Abstract
Cys2His2 zinc finger proteins are important for living organisms, as they—among other functions—specifically recognise DNA when Zn(II) is coordinated to the proteins, stabilising their ββα secondary structure. Therefore, competition with other metal ions may alter their original function. Toxic metal ions such as Cd(II) or Hg(II) might be especially dangerous because of their similar chemical properties to Zn(II). Most competition studies carried out so far have involved small zinc finger peptides. Therefore, we have investigated the interactions of toxic metal ions with a zinc finger proteins consisting of three finger units and the consequences on the DNA binding properties of the protein. Binding of one Cd(II) per finger subunit of the protein was shown by circular dichroism spectroscopy, fluorimetry and electrospray ionisation mass spectrometry. Cd(II) stabilised a similar secondary structure to that of the Zn(II)-bound protein but with a slightly lower affinity. In contrast, Hg(II) could displace Zn(II) quantitatively (logβ′ ≥ 16.7), demolishing the secondary structure, and further Hg(II) binding was also observed. Based on electrophoretic gel mobility shift assays, the Cd(II)-bound zinc finger protein could recognise the specific DNA target sequence similarly to the Zn(II)-loaded form but with a ~0.6 log units lower stability constant, while Hg(II) could destroy DNA binding completely.
1. Introduction
Zinc finger proteins (ZFPs) are present in various living organisms, such as amphibians, reptiles and mammals [1,2,3,4]. ZFPs are involved in DNA transcription, translation, error correction, metabolism, stimulus generation, cell division and cell death by interacting with other proteins, small molecules, RNA and DNA [5,6]. Zinc finger (ZF) motifs of a ZFP are involved in molecular recognition, while the rest of the protein is most commonly responsible for its function [7,8,9,10,11,12,13]. The structure of a ZF motif is stabilised by the tetrahedral coordination of Zn(II) and by the formation of a hydrophobic core [14]. Similar tetrahedral coordination was found in self-assembling peptides offering a Cys2His2 binding site [15]. Cys2His2-type proteins form the most populous family of ZFPs [16]. Their biotechnological significance is highlighted by the fact that a ZF unit recognises three subsequent nucleobases in a double strand (ds) DNA, and several ZF units can be linked together to increase the specificity of the interaction. ZF arrays were first applied as the DNA recognition domains of artificial nucleases linked to the FokI nuclease domain [17]. Since then, numerous gene modification experiments have been performed with nucleases of this type [18,19,20,21,22,23,24]. The recognition sequence can further be extended by the chimera of ZF and other DNA binding motifs [25].
Cys2His2 ZFPs can only bind to DNA specifically in their Zn(II)-bound form. Other metal ions inside the living organism may substitute Zn(II), form mixed complexes and/or promote oxidation of the cysteines. In addition, toxic metal ions may also react with ZFPs, rendering the investigation of these interactions crucial. The coordination chemistry and biophysical properties of ZFPs [11,26,27,28] have been extensively studied, but still there are open questions regarding the stabilities of their complexes and the competition between Zn(II) and non-native metal ions [29].
Cd(II) has a stronger “soft” character than Zn(II). Therefore, it forms the most stable complexes with thiolate ligands, while it can also interact with nitrogen and oxygen donor atoms in biological systems. ZF peptides modelling a Cys2His2 type ZF unit bind Zn(II) about two–three orders of magnitude more strongly than Cd(II). The affinities of the two metal ions are comparable towards the Cys3His binding site, while the Cys4 ZF motifs bind Cd(II) two–three orders of magnitude stronger than Zn(II) [30,31,32,33]. Heinz et al. investigated the metal ion coordination of the consensus peptide 1 (CP1) Cys2His2 ZF model. Starting from the apo-peptide, they reported that a biscomplex forms with Cd(II) when the ligand is in excess through the cysteine thiolates, while Cys2His2 coordination is favoured in the monocomplex formed at a 1:1 initial metal to ligand ratio [34]. Cd(II) may disturb DNA recognition of a ZFP and thus the related biological processes, which is one of the possible mechanisms of its toxic effect [35]. Petering et al. and Hanas et al. found that Cd(II) could inhibit DNA binding of the Zn(II)-bound TFIIIA ZFP [35,36,37]. On the other hand, it was shown that both the high- and low-stability binding sites of TFIIIA bind Cd(II) weaker than Zn(II) by ~2.5 and one order of magnitude, respectively [38]. Investigations with the 3rd ZF subunit of TFIIIA ZFP revealed that once formed, the Cd(II) complex had a similar secondary structure and just a slightly lower DNA binding affinity (not specific) than the Zn(II) complex [39]. The situation is even more complicated in the Sp1 transcription factor consisting of three ZF subunits. In a few publications, Cd(II) has been shown to inhibit DNA binding of Zn(II) saturated Sp1 [40,41,42], while others did not observe such effect [43]. Kuwahara et al. reported that the Cd(II) complex of Sp1 was also capable of recognizing the specific target DNA, but with slightly lower affinity than the Zn(II) complex [44]. Malgieri et al. reported comparable affinity of Ros87, an eukaryotic Cys2His2-type ZFP, towards Cd(II) and Zn(II). The two complexes shared a similar secondary structure based on UV–Vis, CD and NMR measurements. Furthermore, the Cd(II) complex could recognise the same DNA target as the Zn(II)-loaded Ros87. However, it must be emphasised that this protein had only a single ZF unit linked to other protein elements, which also played a role in DNA binding [45]. A similar phenomenon was observed with the Tramtrack ZFP (consisting of two ZF subunits), although the α-helix content and DNA binding affinity of the Cd(II) complex was lower than that of Zn(II) bound protein [46] based on CD and EMSA measurements. The MTF-1 (metal response element-binding transcription factor-1) consists of six Cys2His2 ZF subunits. Cd(II) could inhibit the DNA-binding of this ZFP both when added to the apo-protein or to the Zn(II)-loaded MTF-1 [43,47]. The three unusual C-terminal fingers (4th–6th) of MTF-1 were investigated by Giedroc et al., where NMR and UV–Vis data revealed that although Cd(II) could bind these subunits, the obtained secondary structure differed significantly from the native ββα-type, most probably due to the unusual amino acid composition of the 5th subunit between the two cysteines [48].
The high affinity of Hg(II) towards sulphur donor groups is well known. However, the fact that Hg(II) can form a very stable complex with Cl− ions under physiological conditions (lgβML2 = 13.23) makes it difficult to compare the results of equilibrium studies [49]. Depending on the chloride content of the medium, some studies consider the affinity of Hg(II) and Zn(II) for Cys2His2 ZF motifs to be comparable, while in the absence of Cl− ions, Hg(II) forms more stable complexes [50]. Depending on the applied medium and measurement conditions, it has been shown that Hg(II) could not inhibit the DNA binding of TFIIIA during a DNase I footprinting assay [37], while more studies revealed that the secondary structure of the Zn(II)-loaded ZFPs collapsed in the presence of both organic and inorganic Hg(II), and the Hg(II)-bound ZFPs were unable to recognise DNA target sequences [40,44,51].
Solution equilibria of Cys2His2 zinc finger motifs have been widely investigated using the CP1 model peptide and the metal binding properties of naturally occurring ZFP subunits are usually compared to this model. However, CP1 could only give information regarding the metal binding properties of a single ZF subunit but not the protein–DNA interactions, since one ZF subunit cannot provide significant selectivity and affinity towards DNA. Therefore, the trends predicted on the basis of CP-1 and the behaviour experienced with natural ZFPs are contradictory [35,36,37,40,41,42,43,47].
Recently, we quantitatively characterised the Zn(II) and DNA binding properties of 1MEY#, an artificial ZFP consisting of three CP1-like subunits [52]. The amino acid sequence of 1MEY# can be found in Figure S1. With the knowledge of Zn(II) and DNA affinities, here we investigated the interaction between the 1MEY# artificial ZFP and toxic metal ions by spectrometric and electrophoretic methods. The competition of Cd(II) and Hg(II) with Zn(II) for the ZFP is described quantitatively to better understand the possible mechanisms of toxicity.
2. Results and Discussion
2.1. Interaction of 1MEY# Zinc Finger Protein with Cd(II)
The investigated 1MEY# ZFP consists of three CP1-like ZF subunits. The ZF units in 1MEY# differ only in a few amino acids responsible for DNA recognition. Therefore, it is assumed that they have similar metal binding properties. For this reason, the metal binding affinities of a single “average” 1MEY# binding site, referred to as 1MEY# bs, are presented throughout the text, unless otherwise stated. A Zn(II)-loaded Cys2His2 ZFP displays ββα-type secondary structure, while the apo-protein turns into an unordered structure, both resulting in characteristic circular dichroism spectra [53]. This provides a great opportunity to apply circular dichroism (CD) spectroscopy to investigate the effect of Cd(II) on 1MEY# ZFP. First, Zn(II) was removed from 1MEY# by treatment with ~25× excess of EDTA (Section 3.1). In a subsequent ultrafiltration, the apo-protein was transferred into an EDTA-free buffer. The initial Zn(II)-loaded holo-1MEY# protein displayed an ordered ββα structure as revealed by its CD spectrum with two negative peaks around 220 and 205 nm and a positive one at 190 nm (Figure 1a). The CD spectrum of apo-1MEY# represented an unfolded protein with a single negative peak around 200 nm. By adding three equivalents of Cd(II) (i.e., one equivalent per 1MEY# bs), the CD spectrum of the protein adopted a similar pattern to the Zn(II)-loaded protein. This indicated that Cd(II) could induce folding of the Cys2His2 ZF units into an ordered structure. Furthermore, no additional change in the CD spectra was observed upon an increase in the Cd(II) to protein ratio of up to ~120 fold (40 fold compared to the binding site) (Figure 1b). Cd(II) binding to the thiol groups of the ZFP was observed via ligand-to-metal charge transfer bands [54,55] in the 230–250 nm region of the UV–Vis absorption spectra as well (Figure S2).
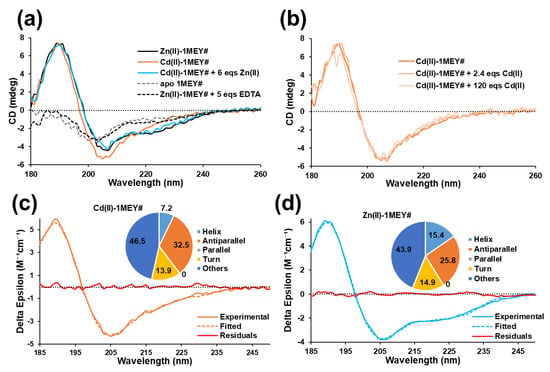
Figure 1.
(a) Circular dichroism spectra of Zn(II)-loaded (black), Cd(II)-loaded (orange) and metal-free (dashed grey) 1MEY# ZFP. The Cd(II)-loaded form in the presence of six equivalents (eqs) Zn(II) per 1MEY# (two eqs per binding site) (light blue) and the Zn(II)-depleted form using five eqs of EDTA per 1MEY# (1.7 eqs per binding site) (dashed black) are also presented. (b) CD spectra of Cd(II)-loaded 1MEY# in the presence of excess Cd(II). All CD spectra were normalised to the intensity of the starting Zn(II)-loaded 1MEY# spectrum recorded at 18.8 µM protein concentration. Measured and fitted CD spectra of (c) Cd(II)-1MEY# and (d) Zn(II)-1MEY# in the 185–250 nm wavelength range. Residual curves showing the differences between the fitted and measured spectra are marked with a red colour. Insets represent the estimated secondary structure composition of the complexes. The fitting was performed by BeStSel program suite [56].
It has to be mentioned that there are slight differences in the CD spectra of the Cd(II)- and Zn(II)-bound 1MEY#. The negative peak around 220 nm disappeared and the intensity of the 205 nm negative peak increased for the Cd(II)-loaded 1MEY#. For a better understanding, we have evaluated the CD spectra using the BeStSel software [56], by means of which the secondary structure compositions of the Zn(II)- and Cd(II)-loaded proteins were obtained (Figure 1c,d). According to the best fit of the data, the percentage of antiparallel β-sheet increased by ~6%, while the percentage of α-helices decreased by ~8.5% in the Cd(II)-bound protein. The cysteines favoured by Cd(II) are located in the antiparallel β-sheet region, and tight binding to these ligands might have caused extension of these structural elements, while the percentage of the helices decreased. Nevertheless, the most significant changes in the spectra occurred in the 220–240 nm region, which may also be attributed to the chiral contribution of the charge transfer transitions [54,55,57]. The programs used for evaluation of the protein CD spectra do not consider these contributions separately; thus, these are finally detected as the change in the secondary structural elements. In our case, this may result in overestimation of the β-sheet content of the protein. Based on the above discussion, we can conclude the 1MEY# ZFP could fold into an ordered secondary structure in the presence of Cd(II), which is most probably similar to that of the Zn(II)-loaded protein. This finding is consistent with the observations of Malgieri et al. for Ros87 ZFP [45], and Krepkiy et al. for the 3rd finger of TFIIIA [39]. In contrast, in case of the Tramtrack ZFP, Roesijadi et al. could only observe the 220–240 nm changes without the intense peak around 190 nm dedicated mostly to the α-helices [46]. The CD spectra of Cd(II)-1MEY# also suggested that under the measurement conditions, binding to the Cys2His2 site was favoured over the Cys3 or Cys4 coordination mode, which could have also been a possibility for 1MEY# containing altogether six cysteines in the three ZF units. The exclusive coordination to the cysteines would, however, result in the collapse of the finger structures, which did not occur.
By adding six eqs of Zn(II) to the Cd(II)-saturated 1MEY# protein (two eqs per 1MEY# bs), the CD spectrum of the initial Zn(II)-loaded ZFP was recovered, indicating that Zn(II) has significantly higher affinity towards the Cys2His2 binding site than Cd(II). The similarity of the resulting spectrum with that of the initial holoprotein also demonstrated that in the series of the above described experiments, no oxidation of the cysteines of 1MEY# occurred.
FluoZin-3 is considered to be a Zn(II) selective fluorescent probe, which can be applied to detect free Zn(II) replaced by Cd(II) in a competition assay with holo-1MEY#. However, Cd(II) also binds to FluoZin-3. Therefore, the stability of this complex was determined first by titrating four samples containing Zn(II) and FluoZin-3 at various molar ratio with Cd(II). Assuming only the formation of a monocomplex and excluding formation of the ternary complex formation, a pH-independent stability constant of logβ = 7.44 ± 0.01 was determined by evaluating the titration curves with the PSEQUAD program [58]. Recalculating this value at pH = 7.0, the obtained value of 7.18 was close to the available literature data for the Cd(II)-FluoZin-3 monocomplex (logβ′ = 6.9 [59]) (Figure S3a).
While starting from the apo-1MEY# ZFP, it was demonstrated that a protein coordinated to three Cd(II) could be established. Competition experiments were also performed to monitor the Cd(II) vs. Zn(II) exchange within 1MEY#. Based on the results of the CD spectroscopic, fluorometric and ESI-MS measurements, it was not possible to completely substitute Zn(II) with Cd(II) in the applied concentration range (Figure 2). A gradual metal–ion exchange was observed in the mass spectra with the subsequent formation of Zn2Cd1 and Zn1Cd2 mixed complexes and the Cd3 species upon an increase in the Cd(II) excess. Since previously we could not distinguish the Zn(II) binding ability of the three subunits of 1MEY# ZFP [52], the ZF units (1MEY# bs) were considered to be identical here as well. The evaluation of the fluorometric titrations revealed that the apparent stability constant of the Cd(II)-1MEY# bs complex is ~2 orders of magnitude lower than that of the Zn(II)-1MEY# bs, characterised by a logβ′Zn(II)-1MEY#bs, pH 7.4 of 12.2 [52]. This is a similar effect to that observed for the CP-1 model ZF peptide, where the stability decreased by ~2.5 orders of magnitude (Table 1) [33]. From the CD titration data, a one order of magnitude higher stability value was calculated, but here, only very small changes were observed in the spectra during the metal–ion exchange, decreasing the sensitivity of the method. By fitting the mass spectrometric data, an average logβ′Cd(II)-1MEY# bs pH 7.4 of 10.75 was obtained. Taking into account that the results of the ESI-MS measurements may not always correlate with the solution equilibria due to the different measurement conditions and the potentially different ionisation rates of different species, this value is in a very good agreement with those determined by fluorometric and CD experiments in Table 1. It is also worth mentioning that average logβ′ values and single binding site models could not be directly used in the calculation of the Cd(II) binding affinity of the protein from the ESI-MS results, since here, the whole protein is observed. Therefore, statistical considerations were applied (Figure 2d) (Supplementary Section S1). A good agreement between the fitted and experimental ESI-MS data supported the hypothesis of the identity of the ZF units within 1MEY#.
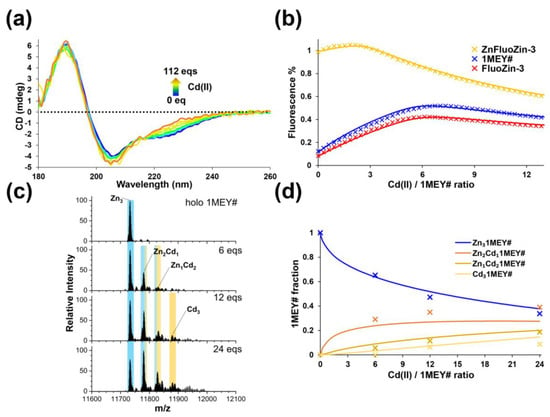
Figure 2.
(a) Measured (full lines) and calculated (dashed lines) CD spectra of 1MEY# in the course of titration with Cd(ClO4)2; c1MEY# = 16.4 µM. The endpoint of the titration (fully Cd(II)-loaded 1MEY#, orange line) was established separately starting from apo-1MEY#. (b) Measured (symbols) and simulated (full lines) relative fluorescence curves obtained by titrating holo-1MEY# (blue) with Cd(II). Reference titrations were simultaneously conducted to obtain data for the relative fluorescence of the system in the absence of 1MEY# (red) or in the presence of an amount of Zn(ClO4)2 equal to the Zn(II) content of 1MEY# (yellow). cFluoZin-3 = 6 µM and c1MEY# = 1 µM. (c) ESI-MS spectra of Zn(II)-loaded 1MEY# in the presence of increasing amounts of Cd(II). Sample amount: 20 µL c1MEY# = 2 µM. (d) The species distribution diagram calculated from the ESI-MS data (separate points).

Table 1.
Apparent average Cd(II) and Zn(II) affinities of some Cys2His2 ZFs and ZFPs in logβ′ units. cITC: competition with complexones monitored by ITC; FTc: competition with fluorescent complexones monitored by fluorescence spectroscopy; rCD: circular dichroism spectroscopy followed reverse titration; ESI-MS: reverse titration followed by ESI-MS; RT: spectroscopic reverse titration; DT: spectroscopic direct titration; ED: equilibrium dialysis.
These results indicated reversible Cd(II)/Zn(II) exchange within the CP-1-based 1MEY# ZFP. No cooperativity was observed during the titrations, suggesting that the ZF subunits behaved independently. Furthermore, the secondary structures of the formed complexes were almost identical (Figure 2) (Scheme 1a). The Cd(II) binding affinity of 1MEY# was found to be the highest among the available literature data with Cys2His2 ZFPs (Table 1), although it was still ~1–2 logβ′ units lower than the Zn(II) binding under similar conditions (logβ′Zn(II) = 12.2) [52]. The difference between the Zn(II) and Cd(II) binding affinity of CP1 and TFIIIA was reported to be 2.5 [33,38], while in case of the Ros87, it was only 1.2 logβ′ units [60].
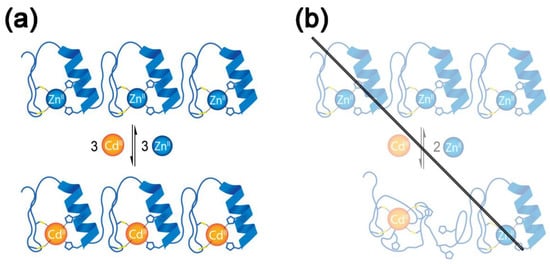
Scheme 1.
(a) Schematic representation of the Zn(II)/Cd(II) exchange in the 1MEY# ZFP. The extended scheme including the microspecies that may form during the replacement of the first and second Zn(II) is included in Supplementary Scheme S1. (b) A hypothetical reaction scheme where Cd(II) coordinates to the cysteine sidechains of multiple ZF subunits inside the ZFP, resulting the collapse of the secondary structure. Such a reaction has been proven not to take place in the case of the highly stable 1MEY#, but might occur in the case of lower stability natural ZFPs such as TFIIIA.
In good correlation with the results of CD measurements, no signals related to Zn1Cd1 or Cd2 species were observed during the analysis of the ESI-MS spectra of 1MEY#. Such species could have been characterised by Cys4 or Cys3 coordination where Cd(II) would bind to the cysteine sidechains of more than one ZF subunit, resulting the collapse of secondary structure and presumably the loss of the DNA-binding function (Scheme 1b). This effect might be responsible for the observed function loss in natural ZFPs with low Zn(II) and Cd(II) affinity towards the Cys2His2 coordination site. For example, in the case of TFIIIA ZFP, out of the nine subunits, only between two and three had higher Zn(II) affinity, and while it was possible to purify a protein with ~9 Zn(II) per ZFP, the purifications in the presence of Cd(II) yielded up to ~4 Cd(II) per ZFP products [35,36,37,38] (Table 1). This might be due to the formation of Cd(II)-Cys3 and Cd(II)-Cys4 coordination where Cd(II) bound the cysteines of multiple ZF subunits [32,36].
2.2. Hg(II) Binding of the 1MEY# Zinc Finger Protein
A perchlorate salt of Hg(II) was used in experiments with 1MEY# ZFP, aiming to avoid the interference with Cl− ions, i.e., to observe the direct interaction of the toxic metal ion with the ZFP. A significant change in the CD spectra of 1MEY# was observed upon addition of three equivalents of Hg(II) (one eq. per 1MEY# bs), referring to the collapse of the ββα secondary structure of the protein, similar to other investigations [51]. The positive peak assigned to the α-helix around 190 nm completely disappeared (Figure 3a,b). Thus, the formed species, assumed to be the Hg31MEY# complex, displayed an unordered structure. Since Hg(II) shows extreme affinity towards the cysteine thiolates, this was the expected outcome, independent of whether Zn(II) was completely displaced from the complex or a ternary species was formed.
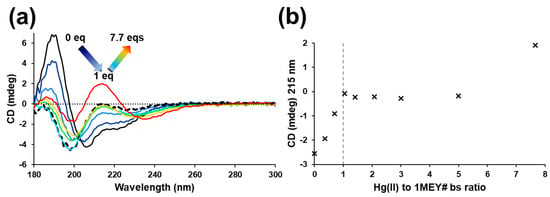
Figure 3.
(a) CD spectra of 1MEY# ZFP in the presence of increasing equivalents of Hg(ClO4)2, starting from the Zn(II)-loaded protein (black full line). The black dashed spectrum belongs to the system containing 1 eq. Hg(II) per 1MEY# bs. (b) The plot of the ellipticity values (×) recorded at 215 nm vs. Hg(II) equivalents per 1MEY# bs. The breakpoint at 1 eq. is indicated by a vertical dashed grey line. c1MEY# = 16.4 µM.
By the addition of further Hg(II), up to ~5 eq per 1MEY# bs, only small, but continuous, changes were observed in the CD spectra, indicating that further thermodynamic events took place. Furthermore, the shape of the CD spectrum changed completely and an intense positive band appeared around 215 nm, most probably due to a charge transfer band related to Hg(II) at ~8 eq Hg(II) per 1MEY# bs [57,62,63].
The sharp breakpoint in the plot of the CD intensity at 215 nm after adding one equivalent of Hg(II) per 1MEY# bs suggested that Hg(II) is indeed a much stronger competitor for ZF subunits than Cd(II) (Figure 3). The sharp breakpoint at one equivalent Hg(II) per ZF binding site also suggests that during the sequential exchange of Zn(II) for Hg(II), the unfolding of one subunit did not weaken the Zn(II) binding (and the secondary structure) of the remaining Zn(II)-bound subunit(s). This again indicated a high degree of independence of the subunits inside 1MEY#.
Hg(II) interactions with 1MEY# ZFP was also investigated by fluorometric titrations. There are no published data characterising the solution equilibria in the Hg(II)–FluoZin-3 system. Therefore, we carried out competitive fluorometric measurements prior to the experiments without protein, titrating the Zn(II)–FluoZin-3 system at various ratios of Hg(II). The best fit of these titration curves was achieved by assuming the formation of mono (HgA logβ = 6.8 ± 0.1), bis (HgA2 logβ = 12.8 ± 0.3) and ternary complexes (HgZnA logβ = 14.0 ± 0.1), although the chemical composition and coordination mode of such species remained ambiguous, but as it turned out there was no need for the use of these constants in further calculations (see below) (Figure S3b).
As the next step, the Hg(II)–holo-1MEY# system was studied in the presence of FluoZin-3. As it turned out, Hg(II) bound very strongly to the 1MEY# ZFP, so that the first three equivalents (one equivalent per binding site) of Hg(II) displaced Zn(II) in the ZFP almost quantitatively, and therefore, did not interact with the fluorophore (Figure 4a). This behaviour indicated that while Cd(II) could not compete efficiently with Zn(II) for 1MEY# ZFP, inorganic Hg(II) is an extremely strong competitor. Another interesting fact is that, contrary to expectations, no increase in fluorescence could be observed even after continuing the titration; the sample practically behaved as if it had been diluted with a buffer. This suggested that the extra added Hg(II) also bound to the 1MEY# ZFP with high affinity. This phenomenon provided an opportunity to estimate the lower limits of affinities of the ZF units as binding sites not only towards the first but also towards additional Hg(II) (Table 2). Based on the obtained stability constants, we could construct a distribution diagram, where the dashed section is based on the estimated limiting constants related to the binding of further Hg(II) (Figure 4b). Although this model may not be accurate in describing metal ion cluster formation, it could be successfully applied to describe the phenomena in this complicated system. Oligomerisation may also occur in these systems, but no species related to oligomers could be identified by ESI-MS.
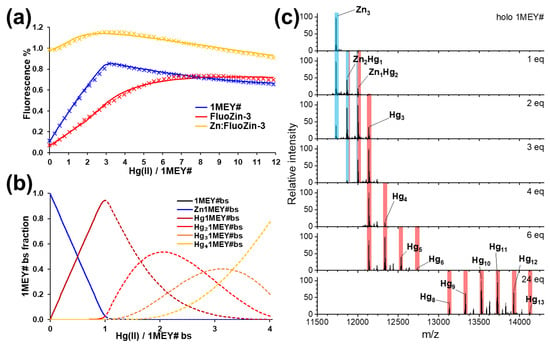
Figure 4.
(a) Measured (symbols) and simulated (continuous line) relative fluorescence obtained during the titration of holo-1MEY# ZFP with Hg(II) (blue). Reference titrations were simultaneously performed to obtain data for the relative fluorescence of the system in the absence of 1MEY# (red) or in the presence of an amount of Zn(ClO4)2 equal to the Zn(II) content of 1MEY# (yellow). cFluoZin-3 = 6 µM and c1MEY# = 1 µM. (b) Distribution diagram obtained by the fitting of the fluorescence titration data. (c) Zn(II)-loaded 1MEY# in the presence of increasing amounts of Hg(II) followed by ESI-MS. Sample amount: 20 µL and c1MEY# = 2 µM.

Table 2.
Estimated stability constants for Hg(II):1MEY# bs system based on the fluorometric method. The presented average constants were calculated for a single subunit of 1MEY# that was assumed to bind 1, 2, 3 and 4 Hg(II) in the 1MEY# protein binding 3, 6, 9 and 12 Hg(II), respectively.
Mass spectrometric measurements confirmed the fluorometric results. A gradual displacement of Zn(II) was detected through the Zn2Hg1MEY#, ZnHg21MEY# and Hg31MEY# complexes. In addition, it was possible to detect the presence of 1MEY# species with even 13 coordinated Hg(II) while increasing the metal ion excess up to 24 eqs (eight eqs per binding site); however, the exact mode of coordination remained unknown (Figure 4c). The stability constant of the monocomplex estimated from ESI-MS data was identical to the value obtained from fluorometric titration (Table 2); however, for the binding of the second Hg(II), a value three orders of magnitude lower was assigned, which is presumably due to the previously mentioned uncertainty of the mass spectrometry. It can be assumed that Hg(II) only coordinates to the cysteines [51]. Thus, in theory, the histidine residues remain available for the coordination of Zn(II). Despite this, no mixed metal species within the same ZF subunit could be seen under the conditions of the ESI-MS measurements (Scheme 2). In the Zn2Hg1MEY# and ZnHg21MEY# ternary complexes, the metal ions bind to different subunits and thus the binding events are independent.
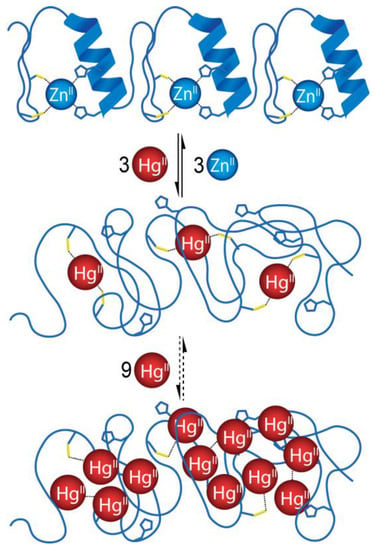
Scheme 2.
Schematic representation of Zn(II)/Hg(II) exchange in 1MEY# ZFP. The extended scheme, including the microspecies that may form during the replacement of the first and second Zn(II), is included in Supplementary Scheme S2.
The obtained conditional stability constant for the binding of the first Hg(II) (logβ′Hg(II)-1MEY# bs ≥ 16.7) is quite a high value on the scale of ZFP metal binding. Among the peptides containing the CXXC-amino acid sequence, only those had a similar or higher affinity for Hg(II), where the cysteines were in favourable position and the secondary structure was not completely unordered [64,65,66,67]. Although the CD spectra of 1MEY# in the presence of one Hg(II) eq. per binding site significantly differed from the unfolded structure (Figure S4), the contribution of the thiolate–Hg(II) charge transfer bands might be significant. Therefore, an accurate evaluation of the secondary structure composition of the Hg(II)-1MEY# complex is not possible [57,62,63]. Nevertheless, based on the NMR analysis of the similarly behaving Ros87 ZFP, it can be assumed that the structure is not completely disordered [51]. Sivo et. al. also determined the Hg(II) affinity of Ros87 using HgCl2 (logβ′Hg(II)-Ros87 = 6.1) [51].
2.3. DNA Binding of 1MEY# ZFP Is Influenced by Toxic Metal Ions
2.3.1. Cd(II)
Previously, we have shown that Zn(II)-saturated 1MEY# could bind DNA by EMSA experiments. A clear difference in the affinity of the protein towards the 34 bp DNA probes with and without the specific 5′-GAGGCAGAA-3′ sequence was observed [52]. Here, we studied the interaction of DNA with the Cd(II)-loaded 1MEY# ZFP obtained from the apo-protein. EMSA experiments revealed a single, well defined shifted DNA band with the nonspecific DNA probe. By fitting the quantified band intensities, a stability constant could be determined as logβ′ = 6.04 ± 0.02 (Figure 5a,c). This corresponds to ~0.2 log units weaker binding compared to the interaction of the Zn(II)-loaded protein with nonspecific DNA. A more significant ~0.6 log unit decrease was found in the affinity of the Cd(II)-loaded 1MEY#, with logβ′ = 7.62 ± 0.04 (Figure 5b,d), compared to that of the Zn(II)-loaded ZFP (logβ′ = 8.20). Nevertheless, the above listed stability constants are still substantially high values. It has to be mentioned that the titration curves for nonspecific DNA binding show a slight sigmoidal pattern instead of the saturation curve expected from the simple binding scheme. This, however, might be attributed to the ambiguity of the gel staining when the amount of bound DNA is too small. It was also visible that the DNA binding of the Cd(II)-loaded 1MEY# resulted in similar changes in the CD spectra of the system around 190 nm, as in case of the Zn(II)-loaded protein. However, the rate of the increase in ellipticity for the Cd(II)-1MEY# was approximately half of that of Zn(II)-1MEY#, which may indicate that the interaction of the Cd(II)-loaded ZFP with DNA induces smaller structural rearrangements, i.e., the process is weaker (Figure 5g).
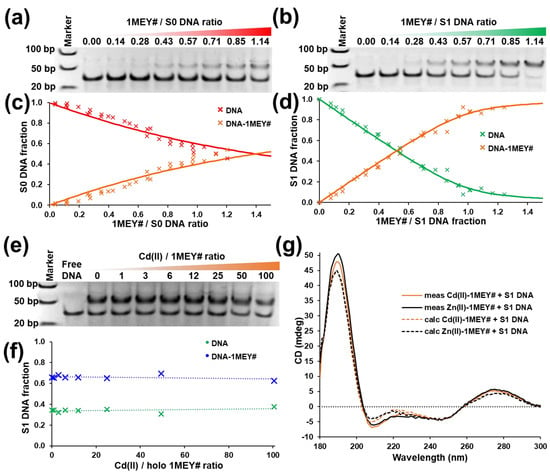
Figure 5.
Representative electrophoretic gel mobility shift assays of (a) nonspecific S0 DNA and (b) specific S1 DNA in the presence of increasing equivalents of Cd(II)-loaded 1MEY# zinc finger protein. cDNA = 0.88–1 µM. (c) Distribution of S0 DNA or (d) S1 DNA in the presence of increasing equivalents of Cd(II)-1MEY# ZFP. DNA fractions (separate points) were calculated based on the intensities of three independent electrophoretic gel mobility shift assays. Band intensity calculations were performed in ImageJ [68]. (e) Electrophoretic gel mobility shift assay of Zn(II)-1MEY# with specific S1 DNA in the presence of increasing equivalents of Cd(II). cDNA = 1 µM, cZn(II)-1MEY# = 0.8 µM. (f) Distribution of S1-DNA among the free and protein-bound forms as calculated from the band intensities of the electrophoretic gel mobility shift assay image. (g) Comparison of the CD spectra of S1 DNA in the presence of 0.5 eq Zn(II)-1MEY# (black) and 0.5 eq Cd(II)-1MEY# (orange). The dashed line represents the CD spectrum calculated by summing the appropriate protein and DNA component spectra. All CD spectra were normalised to the intensity of the starting Zn(II)-loaded 1MEY# spectrum recorded at 18.8 µM protein concentration.
After it was proven that a fully Cd(II)-saturated ZFP can bind its DNA target, competition reactions were performed, where the Zn(II)-loaded 1MEY# ZFP in complex with the specific DNA probe was titrated with increasing amounts of Cd(II). During the process, no change could be seen in the DNA binding ability of the protein, which suggests that if mixed Zn2Cd11MEY# and Zn1Cd21MEY# complexes form, these can also bind DNA with a similar affinity to the Zn(II)-loaded protein (Figure 5e,f). On the other hand, previously we have shown that the interaction with specific DNA increases the stability of the Zn(II) complex of 1MEY# [52]. Therefore, it can be assumed that metal ion exchange is a minor process in this experiment.
Based on these findings, we could assume that the toxicity of cadmium in the living organism cannot be directly attributed to competition with high-stability Cys2His2 ZFPs. If the protein is in the Zn(II)-loaded form, a large excess of Cd(II) would be necessary to compete with Zn(II) and it is unlikely to completely substitute Zn(II). The Cd(II)-loaded 1MEY# and the mixed metal complexes can also bind DNA, the only visible differences during our measurements were in their affinity, similar to the limited literature data [43,44,46]. Although the competition with the high stability Zn(II)-loaded ZFP is not significant, if Cd(II) meets with the apo-ZFP then it can form stable complexes due to its high affinity towards to protein. Regardless, it cannot be ruled out that such a Cd(II)-loaded high stability ZFP may still be able to perform its function, since once it is coordinated in Cys2His2 mode, the structure and function may differ only slightly compared to the Zn(II)-loaded protein. Furthermore, even if the structure of the protein differs, it does not necessarily mean that the DNA binding function is also completely vanished. In the case of the Tramtrack ZFP, where based on CD, the α-helix content of the protein was reduced significantly during Cd(II) binding, the complex could still recognise its target DNA but ~1 order of magnitude more weakly [46].
2.3.2. Hg(II)
Electromobility shift assay titrations revealed that the protein binding to its DNA target had no inhibitory effect on Hg(II) competition. Three equivalents of Hg(II) (one eq. per 1MEY# bs) could completely eliminate the DNA binding of the ZFP (Figure 6a,b). Thus, the effect of Hg(II) was unambiguous in the absence of other competing ligands. By using various buffer conditions (Cl− ions and DTT) this effect can be significantly reduced, yet according to most of the literature data, Hg(II) still could effectively inhibit the DNA binding of a ZFP [40,44,51].
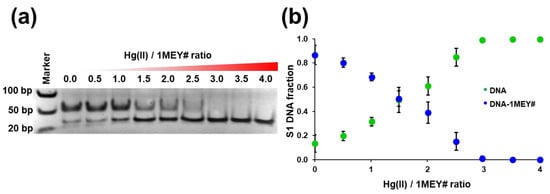
Figure 6.
(a) Representative electrophoretic gel mobility shift assay of Zn(II)-1MEY# with specific S1 DNA in the presence of increasing equivalents of Hg(II). cDNA = 1 µM, cZn(II)-1MEY# = 1 µM. (b) Distribution of S1-DNA based on electrophoretic gel mobility shift assay gel intensities.
3. Materials and Methods
3.1. Protein Expression and Purification
The protein expression and purification steps were described earlier [52]. Briefly, the 1MEY# protein (a consensus peptide-based 1MEY [69,70] derivative) was expressed in E. coli BL21 (DE3) cells from a pETM11 vector, with an N-terminal His-SUMO affinity tag. Ni(II) affinity purification was applied, then the N-terminal affinity and SUMO tag were cleaved specifically using the ULP1 protease [71]. Purification was followed by buffer exchange to 10 mM Cl−-free 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), pH = 7.40, using Amicon 3K 15 mL filters (Merck KGaA, Darmstadt, Germany) at 4000× g 8 × 30 min 15 °C and filtration through 0.22 µm, Ø = 13 mm PES filters (Merck KGaA, Darmstadt, Germany). This process provided the Zn(II)-loaded ZFP; thus, for the Cd(II)-1MEY# experiments, Zn(II) was removed from the protein by 0.5 mM EDTA (~25× excess over 1MEY#) and 0.2 mM TCEP (tris(2-carboxyethyl)phosphine) treatment for 10 min at 25 °C. EDTA was washed away during ultra-filtration (Amicon 3K 0.5 mL filters (Merck KGaA, Darmstadt, Germany), 14,000× g 5 × 5 min 15 °C) with 10 mM HEPES, 0.2 mM TCEP and 50 mM NaClO4 (pH = 7.40) buffer. The TCEP reducing agent was included in order to protect the free cysteines from potential oxidation. After the calculated EDTA content of the protein sample dropped to below 1 μM, 150 μM final concentration Cd(ClO4)2 was added to a portion of the sample and incubated for 5 min at 25 °C. Then, additional buffer exchange was performed with 10 mM HEPES and 50 mM NaClO4 (pH = 7.40) buffer by ultra-filtration as described earlier.
3.2. Mass Spectrometric Analysis of the Protein
Intact protein analysis was performed on an LTQ-Orbitrap Elite (Thermo Scientific, Thousand Oaks, CA, USA) mass spectrometer coupled with a TriVersa NanoMate (Advion, Ithaca, NY, USA) chip-based electrospray ion source. Measurements were carried out in positive mode at 120,000 resolution in 8.2 mM ammonium hydrogen carbonate buffer (pH ~7.8), as described previously [72]. Fitting of the ESI-MS data was performed by the Solver add in of Microsoft Excel based on statistical considerations [73,74].
3.3. CD Spectroscopy
A J-1500 Jasco spectrophotometer was used during spectroscopic measurements under constant nitrogen flow in stepwise scanning mode over the range of 180–330 nm. Synchrotron radiation (SR) CD spectra were recorded at the CD1 beamline of the storage ring ASTRID at the Institute for Storage Ring Facilities (ISA), University of Aarhus, Denmark between the 170 and 330 nm wavelength range [75,76].
All spectra were recorded with 1 nm steps and a dwell time of 2 s per step, using l = 0.2 mm cylindrical quartz cells (SUPRA-SIL, Hellma GmbH, Müllheim, Germany). Each sample containing 8–20 µM protein was prepared separately in 6.6–7.5 mM HEPES buffer (pH = 7.40) and was kept at room temperature for at least 5 min prior to measurement. Samples for CD measurements involving DNA contained 33–45 mM NaClO4 as well. If CD data were fitted, the 215–260 nm wavelength range was selected and the PSEQUAD program was used for calculations [58].
3.4. Electrophoretic Mobility Shift Assay (EMSA)
Electrophoretic mobility shift assays were performed using 34 bp DNA probes containing zero (later referred to as S0 DNA) or one (later referred to as S1 DNA) specific 1MEY# target sequence: 5′–GAGGCAGAA–3′. The S0 DNA probe was obtained by the hybridisation of the Forward-S0: 5′–CTAGTTTGCTGAACTGGGGTCACATAGATTAATA–3′ and Reverse-S0: –5′-TATTAATCTATGTGACCCCAGTTCAGCAAACTAG-3′ oligonucleotides. The S1 DNA probe was obtained by the hybridisation of the Forward-S1: 5′–GAATTCCTGCTGAGAGGCAGAAACATAGGGGTCG–3′ and Reverse-S1: 5′–CGACCCCTATGTTTCTGCCTCTCAGCAGGAATTC–3′ oligonucleotides (target sequence of 1MEY# is underlined). Oligonucleotides were obtained by solid phase synthesis (Invitrogen—Thermo Scientific, CA, USA). EMSA experiments were performed as described earlier in 10 mM HEPES, 150 mM NaClO4 and 10 m/V% glycerol buffer (pH = 7.40) [77]. The FastRuler Ultra Low Range DNA Ladder (Thermo Scientific, CA, USA) served as a reference (marker) and the ImageJ program was used for the quantification of gel intensities [68]. Calculations were performed by the PSEQUAD program [58].
3.5. Fluorimetry
The competition reactions of the Zn(II)-loaded 1MEY# were monitored using the FluoZin-3 fluorescent probe in a CLARIOstar Plus plate reader (BMG Labtech, Ortenberg, Germany). FluoZin-3 has an absorption maximum at 494 nm and exhibits strong fluorescence at 516 nm when binding Zn(II), with a conditional stability constant of logβ′ = 8.04 (pH = 7.40) [78]. In a typical measurement, 480–490 nm extinction and 510–520 nm emission filters were used during the titrations of 200 µL protein-FluoZin-3 sample in 96-well, polystyrene, non-binding, flat-bottom, black microplates (Greiner Bio-One, Kremsmünster, Austria) in 10 mM HEPES and 150 mM NaClO4 buffer (pH = 7.40). An amount of 3 µL titrant (or buffer) was injected to the samples at each titration point by the two built-in injectors of the CLARIOstar Plus plate reader. Each injection was followed by 30 s 150 rpm double orbital shaking of the plate and 5 min incubation at 25 °C to reach equilibrium. The titration process was automated using custom built scripts (Table S1). The FluoZin-3 concentration was determined spectrophotometrically (λmax = 491 nm, εmax = 71,143 M−1cm−1 (pH = 7.40)). During each measurement, additional control samples were prepared containing only FluoZin-3 or FluoZin-3 and the equivalent amount of Zn(II) which can be released from the 1MEY# ZFP during competition reactions, in order to calculate correct relative fluorescence values (Figure S5). Fitting of the fluorometric data was performed by PSEQUAD [58] and by the Solver add in of Microsoft Excel.
4. Conclusions
The interaction of Zn(II)-loaded Cys2His2 ZFPs with other metal ions can decrease, alter or destroy their DNA binding function. Therefore, a better understanding of these systems is essential. Several research works have aimed at studying the interactions between toxic metal ions and ZFPs, but the available data with model peptides and natural ZFPs cannot be directly compared. In a previous publication, we quantitatively characterised the Zn(II) and DNA binding of a ZFP consisting of three CP1-like model peptide subunits [52]. Based on CD measurements, we could prove that Cd(II) binding of 1MEY# resulted in an almost identical secondary structure to the Zn(II) complex. The protein could bind Cd(II) with the highest affinity determined so far for the Cys2Hi2 binding sites. Yet, the Zn(II) binding affinity of 1MEY# is even higher by 1–2 orders of magnitude. Similar tendencies were observed between the Zn(II) and Cd(II) binding of other investigated ZFs [33,38]. Thus, a large excess of Cd(II) would be necessary to compete with Zn(II), while the Cd(II) substituted ZFP can be reverted into Zn(II)-complex by adding two equivalents of Zn(II) per binding site, based on CD measurements. The metal exchange reaction occurred stepwise, with each ZF subunit behaving independently. Formation of Cd(II)-Cys3 or Cd(II)-Cs4 complexes could not be observed by ESI-MS and CD measurements. The protein bound three Cd(II) ions and no further spectral changes were visible, even when applying 120 equivalents of Cd(II). We could demonstrate that the Cd(II) complex was also capable of specific DNA binding by EMSA and CD measurements, although the affinity decreased by logβ′ = 0.6 units compared to the Zn(II) complex. The DNA binding ability of Zn(II)-loaded 1MEY# was not influenced even by ~100 eqs of Cd(II), suggesting that either no exchange occurred or the mixed complexes could recognise the DNA target as well.
Hg(II) behaved in a completely different manner. By using perchlorate salt, we could observe the affinity of Hg(II) towards the 1MEY# ZFP to be logβ′Hg(II)-1MEY# bs ≥ 16.7. During the quantitative exchange of Zn(II) with Hg(II), the well-defined ββα secondary structure of 1MEY# collapsed. Excess of Hg(II) could also bind to 1MEY# with nanomolar affinity. Even a 13 Hg(II)-bound form was visible in ESI-MS, which could also be followed by CD spectroscopy through the charge transfer bands of Hg(II). In contrast, the Hg(II) affinity of the Ros87 ZFP investigated by Sivo et al. using HgCl2 salt was three orders of magnitude weaker than the Zn(II) affinity of the same protein [51]. Hg(II) not only disrupted the structure of 1MEY#, but also destroyed the DNA binding of the ZFP. An investigation into the multi-metal binding sites in the Hg(II)-bound 1MEY# would be of interest.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/inorganics11020064/s1, Section S1. Statistical considerations during ESI-MS measurements. Scheme S1. Schematic representation of the Zn(II)/Cd(II) exchange in the 1MEY# ZFP. In the transition states, Cd(II) can replace Zn(II) of any zinc finger subunit with equal probability. Scheme S2. Schematic representation of the Zn(II)/Hg(II) exchange in the 1MEY# ZFP. In the transition states, Hg(II) can replace Zn(II) of any zinc finger subunit with equal probability. Figure S1: Cartoon representation of the crystal structure of (a) the 1st ZF subunit of the 1MEY ZFP and (b) the whole 1MEY ZFP. ZFP: blue, Zn(II): grey sphere, cysteine thiols: yellow (PyMOL representation of 1MEY PDB [4]). (c) Alignment of the amino acid sequence of 1MEY# ZFP (constructed from 1MEY ZFP [4]) with the 26 amino acid-long consensus Cys2His2 model peptide CP1 and CP1 K/S mutant, established and investigated by Berg et al. [5]. The identical amino acids of 1MEY# compared to CP1 are marked with green, while the ones that differ both compared to the CP1 and CP1 K/S mutant are marked with red. The amino acids differing only compared to CP1 are marked with light red. Figure S2: UV–Vis absorption spectra of 1MEY# in either Zn(II) (blue) or Cd(II) (orange) saturated form. c1MEY# = 13.5 µM in 10 mM HEPES and 50 mM NaClO4 (pH 7.4); l = 1 cm. Figure S3: Measured (separate symbols) and calculated (full lines) relative fluorescence values of Zn(II)–FluoZin-3 systems in the presence of an increasing amount of (a) Cd(ClO4)2; and (b) Hg(ClO4)2. Samples (200 µL) were loaded into the plate wells and titrated with 3 µL aliquots of the titrant at 25 °C. cFluoZin-3 = 3.98 µM, 10 mM HEPES and 150 mM NaClO4 (pH 7.40). The calculations were performed by the PSEQUAD program [6]. Figure S4: Circular dichroism spectra of Zn(II)-loaded 1MEY# (full black line), Hg(II)-loaded (red) and metal-free form using 5 eqs of EDTA per 1MEY# (1.7 eqs per binding site) (dashed black) are also presented. c1MEY# = 16.4 µM in 7.5 mM HEPES (pH = 7.4) buffer. CD1 beamline of the storage ring ASTRID, Aarhus l = 0.2 mm. Figure S5: Fluorometric titration procedure. Baseline fluorescence was determined by applying a 10-fold excess of EDTA over FluoZin-3. The maximal achievable fluorescence value was determined by applying 0.5 eq Zn(II) to FluoZin-3 (‘Max’). A two-fold excess of FluoZin-3 was necessary to make sure 100% of Zn(II) is complexed. A sample containing an identical amount of Zn(II) to the ‘Max’ reference well was titrated with the titrant. The dilution effect during titration was determined by the injection of buffer (instead of the titrant) to the reference wells and to an additional sample well. Table S1: Automated titration script for CLARIOstar Plus plate reader.
Author Contributions
Conceptualisation, B.H. and B.G.; investigation, B.H. and É.H.-G.; data curation, B.H., É.H.-G. and B.G.; writing—original draft preparation, B.H. and B.G.; writing—review and editing, B.H., É.H.-G. and B.G.; funding acquisition, B.H., É.H.-G. and B.G. All authors have read and agreed to the published version of the manuscript.
Funding
Supported by the ÚNKP-22-4-SZTE-491 New National Excellence Program of the Ministry for Culture and Innovation from the source of the National Research, Development and Innovation Fund and by the Hungarian National Research, Development and Innovation Office (GINOP-2.3.2-15-2016-00038, GINOP-2.3.2-15-2016-00001, GINOP-2.3.2-15-2016-00020, 2019-2.1111-TÉT-2019-00089, and K_16/120130) by the EU Horizon 2020 grant no. 739593. This research was partially funded by the CM_SMP_471080_2021 Campus Mundi Student Mobility Traineeship from the Tempus Public Foundation. The support of SRCD measurements from the CALIPSOplus (EU Framework Programme for Research and Innovation HORIZON 2020, grant no. 730872) is also greatly acknowledged.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The authors thank Peter Baker for the development and maintenance of the ELKH Cloud (https://science-cloud.hu/ (accessed on 27 December 2022)) for hosting the ProteinProspector search engine. The authors would like to thank Milan Kožíšek for providing the pETM11-SUMO3 plasmid.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Mackay, J.P.; Crossley, M. Zinc fingers are sticking together. Trends Biochem. Sci. 1998, 23, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Tupler, R.; Perini, G.; Green, M.R. Expressing the human genome. Nature 2001, 409, 832–833. [Google Scholar] [CrossRef] [PubMed]
- Miller, J.; McLachlan, A.; Klug, A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 1985, 4, 1609–1614. [Google Scholar] [CrossRef] [PubMed]
- Klug, A. The Discovery of Zinc Fingers and Their Applications in Gene Regulation and Genome Manipulation. Annu. Rev. Biochem. 2010, 79, 213–231. [Google Scholar] [CrossRef]
- Cassandri, M.; Smirnov, A.; Novelli, F.; Pitolli, C.; Agostini, M.; Malewicz, M.; Melino, G.; Raschellà, G. Zinc-finger proteins in health and disease. Cell Death Discov. 2017, 3, 17071. [Google Scholar] [CrossRef]
- Berg, J.M.; Shi, Y. The Galvanization of Biology: A Growing Appreciation for the Roles of Zinc. Science 1996, 271, 1081–1085. [Google Scholar] [CrossRef]
- Bulaj, G.; Kortemme, T.; Goldenberg, D.P. Ionization−Reactivity Relationships for Cysteine Thiols in Polypeptides. Biochemistry 1998, 37, 8965–8972. [Google Scholar] [CrossRef]
- Maynard, A.T.; Covell, D.G. Reactivity of Zinc Finger Cores: Analysis of Protein Packing and Electrostatic Screening. J. Am. Chem. Soc. 2001, 123, 1047–1058. [Google Scholar] [CrossRef]
- Smith, J.N.; Hoffman, J.T.; Shirin, Z.; Carrano, C.J. H-Bonding Interactions and Control of Thiolate Nucleophilicity and Specificity in Model Complexes of Zinc Metalloproteins. Inorg. Chem. 2005, 44, 2012–2017. [Google Scholar] [CrossRef]
- Lee, Y.-M.; Lim, C. Factors Controlling the Reactivity of Zinc Finger Cores. J. Am. Chem. Soc. 2011, 133, 8691–8703. [Google Scholar] [CrossRef]
- Quintal, S.M.; Depaula, Q.A.; Farrell, N.P. Zinc finger proteins as templates for metal ion exchange and ligand reactivity. Chemical and biological consequences. Metallomics 2011, 3, 121–139. [Google Scholar] [CrossRef] [PubMed]
- Frankel, A.D.; Berg, J.M.; Pabo, C.O. Metal-dependent folding of a single zinc finger from transcription factor IIIA. Proc. Natl. Acad. Sci. USA 1987, 84, 4841–4845. [Google Scholar] [CrossRef] [PubMed]
- Krishna, S.S.; Majumdar, I.; Grishin, N.V. Structural classification of zinc fingers. Nucleic Acids Res. 2003, 31, 532–550. [Google Scholar] [CrossRef] [PubMed]
- Kellis, J.T.; Nyberg, K., Jr.; Šail, D.; Fersht, A.R. Contribution of hydrophobic interactions to protein stability. Nature 1988, 333, 784–786. [Google Scholar] [CrossRef] [PubMed]
- La Gatta, S.; Leone, L.; Maglio, O.; De Fenza, M.; Nastri, F.; Pavone, V.; Chino, M.; Lombardi, A. Unravelling the Structure of the Tetrahedral Metal-Binding Site in METP3 through an Experimental and Computational Approach. Molecules 2021, 26, 5221. [Google Scholar] [CrossRef] [PubMed]
- Lander, E.S.; Linton, L.M.; Birren, B.; Nusbaum, C. Initial sequencing and analysis of the human genome. Nature 2001, 409, 860–921. [Google Scholar] [CrossRef]
- Kim, Y.G.; Cha, J.; Chandrasegaran, S. Hybrid restriction enzymes: Zinc finger fusions to Fok I cleavage domain. Proc. Natl. Acad. Sci. USA 1996, 93, 1156–1160. [Google Scholar] [CrossRef]
- Urnov, F.D.; Rebar, E.J.; Holmes, M.C.; Zhang, H.S.; Gregory, P.D. Genome editing with engineered zinc finger nucleases. Nat. Rev. Genet. 2010, 11, 636–646. [Google Scholar] [CrossRef]
- Gaj, T.; Gersbach, C.A.; Barbas, C.F., III. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2014, 31, 397–405. [Google Scholar] [CrossRef]
- Segal, D.J.; Meckler, J.F.; Carroll, D.; Voytas, D.F.; Porteus, M.; Bickmore, W.A.; Walden, H.; Deans, A.J.; Moldovan, G.-L.; D’Andrea, A.D.; et al. Genome Engineering at the Dawn of the Golden Age. Annu. Rev. Genom. Hum. Genet. 2013, 14, 135–158. [Google Scholar] [CrossRef]
- Urnov, F.D.; Miller, J.C.; Lee, Y.-L.; Beausejour, C.M.; Rock, J.M.; Augustus, S.; Jamieson, A.C.; Porteus, M.H.; Gregory, P.D.; Holmes, M.C. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature 2005, 435, 646–651. [Google Scholar] [CrossRef] [PubMed]
- Passananti, C.; Corbi, N.; Oniri, A.; Di Certo, M.G.; Mattei, E. Transgenic Mice Expressing an Artifical Zinc Finger Regulator Targeting an Endogenous Gene, Engineered Zinc Finger Proteins. Methods Mol. Biol. 2010, 649, 183–206. [Google Scholar] [CrossRef] [PubMed]
- Petolino, J.F. Genome editing in plants via designed zinc finger nucleases. In Vitro Cell. Dev. Biol. Plant 2015, 51, 1–8. [Google Scholar] [CrossRef]
- Ousterout, D.G.; Kabadi, A.M.; Thakore, P.I.; Perez-Pinera, P.; Brown, M.T.; Majoros, W.H.; Reddy, T.E.; Gersbach, C.A. Correction of Dystrophin Expression in Cells from Duchenne Muscular Dystrophy Patients through Genomic Excision of Exon 51 by Zinc Finger Nucleases. Mol. Ther. 2015, 23, 523–532. [Google Scholar] [CrossRef]
- Rodríguez, J.; Mosquera, J.; García-Fandiño, R.; Vázquez, M.E.; Mascareñas, J.L. A designed DNA binding motif that recognizes extended sites and spans two adjacent major grooves. Chem. Sci. 2016, 7, 3298–3303. [Google Scholar] [CrossRef]
- Shi, Y.; Beger, R.D.; Berg, J.M. Metal binding properties of single amino acid deletion mutants of zinc finger peptides: Studies using cobalt(II) as a spectroscopic probe. Biophys. J. 1993, 64, 749–753. [Google Scholar] [CrossRef]
- Hartwig, A.; Asmuss, M.; Ehleben, I.; Herzer, U.; Kostelac, D.; Pelzer, A.; Schwerdtle, T.; Bürkle, A. Interference by toxic metal ions with DNA repair processes and cell cycle control: Molecular mechanisms. Environ. Health Perspect. 2002, 110, 797–799. [Google Scholar] [CrossRef]
- Witkiewiczkucharczyk, A.; Bal, W. Damage of zinc fingers in DNA repair proteins, a novel molecular mechanism in carcinogenesis. Toxicol. Lett. 2006, 162, 29–42. [Google Scholar] [CrossRef]
- Kluska, K.; Adamczyk, J.; Krężel, A. Metal binding properties, stability and reactivity of zinc fingers. Co-ord. Chem. Rev. 2018, 367, 18–64. [Google Scholar] [CrossRef]
- Párraga, G.; Horvath, S.J.; Eisen, A.; Taylor, W.E.; Hood, L.; Young, E.T.; Klevit, R.E. Zinc-dependent structure of a single-finger domain of yeast ADR1. Science 1988, 241, 1489–1492. [Google Scholar] [CrossRef] [PubMed]
- Kopera, E.; Schwerdtle, T.; Hartwig, A.; Bal, W. Co(II) and Cd(II) Substitute for Zn(II) in the Zinc Finger Derived from the DNA Repair Protein XPA, Demonstrating a Variety of Potential Mechanisms of Toxicity. Chem. Res. Toxicol. 2004, 17, 1452–1458. [Google Scholar] [CrossRef] [PubMed]
- Hartwig, A. Zinc Finger Proteins as Potential Targets for Toxic Metal Ions: Differential Effects on Structure and Function. Antioxidants Redox Signal. 2001, 3, 625–634. [Google Scholar] [CrossRef] [PubMed]
- Krizek, B.A.; Merkle, D.L.; Berg, J.M. Ligand variation and metal ion binding specificity in zinc finger peptides. Inorg. Chem. 1993, 32, 937–940. [Google Scholar] [CrossRef]
- Heinz, U.; Hemmingsen, L.; Kiefer, M.; Adolph, H.-W. Structural Adaptability of Zinc Binding Sites: Different Structures in Partially, Fully, and Heavy-Metal Loaded States. Chem. A Eur. J. 2009, 15, 7350–7358. [Google Scholar] [CrossRef] [PubMed]
- Petering, D.H.; Huang, M.; Moteki, S.; Iii, C.F.S. Cadmium and lead interactions with transcription factor IIIA from Xenopus laevis: A model for zinc finger protein reactions with toxic metal ions and metallothionein. Mar. Environ. Res. 2000, 50, 89–92. [Google Scholar] [CrossRef]
- Huang, M.; Krepkiy, D.; Hu, W.; Petering, D.H. Zn-, Cd-, and Pb-transcription factor IIIA: Properties, DNA binding, and comparison with TFIIIA-finger 3 metal complexes. J. Inorg. Biochem. 2004, 98, 775–785. [Google Scholar] [CrossRef]
- Hanas, J.S.; Gunn, C.G. Inhibition of Transcription Factor IIIA-DNA Interactions by Xenobiotic Metal Ions. Nucleic Acids Res. 1996, 24, 924–930. [Google Scholar] [CrossRef]
- Makowski, G.S.; Sunderman, F., Jr. The interactions of zinc, nickel, and cadmium with Xenopus transcription factor IIIA, assessed by equilibrium dialysis. J. Inorg. Biochem. 1992, 48, 107–119. [Google Scholar] [CrossRef]
- Krepkiy, D.; Försterling, F.H.; Petering, D.H. Interaction of Cd2+ with Zn Finger 3 of Transcription Factor IIIA: Structures and Binding to Cognate DNA. Chem. Res. Toxicol. 2004, 17, 863–870. [Google Scholar] [CrossRef]
- Razmiafshari, M.; Zawia, N.H. Utilization of a Synthetic Peptide as a Tool to Study the Interaction of Heavy Metals with the Zinc Finger Domain of Proteins Critical for Gene Expression in the Developing Brain. Toxicol. Appl. Pharmacol. 2000, 166, 1–12. [Google Scholar] [CrossRef]
- Kothinti, R.; Blodgett, A.; Tabatabai, N.M.; Petering, D.H. Zinc Finger Transcription Factor Zn3-Sp1 Reactions with Cd2+. Chem. Res. Toxicol. 2010, 23, 405–412. [Google Scholar] [CrossRef] [PubMed]
- Kothinti, R.K.; Blodgett, A.B.; Petering, D.H.; Tabatabai, N.M. Cadmium down-regulation of kidney Sp1 binding to mouse SGLT1 and SGLT2 gene promoters: Possible reaction of cadmium with the zinc finger domain of Sp1. Toxicol. Appl. Pharmacol. 2010, 244, 254–262. [Google Scholar] [CrossRef] [PubMed]
- Bittel, D.; Dalton, T.; Samson, S.L.-A.; Gedamu, L.; Andrews, G.K. The DNA Binding Activity of Metal Response Element-binding Transcription Factor-1 Is Activated In Vivo and in Vitro by Zinc, but Not by Other Transition Metals. J. Biol. Chem. 1998, 273, 7127–7133. [Google Scholar] [CrossRef] [PubMed]
- Kuwahara, J.; Coleman, J.E. Role of the zinc(II) ions in the structure of the three-finger DNA binding domain of the Sp1 transcription factor. Biochemistry 1990, 29, 8627–8631. [Google Scholar] [CrossRef]
- Malgieri, G.; Palmieri, M.; Esposito, S.; Maione, V.; Russo, L.; Baglivo, I.; de Paola, I.; Milardi, D.; Diana, D.; Zaccaro, L.; et al. Zinc to cadmium replacement in the prokaryotic zinc-finger domain. Metallomics 2014, 6, 96–104. [Google Scholar] [CrossRef]
- Roesijadi, G.; Bogumil, R.; Vasák, M.; Kägi, J.H.R. Modulation of DNA Binding of a Tramtrack Zinc Finger Peptide by the Metallothionein-Thionein Conjugate Pair. J. Biol. Chem. 1998, 273, 17425–17432. [Google Scholar] [CrossRef]
- Zhang, B.; Georgiev, O.; Hagmann, M.; Günes, C.; Cramer, M.; Faller, P.; Vasák, M.; Schaffner, W. Activity of Metal-Responsive Transcription Factor 1 by Toxic Heavy Metals and H2O2 In Vitro Is Modulated by Metallothionein. Mol. Cell. Biol. 2003, 23, 8471–8485. [Google Scholar] [CrossRef]
- Giedroc, D.P.; Chen, X.; Pennella, M.A.; LiWang, A.C. Conformational Heterogeneity in the C-terminal Zinc Fingers of Human MTF-1: An Nmr and Zinc-Binding Study. J. Biol. Chem. 2001, 276, 42322–42332. [Google Scholar] [CrossRef]
- Ciavatta, L.; Grimaldi, M. Equilibrium constants of mercury(II) chloride complexes. J. Inorg. Nucl. Chem. 1968, 30, 197–205. [Google Scholar] [CrossRef]
- Makowski, G.S.; Lin, S.-M.; Brennan, S.M.; Smilowitz, H.M.; Hopfer, S.M.; Sunderman, F.W. Detection of two Zn-finger proteins ofXenopus laevis, TFIIIA, and p43, by probing western blots of ovary cytosol with 65Zn2+, 63Ni2+, or 109Cd2+. Biol. Trace Element Res. 1991, 29, 93–109. [Google Scholar] [CrossRef]
- Sivo, V.; D’Abrosca, G.; Baglivo, I.; Iacovino, R.; Pedone, P.V.; Fattorusso, R.; Russo, L.; Malgieri, G.; Isernia, C. Ni(II), Hg(II), and Pb(II) Coordination in the Prokaryotic Zinc-Finger Ros87. Inorg. Chem. 2019, 58, 1067–1080. [Google Scholar] [CrossRef]
- Hajdu, B.; Hunyadi-Gulyás, É.; Kato, K.; Kawaguchi, A.; Nagata, K.; Gyurcsik, B. Zinc binding of a Cys2His2 type zinc finger protein is enhanced by the interaction with DNA. J. Biol. Inorg. Chem. 2022; accepted for publication. [Google Scholar]
- Sénèque, O.; Latour, J.-M. Coordination Properties of Zinc Finger Peptides Revisited: Ligand Competition Studies Reveal Higher Affinities for Zinc and Cobalt. J. Am. Chem. Soc. 2010, 132, 17760–17774. [Google Scholar] [CrossRef] [PubMed]
- Kägi, J.H.; Vallee, B.L.; Carlson, J.M. Metallothionein: A Cadmium and Zinc-containing Protein from Equine Renal Cortex: II. Physicochemical properties. J. Biol. Chem. 1961, 236, 2435–2442. [Google Scholar] [CrossRef]
- Willner, H.; Vasak, M.; Kaegi, J.H.R. Cadmium-thiolate clusters in metallothionein: Spectrophotometric and spectropolarimetric features. Biochemistry 1987, 26, 6287–6292. [Google Scholar] [CrossRef] [PubMed]
- Micsonai, A.; Moussong, É.; Wien, F.; Boros, E.; Vadászi, H.; Murvai, N.; Lee, Y.-H.; Molnár, T.; Réfrégiers, M.; Goto, Y.; et al. BeStSel: Webserver for secondary structure and fold prediction for protein CD spectroscopy. Nucleic Acids Res. 2022, 50, W90–W98. [Google Scholar] [CrossRef] [PubMed]
- Basile, L.A.; Coleman, J.E. Optical activity associated with the sulfur to metal charge transfer bands of Zn and Cd GAL4. Protein Sci. 1992, 1, 617–624. [Google Scholar] [CrossRef]
- Zékány, L.; Nagypál, I. A Comprehensive Program for the Evaluation of Potentiometric and/or Spectrophotometric Equilibrium Data Using Analytical Derivatives. In Computational Methods for the Determination of Formation Constants; Springer: Boston, MA, USA, 1985; pp. 291–353. [Google Scholar] [CrossRef]
- Malaiyandi, L.M.; Sharthiya, H.; Barakat, A.N.; Edwards, J.R.; Dineley, K.E. Using FluoZin-3 and fura-2 to monitor acute accumulation of free intracellular Cd2+ in a pancreatic beta cell line. Biometals 2019, 32, 951–964. [Google Scholar] [CrossRef]
- Palmieri, M.; Malgieri, G.; Russo, L.; Baglivo, I.; Esposito, S.; Netti, F.; Del Gatto, A.; de Paola, I.; Zaccaro, L.; Pedone, P.V.; et al. Structural Zn(II) Implies a Switch from Fully Cooperative to Partly Downhill Folding in Highly Homologous Proteins. J. Am. Chem. Soc. 2013, 135, 5220–5228. [Google Scholar] [CrossRef]
- Grazioso, R.; García-Viñuales, S.; Russo, L.; D’Abrosca, G.; Esposito, S.; Zaccaro, L.; Iacovino, R.; Milardi, D.; Fattorusso, R.; Malgieri, G.; et al. Substitution of the Native Zn(II) with Cd(II), Co(II) and Ni(II) Changes the Downhill Unfolding Mechanism of Ros87 to a Completely Different Scenario. Int. J. Mol. Sci. 2020, 21, 8285. [Google Scholar] [CrossRef]
- Szunyogh, D.; Gyurcsik, B.; Larsen, F.H.; Stachura, M.; Thulstrup, P.W.; Hemmingsen, L.; Jancsó, A. ZnII and HgII binding to a designed peptide that accommodates different coordination geometries. Dalton Trans. 2015, 44, 12576–12588. [Google Scholar] [CrossRef] [PubMed]
- Kägi, J.H.; Vasák, M.; Lerch, K.; Gilg, D.E.; Hunziker, P.; Bernhard, W.R.; Good, M. Structure of mammalian metallothionein. Environ. Health Perspect. 1984, 54, 93–103. [Google Scholar] [CrossRef] [PubMed]
- Stricks, W.; Kolthoff, I.M. Reactions between Mercuric Mercury and Cysteine and Glutathione. Apparent Dissociation Constants, Heats and Entropies of Formation of Various Forms of Mercuric Mercapto-Cysteine and -Glutathione. J. Am. Chem. Soc. 1953, 75, 5673–5681. [Google Scholar] [CrossRef]
- Pires, S.; Habjanič, J.; Sezer, M.; Soares, C.M.; Hemmingsen, L.; Iranzo, O. Design of a Peptidic Turn with High Affinity for HgII. Inorg. Chem. 2012, 51, 11339–11348. [Google Scholar] [CrossRef] [PubMed]
- DeSilva, T.M.; Veglia, G.; Porcelli, F.; Prantner, A.M.; Opella, S.J. Selectivity in heavy metal- binding to peptides and proteins. Biopolymers 2002, 64, 189–197. [Google Scholar] [CrossRef] [PubMed]
- Rousselot-Pailley, P.; Sénèque, O.; Lebrun, C.; Crouzy, S.; Boturyn, D.; Dumy, P.; Ferrand, M.; Delangle, P. Model Peptides Based on the Binding Loop of the Copper Metallochaperone Atx1: Selectivity of the Consensus Sequence MxCxxC for Metal Ions Hg(II), Cu(I), Cd(II), Pb(II), and Zn(II). Inorg. Chem. 2006, 45, 5510–5520. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 Years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Bjerrum, J. Metal Amine Formation in Aqueous Solution; Haase: Copenhagen, Denmark, 1941. [Google Scholar]
- Beck, M.T.; Nagypál, I. Chemistry of Complex Equilibria; Akadémiai Kiadó: Budapest, Hungary; Ellis Horwood Limited: Chichester, UK, 1990. [Google Scholar]
- Malakhov, M.P.; Mattern, M.R.; Malakhova, O.A.; Drinker, M.; Weeks, S.D.; Butt, T.R. SUMO fusions and SUMO-specific protease for efficient expression and purification of proteins. J. Struct. Funct. Genom. 2004, 5, 75–86. [Google Scholar] [CrossRef]
- Elhameed, H.A.A.; Hajdu, B.; Balogh, R.K.; Hermann, E.; Hunyadi-Gulyás, É.; Gyurcsik, B. Purification of proteins with native terminal sequences using a Ni(II)-cleavable C-terminal hexahistidine affinity tag. Protein Expr. Purif. 2019, 159, 53–59. [Google Scholar] [CrossRef]
- Kim, C.A.; Berg, J.M. A 2.2 Å resolution crystal structure of a designed zinc finger protein bound to DNA. Nat. Struct. Biol. 1996, 3, 940–945. [Google Scholar] [CrossRef]
- Krizek, B.A.; Amann, B.T.; Kilfoil, V.J.; Merkle, D.L.; Berg, J.M. A consensus zinc finger peptide: Design high-affinity metal binding a pH-dependent structure and a His to Cys sequence variant. J. Am. Chem. Soc. 1991, 113, 4518–4523. [Google Scholar] [CrossRef]
- Miles, A.J.; Hoffmann, S.V.; Wallace, B.A.; Tao, Y.; Janes, R.W. Synchrotron Radiation Circular Dichroism (SRCD) spectroscopy: New beamlines and new applications in biology. Spectroscopy 2007, 21, 245–255. [Google Scholar] [CrossRef]
- Miles, A.J.; Janes, R.W.; Brown, A.; Clarke, D.T.; Sutherland, J.C.; Tao, Y.; Wallace, B.A.; Hoffmann, S.V. Light flux density threshold at which protein denaturation is induced by synchrotron radiation circular dichroism beamlines. J. Synchrotron Radiat. 2008, 15, 420–422. [Google Scholar] [CrossRef]
- Kluska, K.; Veronesi, G.; Deniaud, A.; Hajdu, B.; Gyurcsik, B.; Bal, W.; Krężel, A. Structures of Silver Fingers and a Pathway to Their Genotoxicity. Angew. Chem. Int. Ed. 2022, 61, e202116621. [Google Scholar] [CrossRef] [PubMed]
- Marszałek, I.; Krężel, A.; Goch, W.; Zhukov, I.; Paczkowska, I.; Bal, W. Revised stability constant, spectroscopic properties and binding mode of Zn(II) to FluoZin-3, the most common zinc probe in life sciences. J. Inorg. Biochem. 2016, 161, 107–114. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).