Google Scholar] [CrossRef]
- Gomha, S.M.; Muhammad, Z.A.; Abdel-aziz, M.R.; Abdel-aziz, H.M.; Gaber, H.M.; Elaasser, M.M. One-pot synthesis of new thiadiazolyl-pyridines as anticancer and antioxidant agents. J. Heterocycl. Chem. 2018, 55, 530–536. [Google Scholar] [CrossRef]
- Rikagu Oxford Diffraction. CrysAlisPro 1.171.43.100a; Rikagu Oxford Diffraction Inc.: Yarnton, Oxfordshire, UK, 2023. [Google Scholar]
- Sheldrick, G.M. SHELXT—Integrated space-group and crystal-structure determination. Acta Cryst. 2015, A71, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Sheldrick, G.M. Crystal Structure Refinement with SHELXL. Acta Cryst. 2015, C71, 3–8. [Google Scholar]
- Dolomanov, O.V.; Bourhis, L.J.; Gildea, R.J.; Howard, J.A.K.; Puschmann, H.J. OLEX2: A complete structure solution, refinement and analysis program. Appl. Cryst. 2009, 42, 339–341. [Google Scholar] [CrossRef]
- Turner, M.; McKinnon, J.; Wolff, S.; Grimwood, D.; Spackman, P.; Jayatilaka, D.; Spackman, M. CrystalExplorer17; University of Western Australia: Perth, Australia, 2017. [Google Scholar]
- Spackman, M.A.; Jayatilaka, D. Hirshfeld surface analysis. CrystEngComm 2009, 11, 19–32. [Google Scholar] [CrossRef]
- Frisch, M. gaussian 09, Revision d. 01; Gaussian. Inc.: Wallingford, CT, USA, 2009; Volume 201. [Google Scholar]
- Andrienko, G. Chemcraft-Graphical Software for Visualization of Quantum Chemistry Computations. Version 1.8, Build 682. 2010. Available online: https://www.chemcraftprog.com (accessed on 22 July 2020).
- Pritchard, B.P.; Altarawy, D.; Didier, B.; Gibson, T.D.; Windus, T.L. New basis set exchange: An open, up-to-date resource for the molecular sciences community. J. Chem. Inf. Model. 2019, 59, 4814–4820. [Google Scholar] [CrossRef]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]
- Lu, P.-L.; Liu, Y.-C.; Toh, H.-S.; Lee, Y.-L.; Liu, Y.-M.; Ho, C.-M.; Huang, C.-C.; Liu, C.-E.; Ko, W.-C.; Wang, J.-H. Epidemiology and antimicrobial susceptibility profiles of gram-negative bacteria causing urinary tract infections in the Asia-Pacific region: 2009–2010 results from the Study for Monitoring Antimicrobial Resistance Trends (SMART). Int. J. Antimicrob. Agents 2012, 40, S37–S43. [Google Scholar] [CrossRef]

| [ZnL(ONO2)2]; (1) | [ZnL(NCS)2]; (2) | |
|---|---|---|
| CCDC | 2359261 | 2359262 |
| Empirical formula | C18H24N10O8Zn | C20H24N10O2S2Zn |
| Fw | 573.84 | 565.98 |
| Temp (K) | 120.00(10) | 120.00(10) |
| λ (Å) | 1.54184 | 1.54184 |
| Cryst syst | Orthorhombic | Monoclinic |
| Space group | P212121 | P21/c |
| a (Å) | 7.92330(10) | 13.8914(2) |
| b (Å) | 11.89480(10) | 20.5446(2) |
| c (Å) | 24.3806(2) | 8.97020(10) |
| β (deg) | 90 | 108.195(2) |
| V (Å3) | 2297.78(4) | 2432.03(6) |
| Z | 4 | 4 |
| ρcalc (Mg/m3) | 1.659 | 1.546 |
| μ (Mo Kα) (mm−1) | 2.104 | 3.352 |
| No. reflns. | 25,015 | 25,012 |
| Unique reflns. | 4934 | 5007 |
| Completeness to θ = 67.684° | 100% | 99.6% |
| Absolute structure parameter | −0.020(6) | |
| GOOF (F2) | 1.057 | 1.041 |
| Rint | 0.0274 | 0.0246 |
| R1 a (I ≥ 2σ) | 0.0220 | 0.0301 |
| wR2 b (I ≥ 2σ) | 0.0587 | 0.0722 |
| Bond Length | |||
| Zn1-O3 | 2.0448(16) | Zn(1)-N(2) | 2.0443(19) |
| Zn1-O6 | 2.0131(17) | Zn(1)-N(4) | 2.1713(19) |
| Zn1-N1 | 2.1416(19) | ||
| Bond Angle | |||
| O3-Zn1-N1 | 94.98(7) | O(6)-Zn(1)-N(4) | 105.96(7) |
| O3-Zn1-N4 | 101.93(7) | N(1)-Zn(1)-N(4) | 153.23(7) |
| O6-Zn1-O3 | 100.30(7) | N(2)-Zn(1)-O(3) | 136.51(7) |
| O6-Zn1-N1 | 91.02(8) | N(2)-Zn(1)-N(1) | 75.36(8) |
| O6-Zn1-N2 | 121.80(7) | N(2)-Zn(1)-N(4) | 78.09(7) |
| D-H···A | d(D-H) | d(H···A) | d(D···A) | D-H···A |
|---|---|---|---|---|
| N3-H3···O5 a | 0.79(3) | 2.15(3) | 2.915(3) | 164(3) |
| C1-H1···O4 b | 0.95 | 2.36 | 3.109(3) | 135 |
| C2-H2···O7 c | 0.95 | 2.57 | 3.260(3) | 130 |
| C7-H7C···O5 a | 0.98 | 2.56 | 3.495(3) | 160’ |
| C11-H11A···O8 d | 0.99 | 2.53 | 3.216(3) | 127 |
| C13-H13A···O6 e | 0.99 | 2.48 | 3.460(3) | 171 |
| C14-H14A···O4 a | 0.99 | 2.50 | 3.372(3) | 147 |
| C17-H17A···O1 f | 0.99 | 2.49 | 3.327(3) | 142 |
| Bond Length | |||
| Zn1-N4 | 2.0565(14) | Zn1-N3 | 2.1813(14) |
| Zn1-N6 | 2.2696(14) | Zn1-N2 | 1.9466(16) |
| Zn1-N1 | 1.9645(15) | ||
| Bond Angle | |||
| N4-Zn1-N6 | 76.18(5) | N3-Zn1-N6 | 151.30(5) |
| N4-Zn1-N3 | 75.17(5) | N2-Zn1-N4 | 126.46(6) |
| N1-Zn1-N4 | 112.89(6) | N2-Zn1-N6 | 103.14(6) |
| N1-Zn1-N6 | 93.43(6) | N2-Zn1-N1 | 120.51(6) |
| N1-Zn1-N3 | 99.10(6) | N2-Zn1-N3 | 92.58(6) |
| D-H···A | d(D-H) | d(H···A) | d(D···A) | D-H···A |
|---|---|---|---|---|
| N5-H5···N1 a | 0.81(2) | 2.24(3) | 3.008(2) | 159(2) |
| C5-H5A···O1 b | 0.91(3) | 2.50(3) | 3.177(2) | 131(2) |
| C14-H14A···O2 c | 0.99 | 2.38 | 3.371(2) | 174 |
| Interaction | Distance | Interaction | Distance |
|---|---|---|---|
| O1···H17A | 2.416 | C5···O5 | 3.099 |
| O6···H13A | 2.386 | C15···H16A | 2.747 |
| O5···H7C | 2.460 | C9···H16B | 2.724 |
| O4···H14A | 2.424 | C10···H16B | 2.741 |
| O5···H3 | 1.940 | C16···H13A | 2.739 |
| O4···H1 | 2.266 | N5···H16B | 2.549 |
| O7···H2 | 2.486 | C9···C2 | 3.327 |
| O8···H11A | 2.472 | ||
| O2···H3A | 2.596 |
| Species | Charge | Species | Charge |
|---|---|---|---|
| 1 | 2 | ||
| Ligand | 0.325 | Ligand | 0.365 |
| Zn1 | 1.217 | Zn1 | 1.145 |
| NO3− a | −0.764 | NCS− a | −0.753 |
| NO3− b | −0.778 | NCS− b | −0.758 |
| Acceptor NBO | 1 | 2 |
|---|---|---|
| LP*(6) | 0.324 | 0.342 |
| LP*(7) | 0.156 | 0.200 |
| LP*(8) | 0.148 | 0.168 |
| LP*(9) | 0.141 | 0.151 |
| Microorganism | L a | [ZnL(ONO2)2]; 1 | [ZnL(NCS)2]; 2 | [Mn(L)Cl2] a | [Cu(L)Cl2]*H2O a | Control |
|---|---|---|---|---|---|---|
| S. aureus | NA b (ND) c | 14(625) | 15(625) | 11(2500) | 18(625) | 24(78) d |
| B. subtilis | NA b (ND) c | 15(625) | 16(312) | 19(312) | 20(312) | 26(39) d |
| E.coli | NA b (ND) c | NA b (ND) c | NA b (ND) c | 10(2500) | NA(ND) | 30(10) d |
| P.vulgaris | NA b (ND) c | 18(156) | 20(78) | 12(1250) | 14(1250) | 25(5) d |
| A. fumigatus | NA b (ND) c | 10(625) | NA b (ND) c | NA b (ND) c | NA b (ND) c | 17(5) e |
| C. albicans | NA b (ND) c | 9(625) | 10(312) | 8(5000) | NA b (ND) c | 20(5) e |
| Compound | HCT-116 | SI | A-549 | SI | WI-38 |
|---|---|---|---|---|---|
| L | 59.85 ± 2.92 a | 5.4 | 55.84 ± 2.74 a | 4.5 | 104.01 ± 3.61 |
| [ZnL(ONO2)2] | 29.53 ± 1.24 | 1.9 | 35.55 ± 1.69 | 1.6 | 55.95 ± 1.86 |
| [ZnL(NCS)2] | 41.25 ± 2.91 | 1.5 | 55.05 ± 2.87 | 1.1 | 61.57 ± 2.17 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hassan, M.; El-Faham, A.; Barakat, A.; Haukka, M.; Tatikonda, R.; Abu-Youssef, M.A.M.; Soliman, S.M.; Yousri, A. Synthesis, X-ray Structure, Cytotoxic, and Anti-Microbial Activities of Zn(II) Complexes with a Hydrazono s-Triazine Bearing Pyridyl Arm. Inorganics 2024, 12, 176. https://doi.org/10.3390/inorganics12070176
Hassan M, El-Faham A, Barakat A, Haukka M, Tatikonda R, Abu-Youssef MAM, Soliman SM, Yousri A. Synthesis, X-ray Structure, Cytotoxic, and Anti-Microbial Activities of Zn(II) Complexes with a Hydrazono s-Triazine Bearing Pyridyl Arm. Inorganics. 2024; 12(7):176. https://doi.org/10.3390/inorganics12070176
Chicago/Turabian StyleHassan, MennaAllah, Ayman El-Faham, Assem Barakat, Matti Haukka, Rajendhraprasad Tatikonda, Morsy A. M. Abu-Youssef, Saied M. Soliman, and Amal Yousri. 2024. "Synthesis, X-ray Structure, Cytotoxic, and Anti-Microbial Activities of Zn(II) Complexes with a Hydrazono s-Triazine Bearing Pyridyl Arm" Inorganics 12, no. 7: 176. https://doi.org/10.3390/inorganics12070176
APA StyleHassan, M., El-Faham, A., Barakat, A., Haukka, M., Tatikonda, R., Abu-Youssef, M. A. M., Soliman, S. M., & Yousri, A. (2024). Synthesis, X-ray Structure, Cytotoxic, and Anti-Microbial Activities of Zn(II) Complexes with a Hydrazono s-Triazine Bearing Pyridyl Arm. Inorganics, 12(7), 176. https://doi.org/10.3390/inorganics12070176