Excitation-Dependent Fluorescence Helps to Indicate Fungal Contamination of Aquatic Environments and to Differentiate Filamentous Fungi
Abstract
1. Introduction
2. Materials and Methods
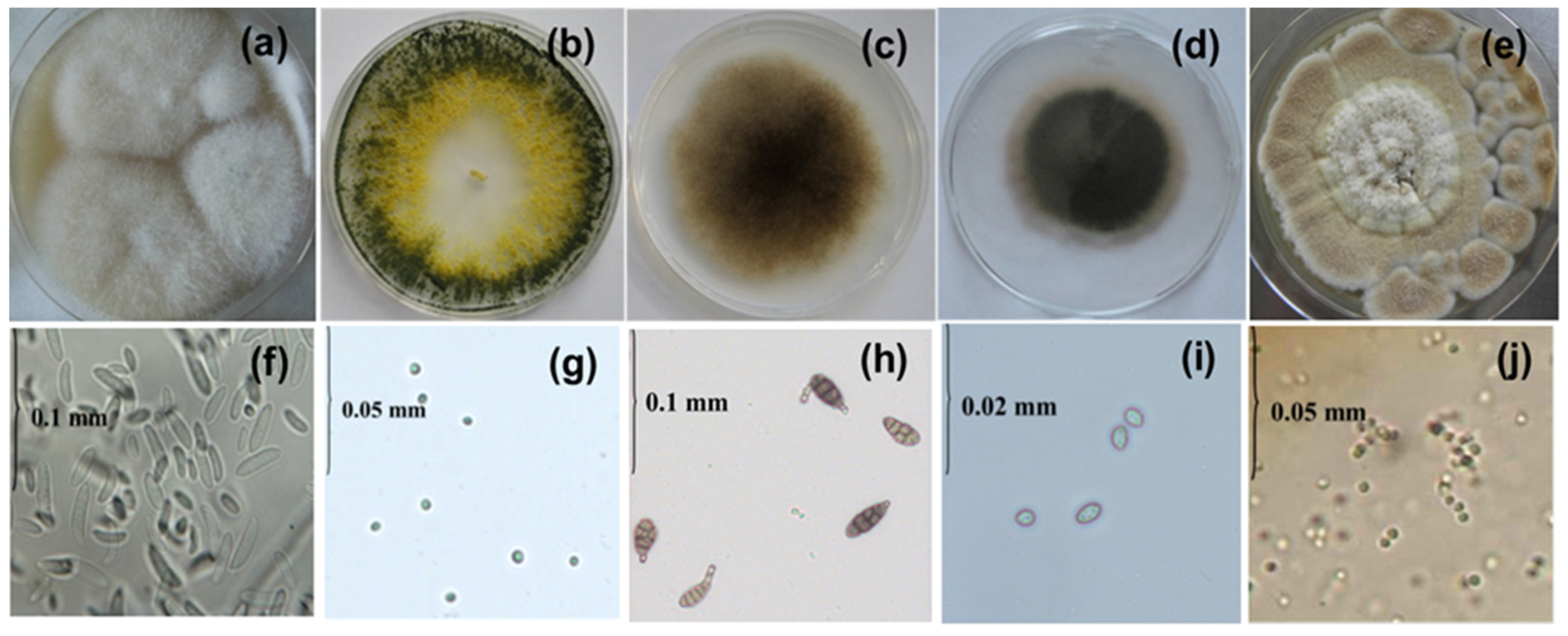
3. Results
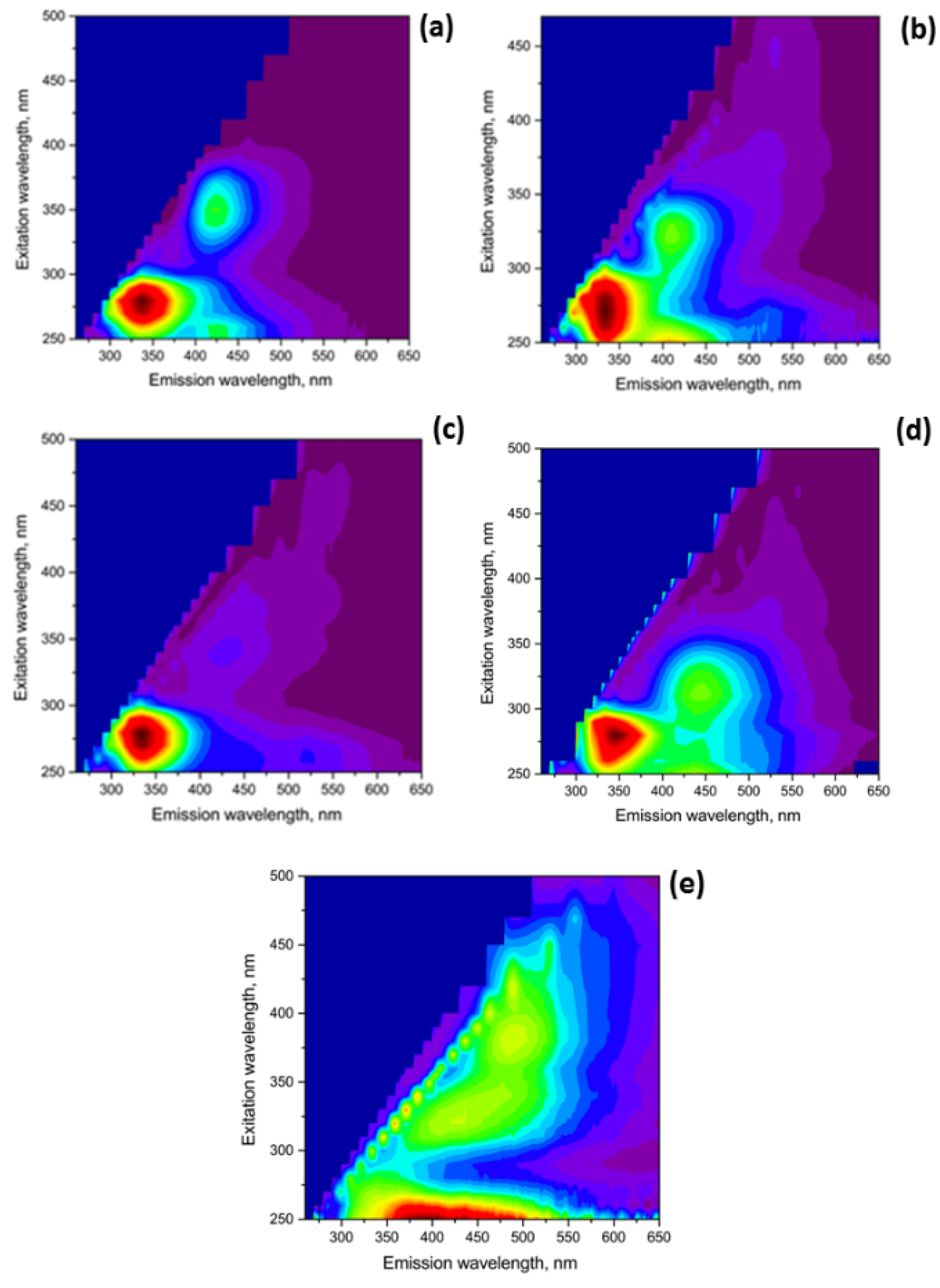
3.1. Fluorescence Excitation/Emission Matrix (EEM) Spectroscopy
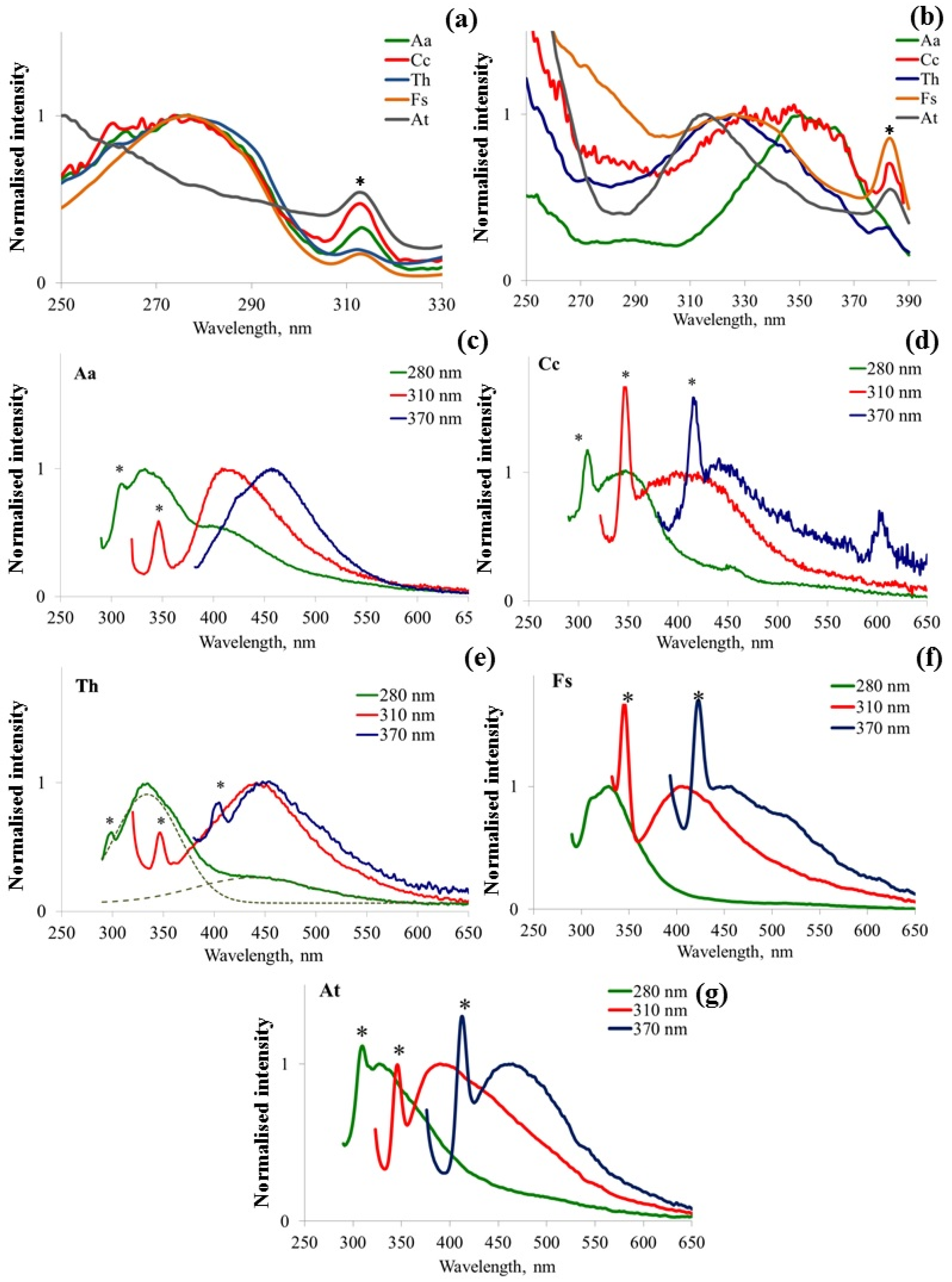
3.2. Fluorescence Spectral Features Analysis of Fungi Samples Cultivated on Agar-Containing Medium
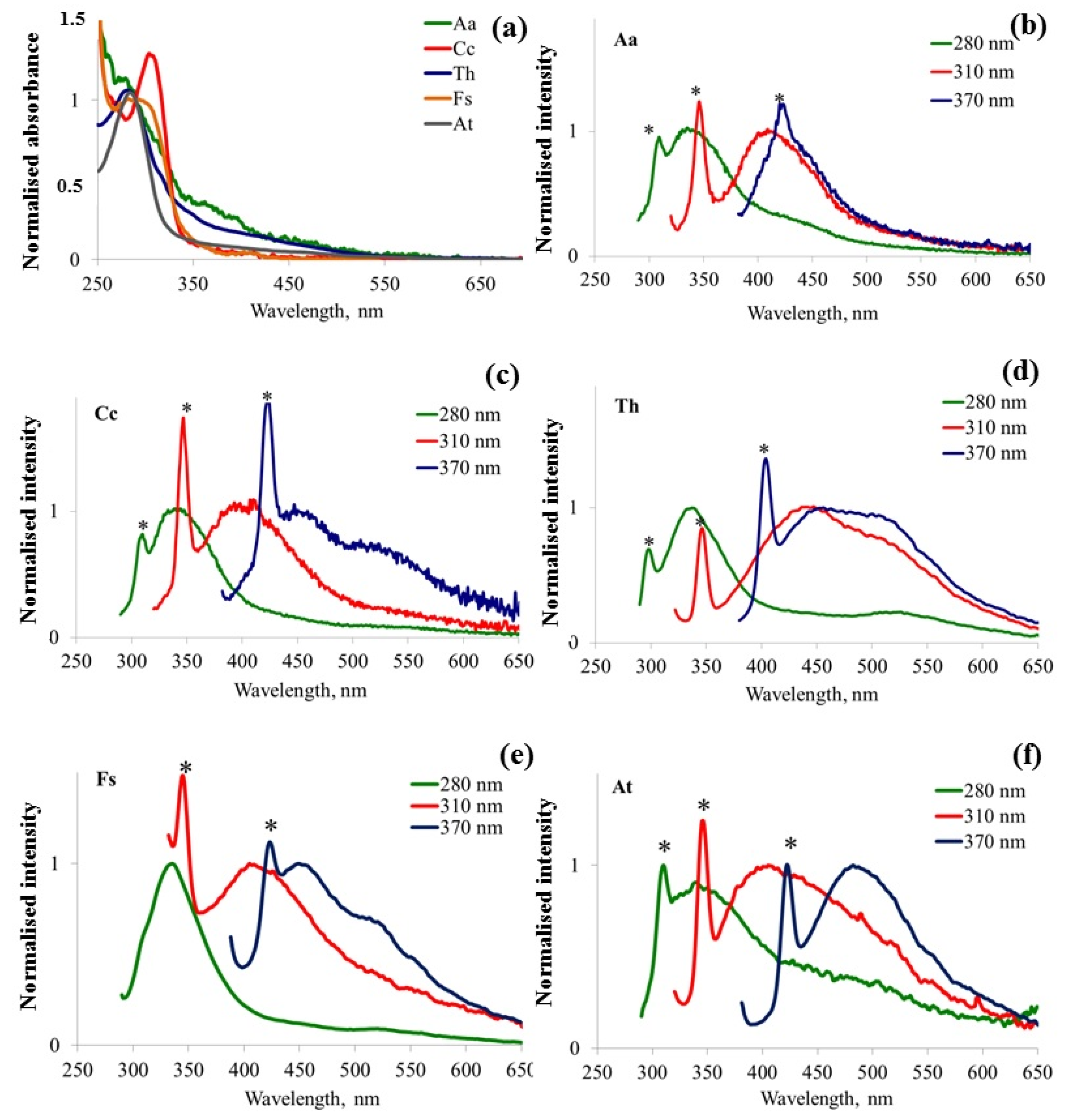
3.3. Fluorescence Spectral Features Analysis of Fungi Samples Cultivated in Liquid Medium
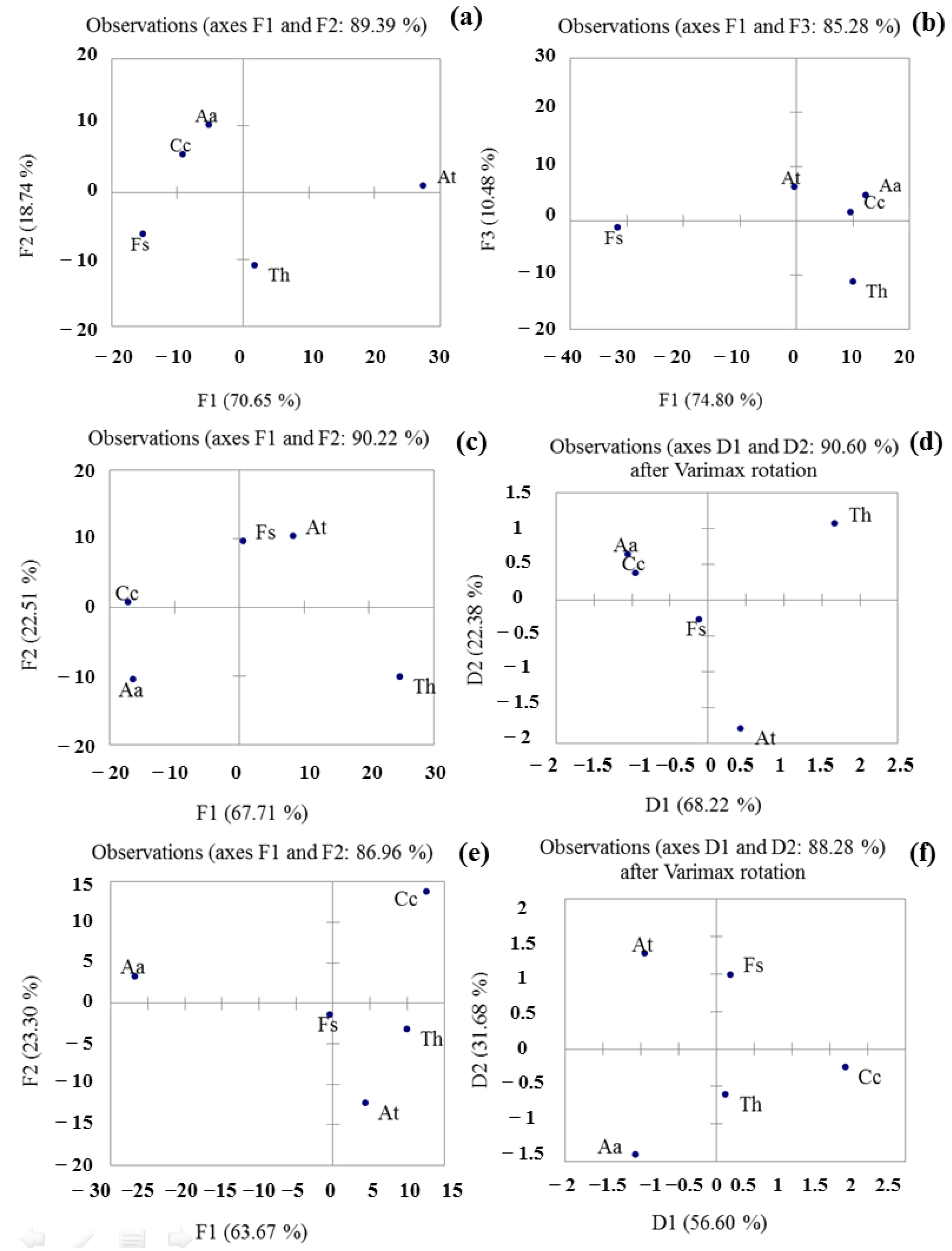
3.4. PCA Analysis to Differentiate of Fungal Samples
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hawksworth, D.L. The magnitude of fungal diversity: The 1.5 million species estimate revisited. Mycol. Res. 2001, 105, 1422–1432. [Google Scholar] [CrossRef]
- Terekhova, V.A. The importance of mycological studies for soil quality control. Eurasian Soil Sci. 2007, 40, 583–587. [Google Scholar] [CrossRef]
- Pivkin, M.V.; Kuznetsova, T.F.; Sova, V.V. Morskie Griby i Ikh Metabolity (Marine Fungi and Their Metabolites); Vladivostok Dalnauka: Vladivostok, Russia, 2006; p. 247. ISBN 5-8044-0539-X. (In Russian)
- Oliveira, B.R.; Barreto Crespo, M.T.; San Romão, M.V.; Benoliel, M.J.; Samson, R.A.; Pereira, V.J. New insights concerning the occurrence of fungi in water sources and their potential pathogenicity. Water Res. 2013, 47, 6338–6347. [Google Scholar] [CrossRef]
- Al-gabr, H.M.; Zheng, T.; Yu, X. Occurrence and quantification of fungi and detection of mycotoxigenic fungi in drinking water in Xiamen City, China. Sci. Total Environ. 2014, 466–467, 1103–1111. [Google Scholar] [CrossRef]
- Graça, M.A.S.; Hyde, K.; Chauvet, E. Aquatic hyphomycetes and litter decomposition in tropical—Subtropical low order streams. Fungal Ecol. 2016, 19, 182–189. [Google Scholar] [CrossRef]
- Mansoldo, F.R.P.; Carta, F.; Angeli, A.; Cardoso, V.S.; Supuran, C.T.; Vermelho, A.B. Chagas disease: Perspectives on the past and present and challenges in drug discovery. Molecules 2020, 25, 5483. [Google Scholar] [CrossRef]
- Wang, J.; Chen, R.; Fan, L.; Cui, L.; Zhang, Y.; Cheng, J.; Wu, X.; Zeng, W.; Tian, Q.; Shen, L. Construction of fungi-microalgae symbiotic system and adsorption study of heavy metal ions. Sep. Purif. Technol. 2021, 268, 118689. [Google Scholar] [CrossRef]
- Pöhlker, C.; Huffman, J.; Pöschl, U. Autofluorescence of atmospheric bioaerosoles fluorescent biomolecules and potential interferences. Atmos. Meas. Technol. 2012, 5, 37–71. [Google Scholar] [CrossRef]
- Pan, Y.-L. Detection and characterization of biological and other organic-carbon aerosol particles in atmosphere using fluorescence. J. Quant. Spectrosc. Radiat. Transf. 2015, 150, 12–35. [Google Scholar] [CrossRef]
- Dyakov, Y.; Dzhavakhiya, V.; Korpela, T. Comprehensive and Molecular Phytopathology; Elsevier: Amsterdam, The Netherlands, 2007. [Google Scholar]
- Kadaifciler, D.G.; Demirel, R. Fungal biodiversity and mycotoxigenic fungi in cooling-tower water systems in Istanbul, Turkey. J. Water Health 2017, 15, 308–320. [Google Scholar] [CrossRef]
- Afonso, T.B.; Chaves, S.L.; Lima, N. Occurrence of filamentous fungi in drinking water: Their role on fungal-bacterial biofilm formation. Res. Microbiol. 2021, 172, 103791. [Google Scholar] [CrossRef]
- Saari, S.; Mensah-Attipoe, J.; Reponen, T.; Veijalainen, A.M.; Salmela, A.; Pasanen, P.; Keskinen, J. Effects of fungal species, cultivation time, growth substrate, and air exposure velocity on the fluorescence properties of airborne fungal spores. Indoor Air 2015, 25, 653–661. [Google Scholar] [CrossRef]
- Petra, P. Identifying fungi spores, yeast, bacteria by opto-electronic imaging and image processing and identification for 0011 protecting human health. Curr. Trends Biomed. Eng. Biosci. 2018, 11, 555806. [Google Scholar] [CrossRef]
- Löbs, N.; Barbosa, C.G.G.; Brill, S.; Walter, D.; Ditas, F.; Sá de Oliveira, M.; de Araújo, A.C.; de Oliveira, R.L.; Godoi, R.H.M.; Wolff, S.; et al. Aerosol measurement methods to quantify spore emissions from fungi and cryptogamic covers in the Amazon. Atmos. Meas. Technol. 2020, 13, 153–164. [Google Scholar] [CrossRef]
- Wang, Y.; Mao, H.; Xu, G.; Zhang, X.; Zhang, Y. A rapid detection method for fungal spores from greenhouse crops based on CMOS image sensors and diffraction fingerprint feature processing. J. Fungi 2022, 8, 374. [Google Scholar] [CrossRef]
- Sarraguca, M.C.; Paulo, A.; Alves, M.M.; Dias, A.M.A.; Lopes, J.A.; Ferreira, E.C. Quantitative monitoring of an activated sludge reactor using on-line UV–vis and near-infrared spectroscopy. Anal. Bioanal. Chem. 2009, 395, 1159–1166. [Google Scholar] [CrossRef]
- Khundzhua, D.A.; Patsaeva, S.V.; Terekhova, V.A.; Yuzhakov, V.I. Spectral characterization of fungal metabolites in aqueous medium with humus substances. J. Spectrosc. 2013, 2013, 538608. [Google Scholar] [CrossRef]
- Singh, G.P.; Goh, S.; Canzoneri, M.; Ram, R.J. Raman spectroscopy of complex defined media. Biopharmaceutical applications. J. Raman Spectrosc. 2015, 46, 545–550. [Google Scholar] [CrossRef]
- Schalk, R.; Geoerg, D.; Staubach, J.; Raedle, M.; Methner, F.-J.; Beuermann, T. Evaluation of a newly developed mid-infrared sensor for real-time monitoring of yeast fermentations. J. Biosci. Bioeng. 2017, 123, 651–657. [Google Scholar] [CrossRef]
- Laptinskiy, K.A.; Burikov, S.A.; Patsaeva, S.V.; Vlasov, I.I.; Shenderova, O.A.; Dolenko, T.A. Absolute luminescence Quantum Yield for Nanosized Carbon Particles in Water as a Function of Excitation Wavelength. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2020, 229, 117879. Available online: https://www.sciencedirect.com/science/article/abs/pii/S1386142519312697 (accessed on 15 March 2020). [CrossRef]
- Zhiltsova, A.A.; Kharcheva, A.V.; Krasnova, E.D.; Lunina, O.N.; Voronov, D.A.; Savvichev, A.S.; Gorshkova, O.M.; Patsaeva, S.V. Spectroscopic study of green sulfur bacteria in stratified water bodies of the Kandalaksha Gulf of the White Sea. Atmos. Ocean. Opt. 2018, 31, 390–396. [Google Scholar] [CrossRef]
- Assawajaruwan, S.; Reinalter, J.; Hitzmann, B. Comparison of methods for wavelength combination selection from multi-wavelength fluorescence spectra for online monitoring of yeast cultivations. Anal. Bioanal. Chem. 2017, 409, 707–717. [Google Scholar] [CrossRef] [PubMed]
- Nakar, A.; Schmilovitch, Z.; Vaizel-Ohayon, D.; Kroupitski, Y.; Borisover, M.; Sela, S.S. Quantification of bacteria in water using PLS analysis of emission spectra of fluorescence and excitation-emission matrices. Water Res. 2019, 1, 115197. [Google Scholar] [CrossRef] [PubMed]
- Toledo, A.V.; Ernesto Franco, M.E.; Yanil Lopez, S.M.; MaríIné, T.; Nazareno Saparrat, M.C.; Balatti, P.A. Melanins in fungi: Types, localization and putative biological roles. Physiol. Mol. Plant Pathol. 2017, 99, 2–6. [Google Scholar] [CrossRef]
- Milyukov, A.S.; Patsaeva, S.V.; Yuzhakov, V.I.; Gorshkova, O.M.; Prashchikina, E.M. Fluorescence of nanoparticles of organic matter dissolved in natural water. Mosc. Univ. Phys. Bull. 2007, 62, 368–372. [Google Scholar] [CrossRef]
- Mao, Y.; Chena, X.-W.; Chen, Z.; Chena, G.-Q.; Lua, Y.; Wu, Y.-H.; Hu, H.-Y. Characterization of bacterial fluorescence: Insight into rapid detection of bacteria in water. Water Reuse 2021, 11, 621–631. [Google Scholar] [CrossRef]
- Zhang, Q.-Q.; Tian, B.-H.; Zhang, X.; Ghulam, A.; Fang, C.-R.; Heet, R. Investigation on characteristics of leachate and concentrated leachate in three landfill leachate treatment plants. Waste Manag. 2013, 33, 2277–2286. [Google Scholar] [CrossRef]
- Zegzouti, Y.; Boutafda, A.; Ezzariai, A.; El Fels, L.; El Hadek, M.; Hassani, L.A.I.; Hafidi, M. Bioremediation of landfill leachate by Aspergillus flavus in submerged culture: Evaluation of the process efficiency by physicochemical methods and 3D fluorescence spectroscopy. J. Environ. Manag. 2020, 255, 109821. [Google Scholar] [CrossRef]
- Muller, M.; Jimenez, J.; Antonini, M.; Dudal, Y.; Latrille, E.; Vedrenne, F.; Steyer, J.-P.; Patureau, D. Combining chemical sequential extractions with 3D fluorescence spectroscopy to characterize sludge organic matter. Waste Manag. 2014, 34, 2572–2580. [Google Scholar] [CrossRef]
- Jimenez, J.; Aemig, Q.; Doussiet, N.; Steyer, J.-P.; Houot, S.; Patureau, D. A new organic matter fractionation methodology for organic wastes: Bioaccessibility and complexity characterization for treatment optimization. Bioresour. Technol. 2015, 194, 344–353. [Google Scholar] [CrossRef]
- Bao, J.; Li, X.-X.; Zhu, K.; He, F.; Wang, Y.-Y.; Yu, J.-H.; Zhang, X.; Zhang, H. Bioactive aromatic butenolides from a mangrove sediment originated fungal species, Aspergillus terreus SCAU011. Fitoterapia 2021, 150, 104856. [Google Scholar] [CrossRef] [PubMed]
- Shubina, D.; Fedoseeva, E.; Gorshkova, O.; Patsaeva, S.; Terekhova, V.; Timofeev, M.; Yuzhakov, V. The «Blue Shift» of Emission Maximum and the Fluorescence Quantum Yield as Quantitative Spectral Characteristics of Dissolved Humic Substances. EARSeL Eproc. 2010, 9, 13–21. Available online: http://www.eproceedings.org/static/vol09_1/09_1_shubina1.pdf (accessed on 1 January 2010).
- Herbrich, S.; Gehder, M.; Krull, R.; Gericke, K.H. Label-free spatial analysis of free and enzyme-bound NAD(P)H in the presence of high concentrations of melanin. J. Fluoresc. 2012, 22, 349–355. [Google Scholar] [CrossRef] [PubMed]
- Knaus, H.; Blab, G.A.; Van Veluw, G.J.; Gerritsen, H.C.; Wösten, H.A.B. Label-free fluorescence microscopy in fungi. Fungal Biol. Rev. 2013, 27, 60–66. [Google Scholar] [CrossRef]
- Saari, S.E.; Putkiranta, M.J.; Keskinen, J. Fluorescence spectroscopy of atmospherically relevant bacterial and fungal spores and potential interferences. Atmos. Environ. 2013, 71, 202–209. [Google Scholar] [CrossRef]
- Llorente, C.; Bárcena, A.; Bahima, J.V.; Saparrat, M.C.N.; Arambarri, A.M.; Rozas, M.F.; Mirífico, M.V.; Balatti, P.A. Cladosporium cladosporioides LPSC 1088 produces the 1,8-dihydroxynaphthalene-melanin-like compound and carries a putative pks gene. Mycopathologia 2012, 174, 397–408. [Google Scholar] [CrossRef]
- Xu, R.-Z.; Cao, J.-S.; Feng, G.; Luo, J.-Y.; Feng, Q.; Ni, B.-J.; Fang, F. Fast identification of fluorescent components in three-dimensional excitation emission matrix fluorescence spectra via deep learning. Chem. Eng. J. 2022, 430, 132893. [Google Scholar] [CrossRef]
- Fedoseeva, E.V.; Patsaeva, S.V.; Khundzhua, D.A.; Pukalchik, M.A.; Terekhova, V.A. Effect of exogenic humic substances on various growth endpoints of Alternaria alternata and Trichoderma harzianum in the experimental conditions. Waste Biomass Valoriz. 2021, 12, 211–222. [Google Scholar] [CrossRef]
- Perna, G.; Palazzo, G.; Mallardi, A.; Capozzi, V. Fluorescence properties of natural eumelanin biopolymer. J. Lumin. 2011, 131, 1584–1588. [Google Scholar] [CrossRef]
- Pavan, M.E.; López, N.I.; Pettinari, M.J. Melanin biosynthesis in bacteria, regulation and production perspectives. Appl. Microbiol. Biotechnol. 2019, 104, 1357–1370. [Google Scholar] [CrossRef]
- Nowicki, S.; Lapworth, D.J.; Ward, J.S.T.; Thomson, P.; Charles, K. Tryptophan-like fluorescence as a measure of microbial contamination risk in groundwater. Sci. Total Environ. 2019, 646, 782–791. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fedoseeva, E.; Patsaeva, S.; Stom, D.; Terekhova, V. Excitation-Dependent Fluorescence Helps to Indicate Fungal Contamination of Aquatic Environments and to Differentiate Filamentous Fungi. Photonics 2022, 9, 692. https://doi.org/10.3390/photonics9100692
Fedoseeva E, Patsaeva S, Stom D, Terekhova V. Excitation-Dependent Fluorescence Helps to Indicate Fungal Contamination of Aquatic Environments and to Differentiate Filamentous Fungi. Photonics. 2022; 9(10):692. https://doi.org/10.3390/photonics9100692
Chicago/Turabian StyleFedoseeva, Elena, Svetlana Patsaeva, Devard Stom, and Vera Terekhova. 2022. "Excitation-Dependent Fluorescence Helps to Indicate Fungal Contamination of Aquatic Environments and to Differentiate Filamentous Fungi" Photonics 9, no. 10: 692. https://doi.org/10.3390/photonics9100692
APA StyleFedoseeva, E., Patsaeva, S., Stom, D., & Terekhova, V. (2022). Excitation-Dependent Fluorescence Helps to Indicate Fungal Contamination of Aquatic Environments and to Differentiate Filamentous Fungi. Photonics, 9(10), 692. https://doi.org/10.3390/photonics9100692




