N-Based Polydentate Ligands and Corresponding Zn(II) Complexes: A Structural and Spectroscopic Study
Abstract
:1. Introduction
2. Results and Discussion
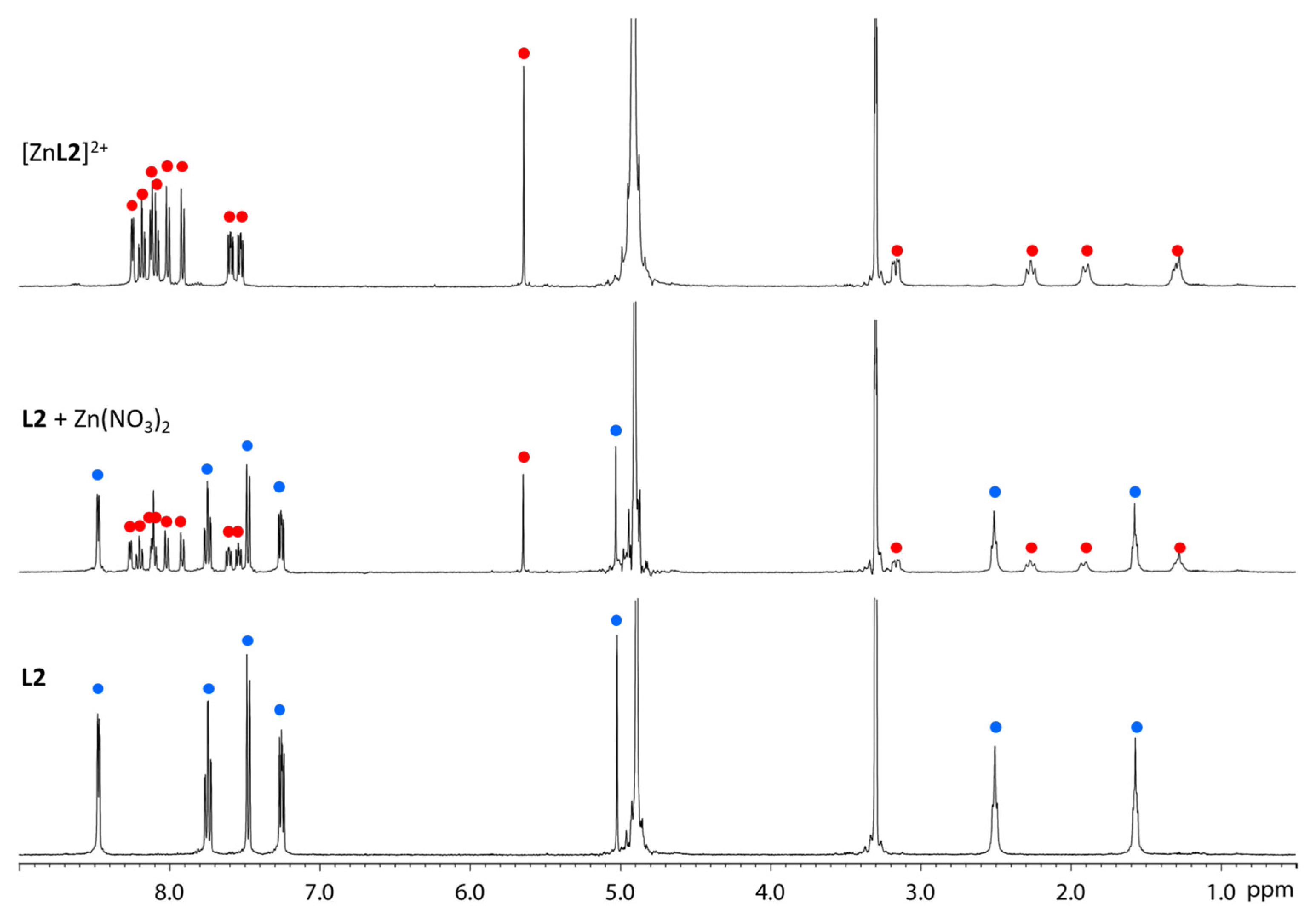
2.1. Complexation Behaviour
2.2. Spectroscopic Characterization
2.3. Mass Spectrometry Characterization
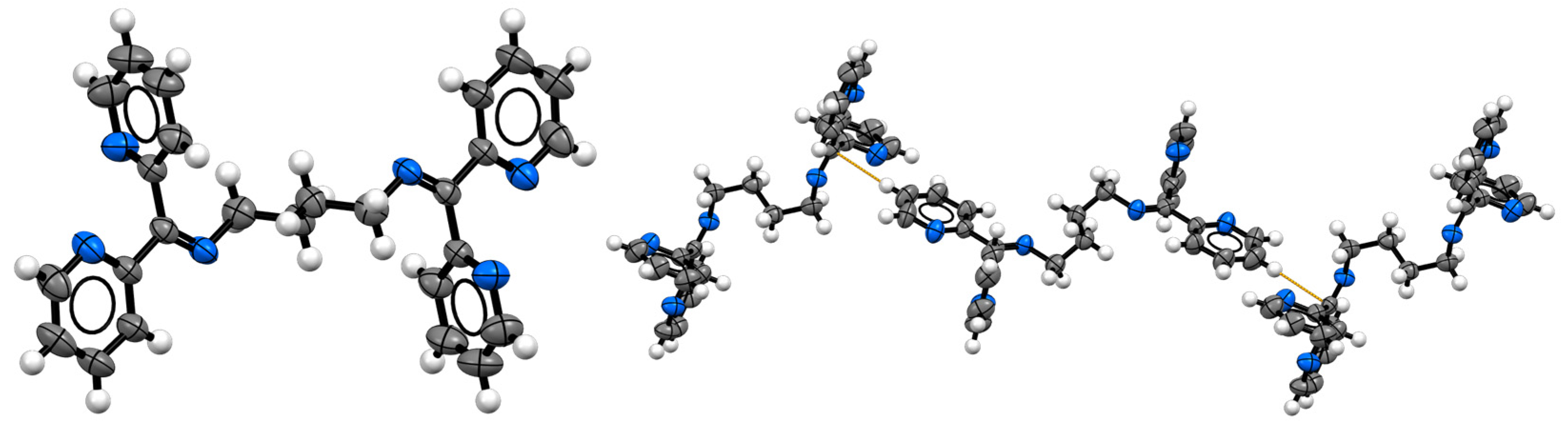
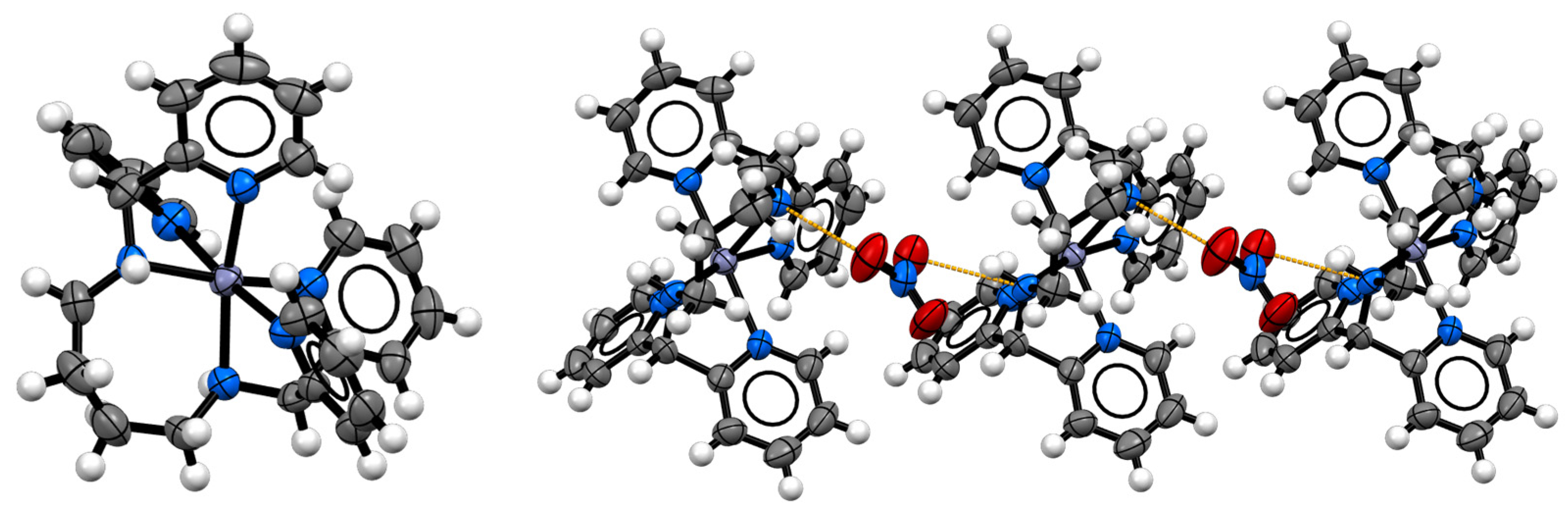
2.4. Structural Characterization
3. Materials and Methods
3.1. Experimental Details
3.2. Synthesis
3.2.1. L1: N,N’-(Butane-1,4-diyl)bis(1,1-di(pyridin-2-yl)methanimine)
3.2.2. L2: N1,N4-Bis(di(pyridin-2-yl)methyl)butane-1,4-diamine
3.2.3. [ZnL1]2+
3.2.4. [ZnL2]2+
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vahidi Ferdowsi, P.; Ng, R.; Adulcikas, J.; Sohal, S.S.; Myers, S. Zinc Modulates Several Transcription-Factor Regulated Pathways in Mouse Skeletal Muscle Cells. Molecules 2020, 25, 5098. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Yin, H.-Y.; Zhang, J.-L. Chapter One—Luminescent Zinc Complexes as Bioprobes for Imaging Molecular Events in Live Cells. In Inorganic and Organometallic Transition Metal Complexes with Biological Molecules and Living Cells; Lo, K.K.-W., Ed.; Academic Press: Cambridge, MA, USA, 2017; pp. 1–53. ISBN 978-0-12-803814-7. [Google Scholar]
- Endo, I.; Nagamune, T. Nano/Micro Biotechnology; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2010; ISBN 978-3-642-14946-7. [Google Scholar]
- Horton, T.M.; Allegretti, P.A.; Lee, S.; Moeller, H.P.; Smith, M.; Annes, J.P. Zinc-Chelating Small Molecules Preferentially Accumulate and Function within Pancreatic β Cells. Cell Chem. Biol. 2019, 26, 213–222.e6. [Google Scholar] [CrossRef] [PubMed]
- Das, R.K.; Pachapur, V.L.; Lonappan, L.; Naghdi, M.; Pulicharla, R.; Maiti, S.; Cledon, M.; Dalila, L.M.A.; Sarma, S.J.; Brar, S.K. Biological Synthesis of Metallic Nanoparticles: Plants, Animals and Microbial Aspects. Nanotechnol. Environ. Eng. 2017, 2, 18. [Google Scholar] [CrossRef]
- Kikuchi, K.; Komatsu, K.; Nagano, T. Zinc Sensing for Cellular Application. Curr. Opin. Chem. Biol. 2004, 8, 182–191. [Google Scholar] [CrossRef]
- Neufeld, E.J. Oral Chelators Deferasirox and Deferiprone for Transfusional Iron Overload in Thalassemia Major: New Data, New Questions. Blood 2006, 107, 3436–3441. [Google Scholar] [CrossRef]
- Purchase, R. The Treatment of Wilson’s Disease, a Rare Genetic Disorder of Copper Metabolism. Sci. Prog. 2013, 96, 19–32. [Google Scholar] [CrossRef]
- Delangle, P.; Mintz, E. Chelation Therapy in Wilson’s Disease: From d-Penicillamine to the Design of Selective Bioinspired Intracellular Cu(i) Chelators. Dalton Trans. 2012, 41, 6359–6370. [Google Scholar] [CrossRef]
- Siegemund, R.; Lößner, J.; Günther, K.; Kühn, H.-J.; Bachmann, H. Mode of Action of Triethylenetetramine Dihydrochloride on Copper Metabolism in Wilson’s Disease. Acta Neurol. Scand. 1991, 83, 364–366. [Google Scholar] [CrossRef]
- Kontoghiorghes, G.J. Advances on Chelation and Chelator Metal Complexes in Medicine. Int. J. Mol. Sci. 2020, 21, 2499. [Google Scholar] [CrossRef]
- Kim, K.; Murray, J.; Selvapalam, N.; Ko, Y.H.; Hwang, I. Cucurbiturils: Chemistry, Supramolecular Chemistry and Applications; WORLD SCIENTIFIC (EUROPE): London, UK, 2018; ISBN 978-1-84816-408-6. [Google Scholar]
- Andreo, L.; Volpi, G.; Rossi, F.; Benzi, P.; Diana, E. Two-Step Synthesis of a New Twenty-Membered Macrocycle: Spectroscopic Characterization and Theoretical Calculations. ChemistrySelect 2022, 7, e202202564. [Google Scholar] [CrossRef]
- Knipe, A.C. Crown Ethers. A New Aid to Synthesis and to Elucidation of Reaction Mechanisms. J. Chem. Educ. 1976, 53, 618. [Google Scholar] [CrossRef]
- Evans, N.H. Chiral Catenanes and Rotaxanes: Fundamentals and Emerging Applications. Chem. Eur. J. 2018, 24, 3101–3112. [Google Scholar] [CrossRef] [PubMed]
- Ji, X.; Ahmed, M.; Long, L.; Khashab, N.M.; Huang, F.; Sessler, J.L. Adhesive Supramolecular Polymeric Materials Constructed from Macrocycle-Based Host–Guest Interactions. Chem. Soc. Rev. 2019, 48, 2682–2697. [Google Scholar] [CrossRef] [PubMed]
- Volpi, G.; Garino, C.; Priola, E.; Diana, E.; Gobetto, R.; Buscaino, R.; Viscardi, G.; Barolo, C. Facile Synthesis of Novel Blue Light and Large Stoke Shift Emitting Tetradentate Polyazines Based on Imidazo[1,5-a]Pyridine—Part 2. Dye. Pigment. 2017, 143, 284–290. [Google Scholar] [CrossRef]
- Nelson, S.M. Developments in the Synthesis and Coordination Chemistry of Macrocyclic Schiff Base Ligands. Pure Appl. Chem. 1980, 52, 2461–2476. [Google Scholar] [CrossRef]
- Wu, J.-R.; Yang, Y.-W. New Opportunities in Synthetic Macrocyclic Arenes. Chem. Commun. 2019, 55, 1533–1543. [Google Scholar] [CrossRef]
- Ball, M.; Zhang, B.; Zhong, Y.; Fowler, B.; Xiao, S.; Ng, F.; Steigerwald, M.; Nuckolls, C. Conjugated Macrocycles in Organic Electronics. Acc. Chem. Res. 2019, 52, 1068–1078. [Google Scholar] [CrossRef]
- Saha, S.K.; Dutta, A.; Ghosh, P.; Sukul, D.; Banerjee, P. Novel Schiff-Base Molecules as Efficient Corrosion Inhibitors for Mild Steel Surface in 1 M HCl Medium: Experimental and Theoretical Approach. Phys. Chem. Chem. Phys. 2016, 18, 17898–17911. [Google Scholar] [CrossRef]
- Buston, J.E.H.; Young, J.R.; Anderson, H.L. Rotaxane-Encapsulated Cyanine Dyes: Enhanced Fluorescence Efficiency and Photostability. Chem. Commun. 2000, 905–906. [Google Scholar] [CrossRef]
- Fujiwara, T.; Muranaka, A.; Nishinaga, T.; Aoyagi, S.; Kobayashi, N.; Uchiyama, M.; Otani, H.; Iyoda, M. Preparation, Spectroscopic Characterization and Theoretical Study of a Three-Dimensional Conjugated 70 π-Electron Thiophene 6-Mer Radical Cation π-Dimer. J. Am. Chem. Soc. 2020, 142, 5933–5937. [Google Scholar] [CrossRef]
- Mako, T.L.; Racicot, J.M.; Levine, M. Supramolecular Luminescent Sensors. Chem. Rev. 2019, 119, 322–477. [Google Scholar] [CrossRef] [PubMed]
- Schettini, R.; Sicignano, M.; De Riccardis, F.; Izzo, I.; Della Sala, G. Macrocyclic Hosts in Asymmetric Phase-Transfer Catalyzed Reactions. Synthesis 2018, 50, 4777–4795. [Google Scholar] [CrossRef]
- Aucagne, V.; Berná, J.; Crowley, J.D.; Goldup, S.M.; Hänni, K.D.; Leigh, D.A.; Lusby, P.J.; Ronaldson, V.E.; Slawin, A.M.Z.; Viterisi, A.; et al. Catalytic “Active-Metal” Template Synthesis of [2]Rotaxanes, [3]Rotaxanes, and Molecular Shuttles, and Some Observations on the Mechanism of the Cu(I)-Catalyzed Azide−Alkyne 1,3-Cycloaddition. J. Am. Chem. Soc. 2007, 129, 11950–11963. [Google Scholar] [CrossRef]
- Peng, R.; Xu, Y.; Cao, Q. Recent Advances in Click-Derived Macrocycles for Ions Recognition. Chin. Chem. Lett. 2018, 29, 1465–1474. [Google Scholar] [CrossRef]
- Mantooth, S.M.; Munoz-Robles, B.G.; Webber, M.J. Dynamic Hydrogels from Host-Guest Supramolecular Interactions. Macromol. Biosci. 2019, 19, 1800281. [Google Scholar] [CrossRef]
- Zhou, T.; Ma, Y.; Kong, X.; Hider, R.C. Design of Iron Chelators with Therapeutic Application. Dalton Trans. 2012, 41, 6371–6389. [Google Scholar] [CrossRef]
- Kim, J.-J.; Kim, Y.-S.; Kumar, V. Heavy Metal Toxicity: An Update of Chelating Therapeutic Strategies. J. Trace Elem. Med. Biol. 2019, 54, 226–231. [Google Scholar] [CrossRef]
- Liu, Z.D.; Hider, R.C. Design of Clinically Useful Iron(III)-Selective Chelators. Med. Res. Rev. 2002, 22, 26–64. [Google Scholar] [CrossRef]
- Roy, M.; Chakravarthi, B.V.S.K.; Jayabaskaran, C.; Karande, A.A.; Chakravarty, A.R. Impact of Metal Binding on the Antitumor Activity and Cellular Imaging of a Metal Chelator Cationic Imidazopyridine Derivative. Dalton Trans. 2011, 40, 4855–4864. [Google Scholar] [CrossRef]
- Liu, Z.D.; Hider, R.C. Design of Iron Chelators with Therapeutic Application. Coord. Chem. Rev. 2002, 232, 151–171. [Google Scholar] [CrossRef]
- Chaudhari, V.; Bagwe-Parab, S.; Buttar, H.S.; Gupta, S.; Vora, A.; Kaur, G. Challenges and Opportunities of Metal Chelation Therapy in Trace Metals Overload-Induced Alzheimer’s Disease. Neurotox. Res. 2023, 41, 270–287. [Google Scholar] [CrossRef] [PubMed]
- Bissani Gasparin, C.; Pilger, D.A. 8-Hydroxyquinoline, Derivatives and Metal-Complexes: A Review of Antileukemia Activities. ChemistrySelect 2023, 8, e202204219. [Google Scholar] [CrossRef]
- Zhao, D.; Moore, J.S. Synthesis and Self-Association of an Imine-Containing m-Phenylene Ethynylene Macrocycle. J. Org. Chem. 2002, 67, 3548–3554. [Google Scholar] [CrossRef] [PubMed]
- Akine, S.; Taniguchi, T.; Nabeshima, T. Synthesis and Crystal Structure of a Novel Triangular Macrocyclic Molecule, Tris(H2saloph), H2saloph=N,N′-Disalicylidene-o-Phenylenediamine. and Its Water Complex. Tetrahedron Lett. 2001, 42, 8861–8864. [Google Scholar] [CrossRef]
- Curtis, N.F. Compounds of Amine-Imine Macrocycles: Syntheses and Structures of Compounds of Aza-Macrocycles with Amine-β-Imine Ring Segments. Inorganica Chim. Acta 2021, 527, 120164. [Google Scholar] [CrossRef]
- Ma, J.; Vannam, R.; Terwilliger, D.W.; Peczuh, M.W. Synthesis, Structure and Reactivity of a Macrocyclic Imine: Aza-[13]-Macrodiolides. Tetrahedron Lett. 2014, 55, 4255–4259. [Google Scholar] [CrossRef]
- Carbajo, D.; Ruiz-Sánchez, A.J.; Nájera, F.; Pérez-Inestrosa, E.; Alfonso, I. Spontaneous Macrocyclization through Multiple Dynamic Cyclic Aminal Formation. Chem. Commun. 2021, 57, 1190–1193. [Google Scholar] [CrossRef]
- Mukhopadhyay, S.; Biswas, A.; Pandey, R.; Gupta, R.K.; Pandey, D.S. A Highly Selective and Femto-Molar Sensitive Fluorescence ‘Turn-on’ Chemodosimeter for Hg2+. Tetrahedron Lett. 2014, 55, 1437–1440. [Google Scholar] [CrossRef]
- Heng, S.; Reineck, P.; Vidanapathirana, A.K.; Pullen, B.J.; Drumm, D.W.; Ritter, L.J.; Schwarz, N.; Bonder, C.S.; Psaltis, P.J.; Thompson, J.G.; et al. Rationally Designed Probe for Reversible Sensing of Zinc and Application in Cells. ACS Omega 2017, 2, 6201–6210. [Google Scholar] [CrossRef]
- Sun, F.; Zhang, G.; Zhang, D.; Xue, L.; Jiang, H. Aqueous Fluorescence Turn-on Sensor for Zn2+ with a Tetraphenylethylene Compound. Org. Lett. 2011, 13, 6378–6381. [Google Scholar] [CrossRef]
- Shellaiah, M.; Wu, Y.-H.; Lin, H.-C. Simple Pyridyl-Salicylimine-Based Fluorescence “Turn-on” Sensors for Distinct Detections of Zn2+, Al3+ and OH− Ions in Mixed Aqueous Media. Analyst 2013, 138, 2931–2942. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Xiong, J.; Qin, Y.; Li, Z.; Pan, C.; Huo, Y.; Zhang, H. A Novel Quinolinyl-Tetraphenylethene-Based Fluorescence “Turn-on” Sensor for Zn2+ with a Large Stokes Shift and Its Applications for Portable Test Strips and Biological Imaging. Mater. Chem. Front. 2020, 4, 3338–3348. [Google Scholar] [CrossRef]
- Pandey, R.; Kumar, P.; Singh, A.K.; Shahid, M.; Li, P.; Singh, S.K.; Xu, Q.; Misra, A.; Pandey, D.S. Fluorescent Zinc(II) Complex Exhibiting “On-Off-On” Switching Toward Cu2+ and Ag+ Ions. Inorg. Chem. 2011, 50, 3189–3197. [Google Scholar] [CrossRef] [PubMed]
- Corrsin, L.; Fax, B.J.; Lord, R.C. The Vibrational Spectra of Pyridine and Pyridine-d5. J. Chem. Phys. 2004, 21, 1170–1176. [Google Scholar] [CrossRef]
- Wong, P.T.T.; Brewer, D.G. Nature of the Coordination Bond in Metal Complexes of Substituted Pyridine Derivatives. IV. Skeletal Vibration Spectra and the Substituent Effect upon the Coordination Bond of the Pyridine Complex of Zinc(II) Ion. Can. J. Chem. 1969, 47, 4589–4597. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H.; et al. Gaussian 16; Revision B.01; Gaussian, Inc.: Wallingford, CT, USA, 2016. [Google Scholar]
- Becke, A.D. Density-functional Thermochemistry. III. The Role of Exact Exchange. J. Chem. Phys. 1993, 98, 5648–5652. [Google Scholar] [CrossRef]
- Lee, C.T.; Yang, W.T.; Parr, R.G. Development of the Colle-Salvetti Correlation-Energy Formula into a Functional of the Electron-Density. Phys. Rev. B 1988, 37, 785–789. [Google Scholar] [CrossRef]
- Cossi, M.; Scalmani, G.; Rega, N.; Barone, V. New Developments in the Polarizable Continuum Model for Quantum Mechanical and Classical Calculations on Molecules in Solution. J. Chem. Phys. 2002, 117, 43–54. [Google Scholar] [CrossRef]
- Miertuš, S.; Scrocco, E.; Tomasi, J. Electrostatic Interaction of a Solute with a Continuum. A Direct Utilizaion of AB Initio Molecular Potentials for the Prevision of Solvent Effects. Chem. Phys. 1981, 55, 117–129. [Google Scholar] [CrossRef]
- McLean, A.D.; Chandler, G.S. Contracted Gaussian Basis Sets for Molecular Calculations. I. Second Row Atoms, Z = 11–18. J. Chem. Phys. 1980, 72, 5639–5648. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Meng, E.C.; Couch, G.S.; Croll, T.I.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Structure Visualization for Researchers, Educators, and Developers. Protein Sci. 2021, 30, 70–82. [Google Scholar] [CrossRef] [PubMed]
- Volpi, G.; Garino, C.; Nervi, C. Exploring Synthetic Pathways to Cationic Heteroleptic Cyclometalated Iridium Complexes Derived from Dipyridylketone. Dalton Trans. 2012, 41, 7098–7108. [Google Scholar] [CrossRef] [PubMed]
- Volpi, G.; Garino, C.; Gobetto, R.; Nervi, C. Dipyridylmethane Ethers as Ligands for Luminescent Ir Complexes. Molecules 2021, 26, 7161. [Google Scholar] [CrossRef] [PubMed]
- Hasan, M.M.; Kubra, K.T.; Hasan, M.N.; Awual, M.E.; Salman, M.S.; Sheikh, M.C.; Rehan, A.I.; Rasee, A.I.; Waliullah, R.M.; Islam, M.S.; et al. Sustainable Ligand-Modified Based Composite Material for the Selective and Effective Cadmium(II) Capturing from Wastewater. J. Mol. Liq. 2023, 371, 121125. [Google Scholar] [CrossRef]
- Montaño-Medina, C.U.; Lopéz-Martínez, L.M.; Ochoa-Terán, A.; López-Maldonado, E.A.; Salazar-Gastelum, M.I.; Trujillo-Navarrete, B.; Pérez-Sicairos, S.; Cornejo-Bravo, J.M. New Pyridyl and Aniline-Functionalized Carbamoylcarboxylic Acids for Removal of Metal Ions from Water by Coagulation-Flocculation Process. Chem. Eng. J. 2023, 451, 138396. [Google Scholar] [CrossRef]
- Hozien, Z.A.; EL-Mahdy, A.F.M.; Ali, L.S.A.; Markeb, A.A.; El-Sherief, H.A.H. One-Pot Synthesis of Some New s-Triazole Derivatives and Their Potential Application for Water Decontamination. ACS Omega 2021, 6, 25574–25584. [Google Scholar] [CrossRef]
- Volpi, G.; Ginepro, M.; Tafur-Marinos, J.; Zelano, V. Pollution Abatement of Heavy Metals in Different Conditions by Water Kefir Grains as a Protective Tool against Toxicity. J. Chem. 2019, 2019, 8763902. [Google Scholar] [CrossRef]
- Costamagna, G.; Volpi, G.; Ghibaudi, E.; Ginepro, M. Quantitative Insights on the Interaction between Metal Ions and Water Kefir Grains: Kinetics Studies and EPR Investigations. Nat. Prod. Res. 2022, 36, 3440–3444. [Google Scholar] [CrossRef]
- Bianco Prevot, A.; Ginepro, M.; Peracaciolo, E.; Zelano, V.; De Luca, D.A. Chemical vs Bio-Mediated Reduction of Hexavalent Chromium. An In-Vitro Study for Soil and Deep Waters Remediation. Geoderma 2018, 312, 17–23. [Google Scholar] [CrossRef]
- Gul, Z.; Salman, M.; Khan, S.; Shehzad, A.; Ullah, H.; Irshad, M.; Zeeshan, M.; Batool, S.; Ahmed, M.; Altaf, A.A. Single Organic Ligands Act as a Bifunctional Sensor for Subsequent Detection of Metal and Cyanide Ions, a Statistical Approach toward Coordination and Sensitivity. Crit. Rev. Anal. Chem. 2023, 1–17. [Google Scholar] [CrossRef]


| Compound | λabs (nm) | log ε (L mol−1 cm−1) | λem (nm) | Stokes Shift (cm−1) |
|---|---|---|---|---|
| L1 | 238 267 | 4.17 4.20 | 407 | 12,883 |
| [ZnL1]2+ | 278 | 4.15 | 418 | 12,048 |
| L2 | 257 (sh) 262 272 (sh) | 4.10 4.12 4.03 | 406 | 13,538 |
| [ZnL2]2+ | 257 266 272 | 4.10 4.10 4.08 | 406 | 12,134 |
| d(Zn-N) | Distance (Å) |
|---|---|
| Zn-N1 | 2.231(4) |
| Zn-N2 | 2.105(4) |
| Zn-N3 | 2.160(5) |
| Zn-N4 | 2.126(4) |
| Zn-N5 | 2.143(4) |
| Zn-N6 | 2.240(4) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Volpi, G.; Zago, S.; Rabezzana, R.; Diana, E.; Priola, E.; Garino, C.; Gobetto, R. N-Based Polydentate Ligands and Corresponding Zn(II) Complexes: A Structural and Spectroscopic Study. Inorganics 2023, 11, 435. https://doi.org/10.3390/inorganics11110435
Volpi G, Zago S, Rabezzana R, Diana E, Priola E, Garino C, Gobetto R. N-Based Polydentate Ligands and Corresponding Zn(II) Complexes: A Structural and Spectroscopic Study. Inorganics. 2023; 11(11):435. https://doi.org/10.3390/inorganics11110435
Chicago/Turabian StyleVolpi, Giorgio, Stefano Zago, Roberto Rabezzana, Eliano Diana, Emanuele Priola, Claudio Garino, and Roberto Gobetto. 2023. "N-Based Polydentate Ligands and Corresponding Zn(II) Complexes: A Structural and Spectroscopic Study" Inorganics 11, no. 11: 435. https://doi.org/10.3390/inorganics11110435
APA StyleVolpi, G., Zago, S., Rabezzana, R., Diana, E., Priola, E., Garino, C., & Gobetto, R. (2023). N-Based Polydentate Ligands and Corresponding Zn(II) Complexes: A Structural and Spectroscopic Study. Inorganics, 11(11), 435. https://doi.org/10.3390/inorganics11110435









