Probing G-Quadruplexes Conformational Dynamics and Nano-Mechanical Interactions at the Single Molecule Level: Techniques and Perspectives
Abstract
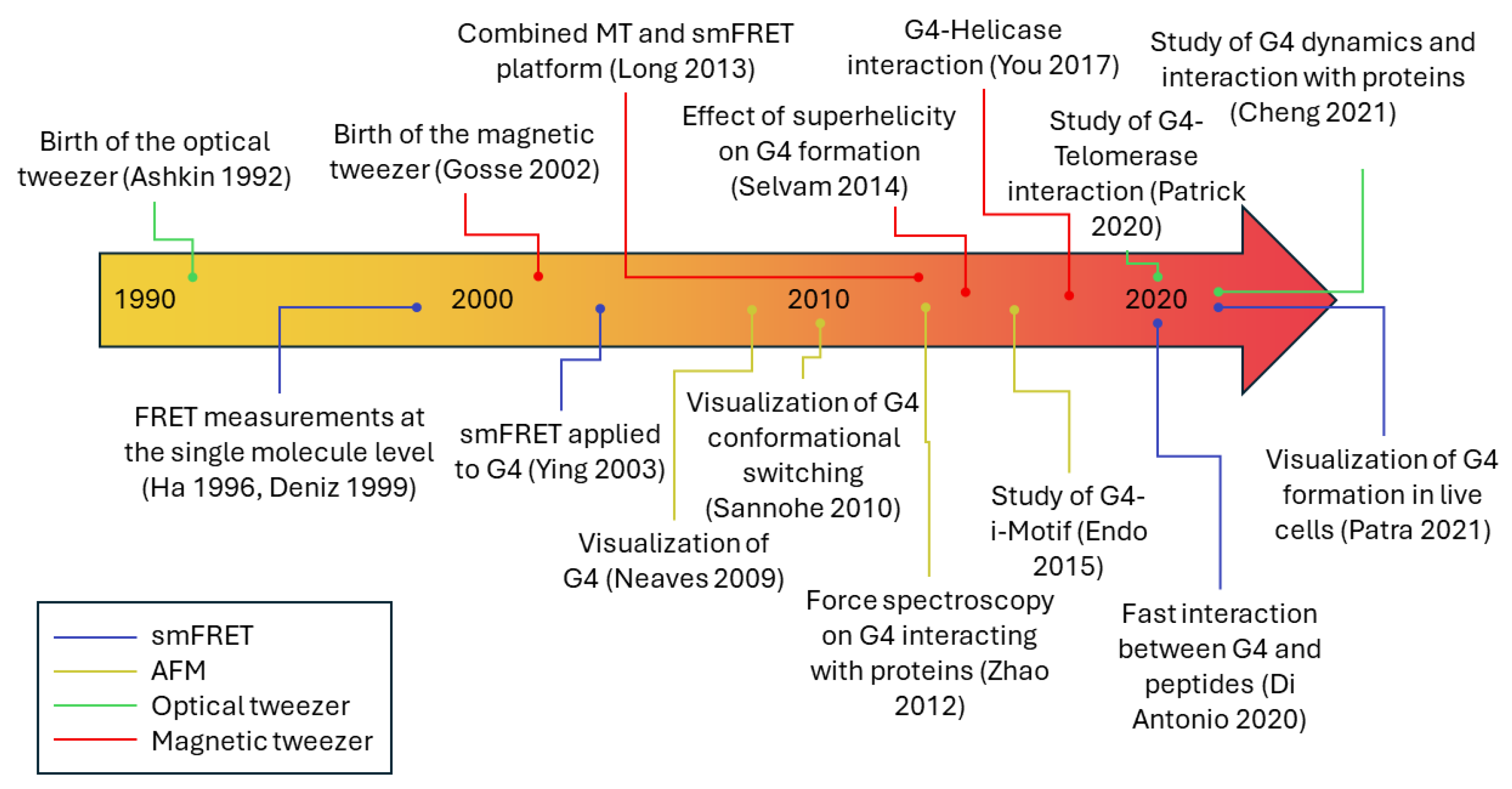
1. Introduction
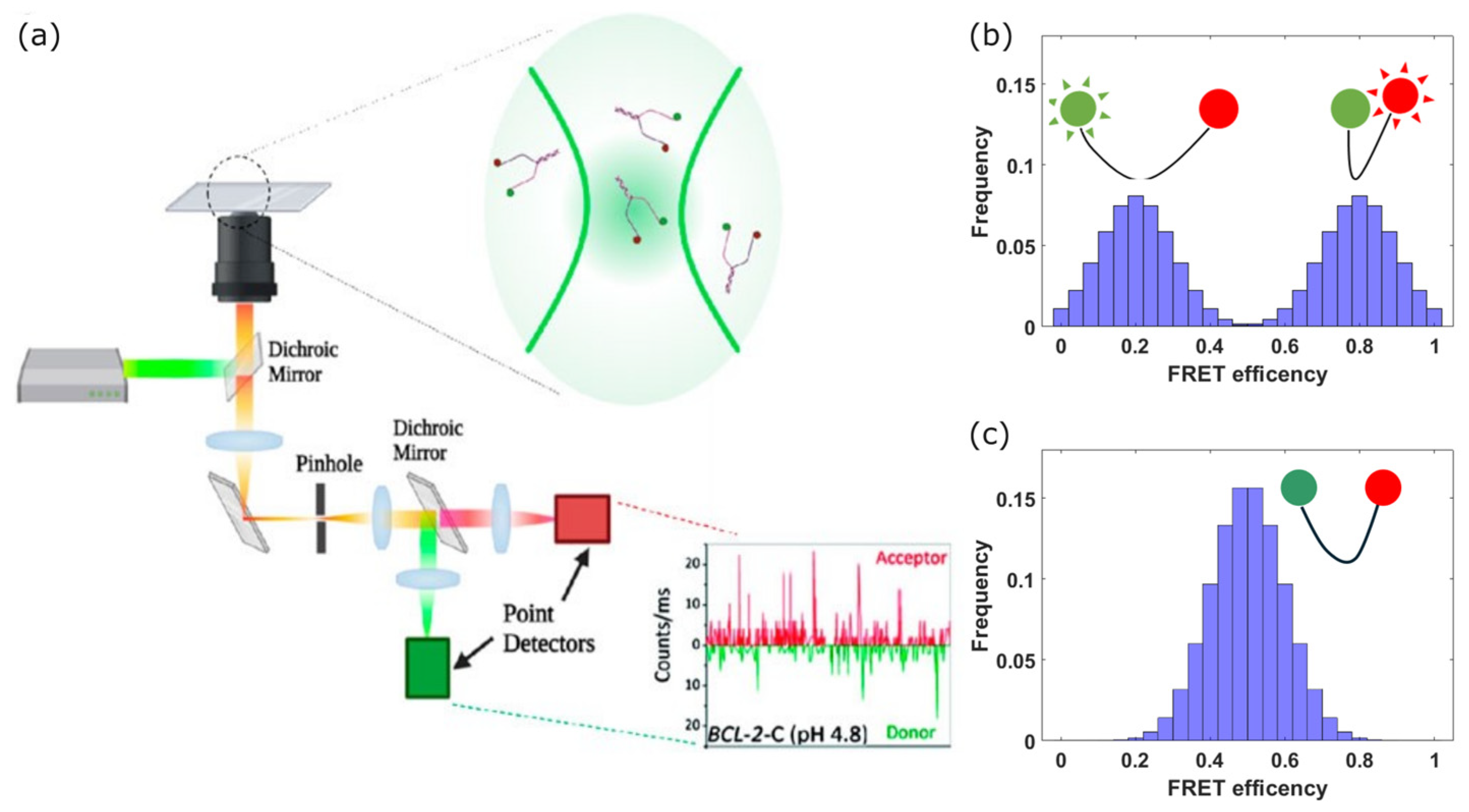
2. Conformational Studies — Single-Molecule Fluorescence Resonance Energy Transfer
3. Probing the G4 Mechanical Stability and Mimicking Traction and Torsion Stresses Exerted on G4 by Proteins in Physiological Environment
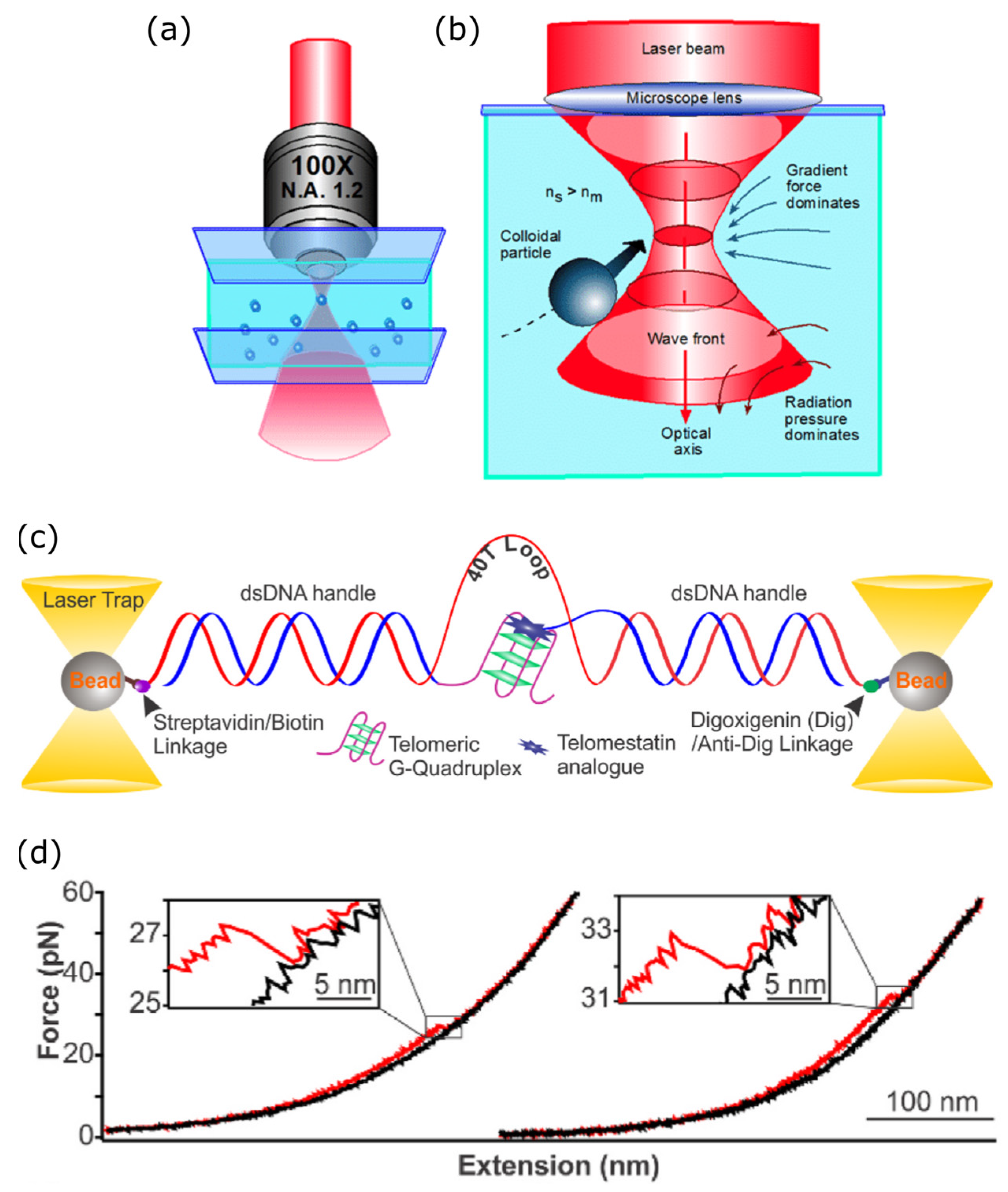
3.1. Optical Tweezers
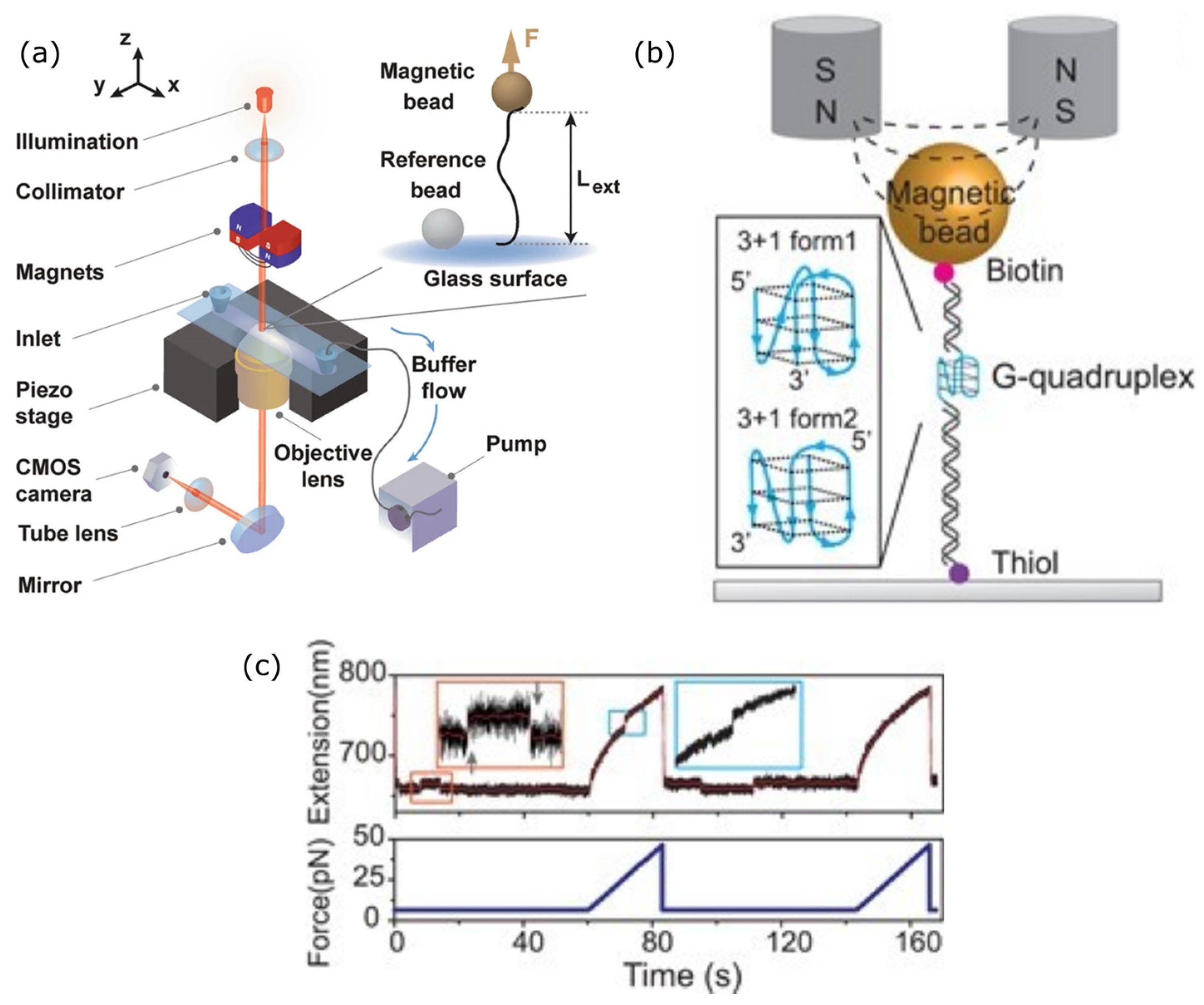
3.2. Magnetic Tweezers
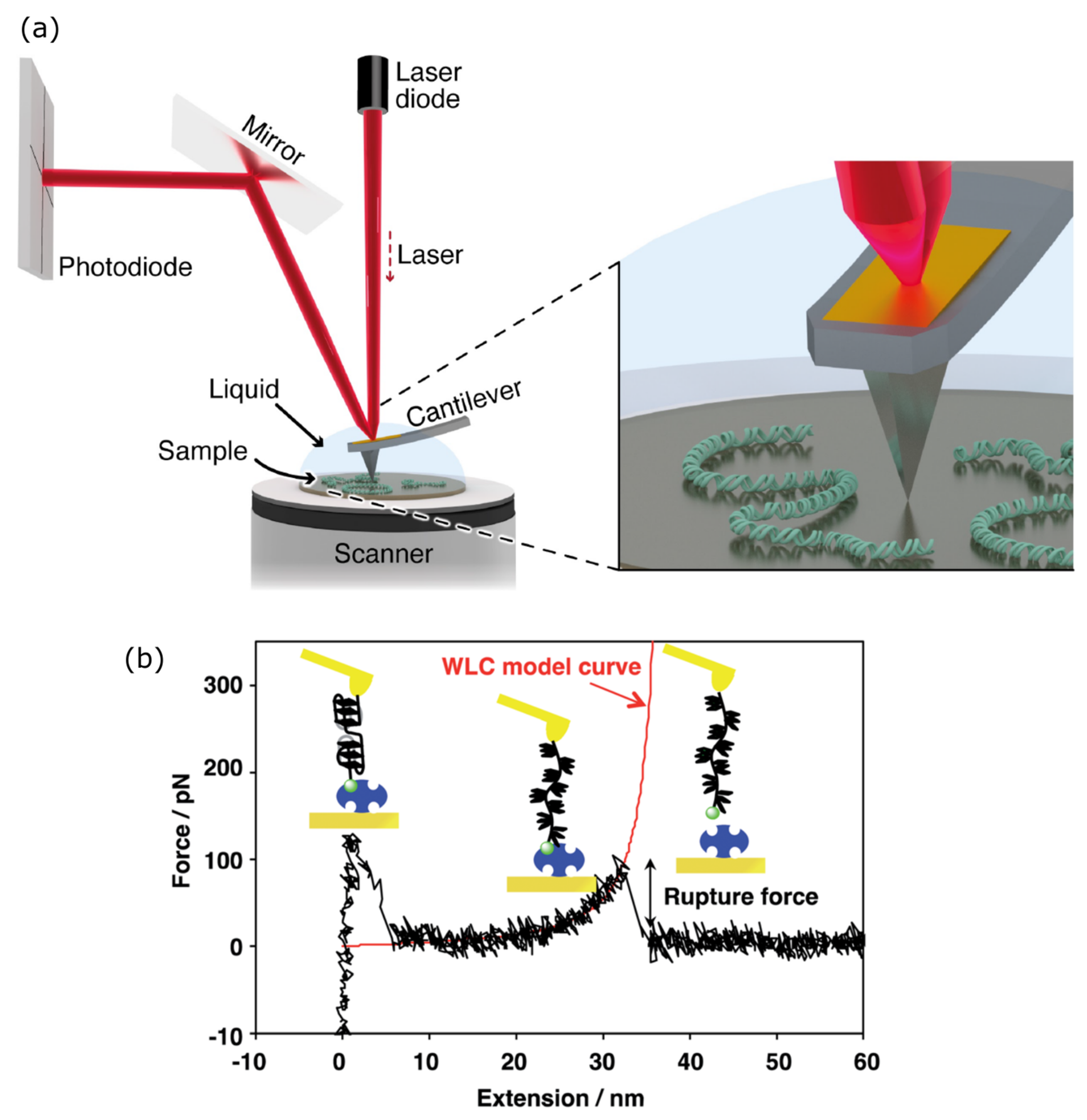
3.3. Atomic Force Microscopy
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Figueiredo, J.; Mergny, J.-L.; Cruz, C. G-Quadruplex Ligands in Cancer Therapy: Progress, Challenges, and Clinical Perspectives. Life Sci. 2024, 340, 122481. [Google Scholar] [CrossRef]
- Liano, D.; Monti, L.; Chowdhury, S.; Raguseo, F.; Antonio, M.D. Long-Range DNA Interactions: Inter-Molecular G-Quadruplexes and Their Potential Biological Relevance. Chem. Commun. 2022, 58, 12753–12762. [Google Scholar] [CrossRef] [PubMed]
- Varshney, D.; Spiegel, J.; Zyner, K.; Tannahill, D.; Balasubramanian, S. The Regulation and Functions of DNA and RNA G-Quadruplexes. Nat. Rev. Mol. Cell Biol. 2020, 21, 459–474. [Google Scholar] [CrossRef]
- Wu, S.; Jiang, L.; Lei, L.; Fu, C.; Huang, J.; Hu, Y.; Dong, Y.; Chen, J.; Zeng, Q. Crosstalk between G-Quadruplex and ROS. Cell Death Dis. 2023, 14, 37. [Google Scholar] [CrossRef] [PubMed]
- Sato, K.; Knipscheer, P. G-Quadruplex Resolution: From Molecular Mechanisms to Physiological Relevance. DNA Repair 2023, 130, 103552. [Google Scholar] [CrossRef]
- Linke, R.; Limmer, M.; Juranek, S.A.; Heine, A.; Paeschke, K. The Relevance of G-Quadruplexes for DNA Repair. Int. J. Mol. Sci. 2021, 22, 12599. [Google Scholar] [CrossRef] [PubMed]
- Gorini, F.; Ambrosio, S.; Lania, L.; Majello, B.; Amente, S. The Intertwined Role of 8-oxodG and G4 in Transcription Regulation. Int. J. Mol. Sci. 2023, 24, 2031. [Google Scholar] [CrossRef]
- Huppert, J.L. Four-Stranded Nucleic Acids: Structure, Function and Targeting of G-Quadruplexes. Chem. Soc. Rev. 2008, 37, 1375–1384. [Google Scholar] [CrossRef]
- Lightfoot, H.L.; Hagen, T.; Tatum, N.J.; Hall, J. The Diverse Structural Landscape of Quadruplexes. FEBS Lett. 2019, 593, 2083–2102. [Google Scholar] [CrossRef]
- Burge, S.; Parkinson, G.N.; Hazel, P.; Todd, A.K.; Neidle, S. Quadruplex DNA: Sequence, Topology and Structure. Nucleic Acids Res. 2006, 34, 5402–5415. [Google Scholar] [CrossRef]
- Dettler, J.M.; Buscaglia, R.; Le, V.H.; Lewis, E.A. DSC Deconvolution of the Structural Complexity of C-MYC P1 Promoter G-Quadruplexes. Biophys. J. 2011, 100, 1517–1525. [Google Scholar] [CrossRef] [PubMed]
- Harkness, R.W.; Mittermaier, A.K. G-Quadruplex Dynamics. Biochim. Biophys. Acta (BBA)—Proteins Proteom. 2017, 1865, 1544–1554. [Google Scholar] [CrossRef] [PubMed]
- Dai, J.; Carver, M.; Punchihewa, C.; Jones, R.A.; Yang, D. Structure of the Hybrid-2 Type Intramolecular Human Telomeric G-Quadruplex in K+ Solution: Insights into Structure Polymorphism of the Human Telomeric Sequence. Nucleic Acids Res. 2007, 35, 4927–4940. [Google Scholar] [CrossRef] [PubMed]
- Noer, S.L.; Preus, S.; Gudnason, D.; Aznauryan, M.; Mergny, J.-L.; Birkedal, V. Folding Dynamics and Conformational Heterogeneity of Human Telomeric G-Quadruplex Structures in Na+ Solutions by Single Molecule FRET Microscopy. Nucleic Acids Res. 2016, 44, 464–471. [Google Scholar] [CrossRef]
- Troisi, R.; Sica, F. Structural Overview of DNA and RNA G-Quadruplexes in Their Interaction with Proteins. Curr. Opin. Struct. Biol. 2024, 87, 102846. [Google Scholar] [CrossRef]
- Sahoo, B.R.; Kocman, V.; Clark, N.; Myers, N.; Deng, X.; Wong, E.L.; Yang, H.J.; Kotar, A.; Guzman, B.B.; Dominguez, D.; et al. Protein G-Quadruplex Interactions and Their Effects on Phase Transitions and Protein Aggregation. Nucleic Acids Res. 2024, 52, 4702–4722. [Google Scholar] [CrossRef]
- Lu, Z.; Xie, S.; Su, H.; Han, S.; Huang, H.; Zhou, X. Identification of G-Quadruplex-Interacting Proteins in Living Cells Using an Artificial G4-Targeting Biotin Ligase. Nucleic Acids Res. 2024, 52, e37. [Google Scholar] [CrossRef]
- Sengupta, A.; Ganguly, A.; Chowdhury, S. Promise of G-Quadruplex Structure Binding Ligands as Epigenetic Modifiers with Anti-Cancer Effects. Molecules 2019, 24, 582. [Google Scholar] [CrossRef]
- Che, T.; Wang, Y.-Q.; Huang, Z.-L.; Tan, J.-H.; Huang, Z.-S.; Chen, S.-B. Natural Alkaloids and Heterocycles as G-Quadruplex Ligands and Potential Anticancer Agents. Molecules 2018, 23, 493. [Google Scholar] [CrossRef]
- Ruggiero, E.; Richter, S.N. G-Quadruplexes and G-Quadruplex Ligands: Targets and Tools in Antiviral Therapy. Nucleic Acids Res. 2018, 46, 3270–3283. [Google Scholar] [CrossRef]
- Asamitsu, S.; Obata, S.; Yu, Z.; Bando, T.; Sugiyama, H. Recent Progress of Targeted G-Quadruplex-Preferred Ligands Toward Cancer Therapy. Molecules 2019, 24, 429. [Google Scholar] [CrossRef] [PubMed]
- Balasubramanian, S.; Hurley, L.H.; Neidle, S. Targeting G-quadruplexes in gene promoters: A novel anticancer strategy? Nat. Rev. Drug Discov. 2011, 10, 261–275. [Google Scholar] [CrossRef] [PubMed]
- Debbarma, S.; Acharya, P.C. Targeting G-Quadruplex DNA for Cancer Chemotherapy. Curr. Drug Discov. Technol. 2022, 19, 13–25. [Google Scholar] [CrossRef]
- Iachettini, S.; Biroccio, A.; Zizza, P. Therapeutic Use of G4-Ligands in Cancer: State-of-the-Art and Future Perspectives. Pharmaceuticals 2024, 17, 771. [Google Scholar] [CrossRef] [PubMed]
- Neidle, S. A Phenotypic Approach to the Discovery of Potent G-Quadruplex Targeted Drugs. Molecules 2024, 29, 3653. [Google Scholar] [CrossRef]
- Shu, H.; Zhang, R.; Xiao, K.; Yang, J.; Sun, X. G-Quadruplex-Binding Proteins: Promising Targets for Drug Design. Biomolecules 2022, 12, 648. [Google Scholar] [CrossRef]
- Frasson, I.; Pirota, V.; Richter, S.N.; Doria, F. Multimeric G-Quadruplexes: A Review on Their Biological Roles and Targeting. Int. J. Biol. Macromol. 2022, 204, 89–102. [Google Scholar] [CrossRef]
- Choudhury, S.D.; Kumar, P.; Choudhury, D. Bioactive Nutraceuticals as G4 Stabilizers: Potential Cancer Prevention and Therapy—A Critical Review. Naunyn Schmiedebergs Arch. Pharmacol. 2024, 397, 3585–3616. [Google Scholar] [CrossRef]
- Förster, T. Experimentelle Und Theoretische Untersuchung Des Zwischenmolekularen Übergangs von Elektronenanregungsenergie. Z. Naturforschung A 1949, 4, 321–327. [Google Scholar] [CrossRef]
- Stryer, L. Fluorescence Energy Transfer as a Spectroscopic Ruler. Annu. Rev. Biochem. 1978, 47, 819–846. [Google Scholar] [CrossRef]
- Bandyopadhyay, D.; Mishra, P.P. Decoding the Structural Dynamics and Conformational Alternations of DNA Secondary Structures by Single-Molecule FRET Microspectroscopy. Front. Mol. Biosci. 2021, 8, 725541. [Google Scholar] [CrossRef] [PubMed]
- Clegg, R.M. Fluorescence Resonance Energy Transfer. Curr. Opin. Biotechnol. 1995, 6, 103–110. [Google Scholar] [CrossRef] [PubMed]
- Selvin, P.R. The Renaissance of Fluorescence Resonance Energy Transfer. Nat. Struct. Mol. Biol. 2000, 7, 730–734. [Google Scholar] [CrossRef] [PubMed]
- Weiss, S. Fluorescence Spectroscopy of Single Biomolecules. Science 1999, 283, 1676–1683. [Google Scholar] [CrossRef]
- Ha, T.; Enderle, T.; Ogletree, D.F.; Chemla, D.S.; Selvin, P.R.; Weiss, S. Probing the Interaction between Two Single Molecules: Fluorescence Resonance Energy Transfer between a Single Donor and a Single Acceptor. Proc. Natl. Acad. Sci. USA 1996, 93, 6264–6268. [Google Scholar] [CrossRef]
- Deniz, A.A.; Dahan, M.; Grunwell, J.R.; Ha, T.; Faulhaber, A.E.; Chemla, D.S.; Weiss, S.; Schultz, P.G. Single-Pair Fluorescence Resonance Energy Transfer on Freely Diffusing Molecules: Observation of Förster Distance Dependence and Subpopulations. Proc. Natl. Acad. Sci. USA 1999, 96, 3670–3675. [Google Scholar] [CrossRef]
- Deniz, A.A.; Laurence, T.A.; Dahan, M.; Chemla, D.S.; Schultz, P.G.; Weiss, S. Ratiometric Single-Molecule Studies of Freely Diffusing Biomolecules. Annu. Rev. Phys. Chem. 2001, 52, 233–253. [Google Scholar] [CrossRef]
- Murphy, M.C.; Rasnik, I.; Cheng, W.; Lohman, T.M.; Ha, T. Probing Single-Stranded DNA Conformational Flexibility Using Fluorescence Spectroscopy. Biophys. J. 2004, 86, 2530–2537. [Google Scholar] [CrossRef]
- Schuler, B.; Lipman, E.A.; Eaton, W.A. Probing the Free-Energy Surface for Protein Folding with Single-Molecule Fluorescence Spectroscopy. Nature 2002, 419, 743–747. [Google Scholar] [CrossRef]
- Nettels, D.; Gopich, I.V.; Hoffmann, A.; Schuler, B. Ultrafast Dynamics of Protein Collapse from Single-Molecule Photon Statistics. Proc. Natl. Acad. Sci. USA 2007, 104, 2655–2660. [Google Scholar] [CrossRef]
- Schuler, B.; Eaton, W.A. Protein Folding Studied by Single-Molecule FRET. Curr. Opin. Struct. Biol. 2008, 18, 16–26. [Google Scholar] [CrossRef] [PubMed]
- Borgia, A.; Borgia, M.B.; Bugge, K.; Kissling, V.M.; Heidarsson, P.O.; Fernandes, C.B.; Sottini, A.; Soranno, A.; Buholzer, K.J.; Nettels, D.; et al. Extreme Disorder in an Ultrahigh-Affinity Protein Complex. Nature 2018, 555, 61–66. [Google Scholar] [CrossRef] [PubMed]
- Nüesch, M.F.; Ivanović, M.T.; Claude, J.-B.; Nettels, D.; Best, R.B.; Wenger, J.; Schuler, B. Single-Molecule Detection of Ultrafast Biomolecular Dynamics with Nanophotonics. J. Am. Chem. Soc. 2022, 144, 52–56. [Google Scholar] [CrossRef] [PubMed]
- Phelps, C.; Lee, W.; Jose, D.; Von Hippel, P.H.; Marcus, A.H. Single-Molecule FRET and Linear Dichroism Studies of DNA Breathing and Helicase Binding at Replication Fork Junctions. Proc. Natl. Acad. Sci. USA 2013, 110, 17320–17325. [Google Scholar] [CrossRef]
- Šponer, J.; Islam, B.; Stadlbauer, P.; Haider, S. Chapter Seven—Molecular Dynamics Simulations of G-Quadruplexes: The Basic Principles and Their Application to Folding and Ligand Binding. In Annual Reports in Medicinal Chemistry; Neidle, S., Ed.; Quadruplex Nucleic Acids as Targets For Medicinal Chemistry; Academic Press: Cambridge, MA, USA, 2020; Volume 54, pp. 197–241. [Google Scholar]
- Roy, S.; Ali, A.; Bhattacharya, S. Theoretical Insight into the Library Screening Approach for Binding of Intermolecular G-Quadruplex RNA and Small Molecules through Docking and Molecular Dynamics Simulation Studies. J. Phys. Chem. B 2021, 125, 5489–5501. [Google Scholar] [CrossRef]
- Lemkul, J.A. Same Fold, Different Properties: Polarizable Molecular Dynamics Simulations of Telomeric and TERRA G-Quadruplexes. Nucleic Acids Res. 2020, 48, 561–575. [Google Scholar] [CrossRef]
- Jurkowski, M.; Kogut, M.; Czub, J. Stability Differences between Right- and Left-Handed G-Quadruplexes—Explained by Molecular Dynamics Simulations. Biophys. J. 2024, 123, 80a. [Google Scholar] [CrossRef]
- Ying, L.; Green, J.J.; Li, H.; Klenerman, D.; Balasubramanian, S. Studies on the Structure and Dynamics of the Human Telomeric G Quadruplex by Single-Molecule Fluorescence Resonance Energy Transfer. Proc. Natl. Acad. Sci. USA 2003, 100, 14629–14634. [Google Scholar] [CrossRef]
- Lee, J.Y.; Okumus, B.; Kim, D.S.; Ha, T. Extreme Conformational Diversity in Human Telomeric DNA. Proc. Natl. Acad. Sci. USA 2005, 102, 18938–18943. [Google Scholar] [CrossRef]
- Patra, S.; Claude, J.-B.; Naubron, J.-V.; Wenger, J. Fast Interaction Dynamics of G-Quadruplex and RGG-Rich Peptides Unveiled in Zero-Mode Waveguides. Nucleic Acids Res. 2021, 49, 12348–12357. [Google Scholar] [CrossRef]
- Paudel, B.P.; Moye, A.L.; Abou Assi, H.; El-Khoury, R.; Cohen, S.B.; Holien, J.K.; Birrento, M.L.; Samosorn, S.; Intharapichai, K.; Tomlinson, C.G.; et al. A Mechanism for the Extension and Unfolding of Parallel Telomeric G-Quadruplexes by Human Telomerase at Single-Molecule Resolution. eLife 2020, 9, e56428. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.-T.; Sanford, S.; Paul, T.; Choe, J.; Bose, A.; Opresko, P.L.; Myong, S. Position-Dependent Effect of Guanine Base Damage and Mutations on Telomeric G-Quadruplex and Telomerase Extension. Biochemistry 2020, 59, 2627–2639. [Google Scholar] [CrossRef] [PubMed]
- Ray, S.; Bandaria, J.N.; Qureshi, M.H.; Yildiz, A.; Balci, H. G-Quadruplex Formation in Telomeres Enhances POT1/TPP1 Protection against RPA Binding. Proc. Natl. Acad. Sci. USA 2014, 111, 2990–2995. [Google Scholar] [CrossRef]
- Lee, J.Y.; Kim, D.S. Dramatic Effect of Single-Base Mutation on the Conformational Dynamics of Human Telomeric G-Quadruplex. Nucleic Acids Res. 2009, 37, 3625–3634. [Google Scholar] [CrossRef]
- Jena, P.V.; Shirude, P.S.; Okumus, B.; Laxmi-Reddy, K.; Godde, F.; Huc, I.; Balasubramanian, S.; Ha, T. G-Quadruplex DNA Bound by a Synthetic Ligand Is Highly Dynamic. J. Am. Chem. Soc. 2009, 131, 12522–12523. [Google Scholar] [CrossRef]
- Okamoto, K.; Sannohe, Y.; Mashimo, T.; Sugiyama, H.; Terazima, M. G-Quadruplex Structures of Human Telomere DNA Examined by Single Molecule FRET and BrG-Substitution. Bioorganic Med. Chem. 2008, 16, 6873–6879. [Google Scholar] [CrossRef] [PubMed]
- Long, X.; Stone, M.D. Kinetic Partitioning Modulates Human Telomere DNA G-Quadruplex Structural Polymorphism. PLoS ONE 2013, 8, e83420. [Google Scholar] [CrossRef]
- Shirude, P.S.; Okumus, B.; Ying, L.; Ha, T.; Balasubramanian, S. Single-Molecule Conformational Analysis of G-Quadruplex Formation in the Promoter DNA Duplex of the Proto-Oncogene C-Kit. J. Am. Chem. Soc. 2007, 129, 7484–7485. [Google Scholar] [CrossRef]
- Fegan, A.; Shirude, P.S.; Ying, L.; Balasubramanian, S. Ensemble and Single Molecule FRET Analysis of the Structure and Unfolding Kinetics of the C-Kit Promoter Quadruplexes. Chem. Commun. 2010, 46, 946–948. [Google Scholar] [CrossRef]
- Tippana, R.; Xiao, W.; Myong, S. G-Quadruplex Conformation and Dynamics Are Determined by Loop Length and Sequence. Nucleic Acids Res. 2014, 42, 8106–8114. [Google Scholar] [CrossRef]
- Tippana, R.; Hwang, H.; Opresko, P.L.; Bohr, V.A.; Myong, S. Single-Molecule Imaging Reveals a Common Mechanism Shared by G-Quadruplex–Resolving Helicases. Proc. Natl. Acad. Sci. USA 2016, 113, 8448–8453. [Google Scholar] [CrossRef] [PubMed]
- Vesco, G.; Lualdi, M.; Fasano, M.; Nardo, L.; Alberio, T. Demonstration of Fibrinogen-FcRn Binding at Acidic pH by Means of Fluorescence Correlation Spectroscopy. Biochem. Biophys. Res. Commun. 2021, 536, 32–37. [Google Scholar] [CrossRef] [PubMed]
- Shirude, P.S.; Balasubramanian, S. Single Molecule Conformational Analysis of DNA G-Quadruplexes. Biochimie 2008, 90, 1197–1206. [Google Scholar] [CrossRef] [PubMed]
- Ray, S.; Qureshi, M.H.; Malcolm, D.W.; Budhathoki, J.B.; Çelik, U.; Balci, H. RPA-Mediated Unfolding of Systematically Varying G-Quadruplex Structures. Biophys. J. 2013, 104, 2235–2245. [Google Scholar] [CrossRef]
- Roy, R.; Hohng, S.; Ha, T. A Practical Guide to Single-Molecule FRET. Nat. Methods 2008, 5, 507–516. [Google Scholar] [CrossRef]
- Joo, C.; Ha, T. Single-Molecule FRET with Total Internal Reflection Microscopy. Cold Spring Harb. Protoc. 2012, 2012, pdb.top072058. [Google Scholar] [CrossRef]
- Ha, T. Single-Molecule Fluorescence Resonance Energy Transfer. Methods 2001, 25, 78–86. [Google Scholar] [CrossRef]
- Ha, T. Single-Molecule Fluorescence Methods for the Study of Nucleic Acids. Curr. Opin. Struct. Biol. 2001, 11, 287–292. [Google Scholar] [CrossRef]
- Wang, Q.; Goldsmith, R.H.; Jiang, Y.; Bockenhauer, S.D.; Moerner, W.E. Probing Single Biomolecules in Solution Using the Anti-Brownian Electrokinetic (ABEL) Trap. Acc. Chem. Res. 2012, 45, 1955–1964. [Google Scholar] [CrossRef]
- Kim, J.; Doose, S.; Neuweiler, H.; Sauer, M. The Initial Step of DNA Hairpin Folding: A Kinetic Analysis Using Fluorescence Correlation Spectroscopy. Nucleic Acids Res. 2006, 34, 2516–2527. [Google Scholar] [CrossRef]
- Nardo, L.; Lamperti, M.; Salerno, D.; Cassina, V.; Missana, N.; Bondani, M.; Tempestini, A.; Mantegazza, F. Effects of Non-CpG Site Methylation on DNA Thermal Stability: A Fluorescence Study. Nucleic Acids Res. 2015, 43, 10722–10733. [Google Scholar] [CrossRef] [PubMed]
- Nag, S.; Chen, J.; Irudayaraj, J.; Maiti, S. Measurement of the Attachment and Assembly of Small Amyloid-β Oligomers on Live Cell Membranes at Physiological Concentrations Using Single-Molecule Tools. Biophys. J. 2010, 99, 1969–1975. [Google Scholar] [CrossRef]
- Nardo, L.; Re, F.; Brioschi, S.; Cazzaniga, E.; Orlando, A.; Minniti, S.; Lamperti, M.; Gregori, M.; Cassina, V.; Brogioli, D.; et al. Fluorimetric Detection of the Earliest Events in Amyloid β Oligomerization and Its Inhibition by Pharmacologically Active Liposomes. Biochim. Biophys. Acta (BBA)—Gen. Subj. 2016, 1860, 746–756. [Google Scholar] [CrossRef] [PubMed]
- Strömqvist, J.; Nardo, L.; Broekmans, O.; Kohn, J.; Lamperti, M.; Santamato, A.; Shalaby, M.; Sharma, G.; Di Trapani, P.; Bondani, M.; et al. Binding of Biotin to Streptavidin: A Combined Fluorescence Correlation Spectroscopy and Time-Resolved Fluorescence Study. Eur. Phys. J. Spec. Top. 2011, 199, 181–194. [Google Scholar] [CrossRef][Green Version]
- Rigo, R.; Dean, W.L.; Gray, R.D.; Chaires, J.B.; Sissi, C. Conformational Profiling of a G-Rich Sequence within the c-KIT Promoter. Nucleic Acids Res. 2017, 45, 13056–13067. [Google Scholar] [CrossRef]
- Vesco, G.; Lamperti, M.; Salerno, D.; Marrano, C.A.; Cassina, V.; Rigo, R.; Buglione, E.; Bondani, M.; Nicoletto, G.; Mantegazza, F.; et al. Double-Stranded Flanking Ends Affect the Folding Kinetics and Conformational Equilibrium of G-Quadruplexes Forming Sequences within the Promoter of KIT Oncogene. Nucleic Acids Res. 2021, 49, 9724–9737. [Google Scholar] [CrossRef]
- Sabanayagam, C.R.; Eid, J.S.; Meller, A. High-Throughput Scanning Confocal Microscope for Single Molecule Analysis. Appl. Phys. Lett. 2004, 84, 1216–1218. [Google Scholar] [CrossRef]
- Kapanidis, A.N.; Weiss, S. Fluorescent Probes and Bioconjugation Chemistries for Single-Molecule Fluorescence Analysis of Biomolecules. J. Chem. Phys. 2002, 117, 10953–10964. [Google Scholar] [CrossRef]
- Eggeling, C.; Widengren, J.; Rigler, R.; Seidel, C.A.M. Photobleaching of Fluorescent Dyes under Conditions Used for Single-Molecule Detection: Evidence of Two-Step Photolysis. Anal. Chem. 1998, 70, 2651–2659. [Google Scholar] [CrossRef]
- Kudalkar, E.M.; Deng, Y.; Davis, T.N.; Asbury, C.L. Coverslip Cleaning and Functionalization for Total Internal Reflection Fluorescence Microscopy. Cold Spring Harb. Protoc. 2016, 2016, pdb.prot085548. [Google Scholar] [CrossRef][Green Version]
- Di Antonio, M.; Ponjavic, A.; Radzevičius, A.; Ranasinghe, R.T.; Catalano, M.; Zhang, X.; Shen, J.; Needham, L.-M.; Lee, S.F.; Klenerman, D.; et al. Single-Molecule Visualization of DNA G-Quadruplex Formation in Live Cells. Nat. Chem. 2020, 12, 832–837. [Google Scholar] [CrossRef] [PubMed]
- Cras, J.J.; Rowe-Taitt, C.A.; Nivens, D.A.; Ligler, F.S. Comparison of Chemical Cleaning Methods of Glass in Preparation for Silanization. Biosens. Bioelectron. 1999, 14, 683–688. [Google Scholar] [CrossRef]
- Wang, L.; Gaigalas, A.K.; Reipa, V. Optical Properties of AlexaTM 488 and CyTM5 Immobilized on a Glass Surface. BioTechniques 2005, 38, 127–132. [Google Scholar] [CrossRef]
- Axelrod, D.; Hellen, E.H. Chapter 15 Emission of Fluorescence at an Interface. In Methods in Cell Biology; Taylor, D.L., Wang, Y.-L., Eds.; Fluorescence Microscopy of Living Cells in Culture Part B. Quantitative Fluorescence Microscopy—Imaging and Spectroscopy; Academic Press: Cambridge, MA, USA, 1989; Volume 30, pp. 399–416. [Google Scholar]
- Aznauryan, M.; Noer, S.L.; Pedersen, C.W.; Mergny, J.-L.; Teulade-Fichou, M.-P.; Birkedal, V. Ligand Binding to Dynamically Populated G-Quadruplex DNA. ChemBioChem 2021, 22, 1811–1817. [Google Scholar] [CrossRef]
- Xue, Z.-Y.; Wu, W.-Q.; Zhao, X.-C.; Kumar, A.; Ran, X.; Zhang, X.-H.; Zhang, Y.; Guo, L.-J. Single-Molecule Probing the Duplex and G4 Unwinding Patterns of a RecD Family Helicase. Int. J. Biol. Macromol. 2020, 164, 902–910. [Google Scholar] [CrossRef]
- Lim, G.; Hohng, S. Single-Molecule Fluorescence Studies on Cotranscriptional G-Quadruplex Formation Coupled with R-Loop Formation. Nucleic Acids Res. 2020, 48, 9195–9203. [Google Scholar] [CrossRef]
- Wu, C.G.; Spies, M. G-Quadruplex Recognition and Remodeling by the FANCJ Helicase. Nucleic Acids Res. 2016, 44, 8742–8753. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, S.; Zagelbaum, J.; Savitsky, P.; Sturzenegger, A.; Huttner, D.; Janscak, P.; Hickson, I.D.; Gileadi, O.; Rothenberg, E. Mechanistic Insight into the Interaction of BLM Helicase with Intra-Strand G-Quadruplex Structures. Nat. Commun. 2014, 5, 5556. [Google Scholar] [CrossRef]
- Qureshi, M.H.; Ray, S.; Sewell, A.L.; Basu, S.; Balci, H. Replication Protein A Unfolds G-Quadruplex Structures with Varying Degrees of Efficiency. J. Phys. Chem. B 2012, 116, 5588–5594. [Google Scholar] [CrossRef]
- Kreig, A.; Calvert, J.; Sanoica, J.; Cullum, E.; Tipanna, R.; Myong, S. G-Quadruplex Formation in Double Strand DNA Probed by NMM and CV Fluorescence. Nucleic Acids Res. 2015, 43, 7961–7970. [Google Scholar] [CrossRef]
- Chen, J.; Cheng, M.; Salgado, G.F.; Stadlbauer, P.; Zhang, X.; Amrane, S.; Guédin, A.; He, F.; Šponer, J.; Ju, H.; et al. The Beginning and the End: Flanking Nucleotides Induce a Parallel G-Quadruplex Topology. Nucleic Acids Res. 2021, 49, 9548–9559. [Google Scholar] [CrossRef] [PubMed]
- Neuman, K.C.; Nagy, A. Single-Molecule Force Spectroscopy: Optical Tweezers, Magnetic Tweezers and Atomic Force Microscopy. Nat. Methods 2008, 5, 491–505. [Google Scholar] [CrossRef] [PubMed]
- Woodside, M.T.; Block, S.M. Reconstructing Folding Energy Landscapes by Single-Molecule Force Spectroscopy. Annu. Rev. Biophys. 2014, 43, 19–39. [Google Scholar] [CrossRef]
- Pesce, G.; Jones, P.H.; Maragò, O.M.; Volpe, G. Optical Tweezers: Theory and Practice. Eur. Phys. J. Plus 2020, 135, 949. [Google Scholar] [CrossRef]
- Molloy, J.E.; Padgett, M.J. Lights, Action: Optical Tweezers. Contemp. Phys. 2002, 43, 241–258. [Google Scholar] [CrossRef]
- Ashkin, A.; Dziedzic, J.M.; Bjorkholm, J.E.; Chu, S. Observation of a Single-Beam Gradient Force Optical Trap for Dielectric Particles. Opt. Lett. OL 1986, 11, 288–290. [Google Scholar] [CrossRef]
- Ashkin, A. Forces of a Single-Beam Gradient Laser Trap on a Dielectric Sphere in the Ray Optics Regime. Biophys. J. 1992, 61, 569–582. [Google Scholar] [CrossRef] [PubMed]
- Bustamante, C.; Alexander, L.; Maciuba, K.; Kaiser, C.M. Single-Molecule Studies of Protein Folding with Optical Tweezers. Annu. Rev. Biochem. 2020, 89, 443–470. [Google Scholar] [CrossRef]
- Bustamante, C.J.; Chemla, Y.R.; Liu, S.; Wang, M.D. Optical Tweezers in Single-Molecule Biophysics. Nat. Rev. Methods Primers 2021, 1, 25. [Google Scholar] [CrossRef]
- Wang, M.D.; Yin, H.; Landick, R.; Gelles, J.; Block, S.M. Stretching DNA with Optical Tweezers. Biophys. J. 1997, 72, 1335–1346. [Google Scholar] [CrossRef]
- McCauley, M.J.; Williams, M.C. Mechanisms of DNA Binding Determined in Optical Tweezers Experiments. Biopolymers 2007, 85, 154–168. [Google Scholar] [CrossRef] [PubMed]
- Bockelmann, U.; Thomen, P.; Essevaz-Roulet, B.; Viasnoff, V.; Heslot, F. Unzipping DNA with Optical Tweezers: High Sequence Sensitivity and Force Flips. Biophys. J. 2002, 82, 1537–1553. [Google Scholar] [CrossRef] [PubMed]
- Heller, I.; Hoekstra, T.P.; King, G.A.; Peterman, E.J.G.; Wuite, G.J.L. Optical Tweezers Analysis of DNA–Protein Complexes. Chem. Rev. 2014, 114, 3087–3119. [Google Scholar] [CrossRef] [PubMed]
- King, G.A.; Spakman, D.; Peterman, E.J.G.; Wuite, G.J.L. Generating Negatively Supercoiled DNA Using Dual-Trap Optical Tweezers. In Optical Tweezers: Methods and Protocols; Gennerich, A., Ed.; Methods in Molecular Biology; Springer: New York, NY, USA, 2022; pp. 243–272. ISBN 978-1-07-162229-2. [Google Scholar]
- Choudhary, D.; Mossa, A.; Jadhav, M.; Cecconi, C. Bio-Molecular Applications of Recent Developments in Optical Tweezers. Biomolecules 2019, 9, 23. [Google Scholar] [CrossRef]
- Cheng, Y.; Zhang, Y.; You, H. Characterization of G-Quadruplexes Folding/Unfolding Dynamics and Interactions with Proteins from Single-Molecule Force Spectroscopy. Biomolecules 2021, 11, 1579. [Google Scholar] [CrossRef]
- Yu, Z.; Schonhoft, J.D.; Dhakal, S.; Bajracharya, R.; Hegde, R.; Basu, S.; Mao, H. ILPR G-Quadruplexes Formed in Seconds Demonstrate High Mechanical Stabilities. J. Am. Chem. Soc. 2009, 131, 1876–1882. [Google Scholar] [CrossRef]
- Dhakal, S.; Yu, Z.; Konik, R.; Cui, Y.; Koirala, D.; Mao, H. G-Quadruplex and i-Motif Are Mutually Exclusive in ILPR Double-Stranded DNA. Biophys. J. 2012, 102, 2575–2584. [Google Scholar] [CrossRef]
- de Messieres, M.; Chang, J.-C.; Brawn-Cinani, B.; La Porta, A. Single-Molecule Study of $G$-Quadruplex Disruption Using Dynamic Force Spectroscopy. Phys. Rev. Lett. 2012, 109, 058101. [Google Scholar] [CrossRef]
- Schonhoft, J.D.; Bajracharya, R.; Dhakal, S.; Yu, Z.; Mao, H.; Basu, S. Direct Experimental Evidence for Quadruplex–Quadruplex Interaction within the Human ILPR. Nucleic Acids Res. 2009, 37, 3310–3320. [Google Scholar] [CrossRef]
- Abraham Punnoose, J.; Ma, Y.; Li, Y.; Sakuma, M.; Mandal, S.; Nagasawa, K.; Mao, H. Adaptive and Specific Recognition of Telomeric G-Quadruplexes via Polyvalency Induced Unstacking of Binding Units. J. Am. Chem. Soc. 2017, 139, 7476–7484. [Google Scholar] [CrossRef]
- Pokhrel, P.; Sasaki, S.; Hu, C.; Karna, D.; Pandey, S.; Ma, Y.; Nagasawa, K.; Mao, H. Single-Molecule Displacement Assay Reveals Strong Binding of Polyvalent Dendrimer Ligands to Telomeric G-Quadruplex. Anal. Biochem. 2022, 649, 114693. [Google Scholar] [CrossRef] [PubMed]
- Koirala, D.; Mashimo, T.; Sannohe, Y.; Yu, Z.; Mao, H.; Sugiyama, H. Intramolecular Folding in Three Tandem Guanine Repeats of Human Telomeric DNA. Chem. Commun. 2012, 48, 2006–2008. [Google Scholar] [CrossRef] [PubMed]
- Koirala, D.; Ghimire, C.; Bohrer, C.; Sannohe, Y.; Sugiyama, H.; Mao, H. Long-Loop G-Quadruplexes Are Misfolded Population Minorities with Fast Transition Kinetics in Human Telomeric Sequences. J. Am. Chem. Soc. 2013, 135, 2235–2241. [Google Scholar] [CrossRef] [PubMed]
- Ghimire, C.; Park, S.; Iida, K.; Yangyuoru, P.; Otomo, H.; Yu, Z.; Nagasawa, K.; Sugiyama, H.; Mao, H. Direct Quantification of Loop Interaction and π–π Stacking for G-Quadruplex Stability at the Submolecular Level. J. Am. Chem. Soc. 2014, 136, 15537–15544. [Google Scholar] [CrossRef]
- Yangyuoru, P.M.; Zhang, A.Y.Q.; Shi, Z.; Koirala, D.; Balasubramanian, S.; Mao, H. Mechanochemical Properties of Individual Human Telomeric RNA (TERRA) G-Quadruplexes. ChemBioChem 2013, 14, 1931–1935. [Google Scholar] [CrossRef]
- Koirala, D.; Dhakal, S.; Ashbridge, B.; Sannohe, Y.; Rodriguez, R.; Sugiyama, H.; Balasubramanian, S.; Mao, H. A Single-Molecule Platform for Investigation of Interactions between G-Quadruplexes and Small-Molecule Ligands. Nat. Chem. 2011, 3, 782–787. [Google Scholar] [CrossRef]
- Jonchhe, S.; Ghimire, C.; Cui, Y.; Sasaki, S.; McCool, M.; Park, S.; Iida, K.; Nagasawa, K.; Sugiyama, H.; Mao, H. Binding of a Telomestatin Derivative Changes the Mechanical Anisotropy of a Human Telomeric G-Quadruplex. Angew. Chem. Int. Ed. 2019, 58, 877–881. [Google Scholar] [CrossRef]
- Mandal, S.; Hoque, M.E.; Mao, H. Single-Molecule Investigations of G-Quadruplex. In G-Quadruplex Nucleic Acids: Methods and Protocols; Yang, D., Lin, C., Eds.; Methods in Molecular Biology; Springer: New York, NY, USA, 2019; pp. 275–298. ISBN 978-1-4939-9666-7. [Google Scholar]
- Pandey, S.; Li, Y.; Young, M.D.; Mandal, S.; Lu, L.; Shelley, J.T.; Mao, H. Cooperative Heteroligand Interaction with G-Quadruplexes Shows Evidence of Allosteric Binding. Biochemistry 2020, 59, 3438–3446. [Google Scholar] [CrossRef]
- Yangyuoru, P.M.; Di Antonio, M.; Ghimire, C.; Biffi, G.; Balasubramanian, S.; Mao, H. Dual Binding of an Antibody and a Small Molecule Increases the Stability of TERRA G-Quadruplex. Angew. Chem. Int. Ed. 2015, 54, 910–913. [Google Scholar] [CrossRef]
- Yu, Z.; Gaerig, V.; Cui, Y.; Kang, H.; Gokhale, V.; Zhao, Y.; Hurley, L.H.; Mao, H. Tertiary DNA Structure in the Single-Stranded hTERT Promoter Fragment Unfolds and Refolds by Parallel Pathways via Cooperative or Sequential Events. J. Am. Chem. Soc. 2012, 134, 5157–5164. [Google Scholar] [CrossRef]
- Selvam, S.; Yu, Z.; Mao, H. Exploded View of Higher Order G-Quadruplex Structures through Click-Chemistry Assisted Single-Molecule Mechanical Unfolding. Nucleic Acids Res. 2016, 44, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, P.; Jonchhe, S.; Emura, T.; Hidaka, K.; Endo, M.; Sugiyama, H.; Mao, H. Confined Space Facilitates G-Quadruplex Formation. Nat. Nanotech. 2017, 12, 582–588. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez, I.; Garavís, M.; de Lorenzo, S.; Villasante, A.; González, C.; Arias-Gonzalez, J.R. Single-Stranded Condensation Stochastically Blocks G-Quadruplex Assembly in Human Telomeric RNA. J. Phys. Chem. Lett. 2018, 9, 2498–2503. [Google Scholar] [CrossRef]
- Patrick, E.M.; Slivka, J.D.; Payne, B.; Comstock, M.J.; Schmidt, J.C. Observation of Processive Telomerase Catalysis Using High-Resolution Optical Tweezers. Nat. Chem. Biol. 2020, 16, 801–809. [Google Scholar] [CrossRef]
- Hernandez, R.J.H. Optical Trapping and Manipulation Exploiting Liquid Crystalline Systems. Ph. D. Thesis, Università della Calabria, Arcavacata, Italy, 2012. [Google Scholar]
- Gosse, C.; Croquette, V. Magnetic Tweezers: Micromanipulation and Force Measurement at the Molecular Level. Biophys. J. 2002, 82, 3314–3329. [Google Scholar] [CrossRef]
- Kilinc, D.; Lee, G.U. Advances in Magnetic Tweezers for Single Molecule and Cell Biophysics. Integr. Biol. 2014, 6, 27–34. [Google Scholar] [CrossRef]
- Sun, D.; Hurley, L.H. The Importance of Negative Superhelicity in Inducing the Formation of G-Quadruplex and i-Motif Structures in the c-Myc Promoter: Implications for Drug Targeting and Control of Gene Expression. J. Med. Chem. 2009, 52, 2863–2874. [Google Scholar] [CrossRef] [PubMed]
- Selvam, S.; Koirala, D.; Yu, Z.; Mao, H. Quantification of Topological Coupling between DNA Superhelicity and G-Quadruplex Formation. J. Am. Chem. Soc. 2014, 136, 13967–13970. [Google Scholar] [CrossRef] [PubMed]
- Selvam, S.; Mandal, S.; Mao, H. Quantification of Chemical and Mechanical Effects on the Formation of the G-Quadruplex and i-Motif in Duplex DNA. Biochemistry 2017, 56, 4616–4625. [Google Scholar] [CrossRef]
- Dulin, D. An Introduction to Magnetic Tweezers. In Single Molecule Analysis: Methods and Protocols; Heller, I., Dulin, D., Peterman, E.J.G., Eds.; Springer: New York, NY, USA, 2024; pp. 375–401. ISBN 978-1-07-163377-9. [Google Scholar]
- You, H.; Zeng, X.; Xu, Y.; Lim, C.J.; Efremov, A.K.; Phan, A.T.; Yan, J. Dynamics and Stability of Polymorphic Human Telomeric G-Quadruplex under Tension. Nucleic Acids Res. 2014, 42, 8789–8795. [Google Scholar] [CrossRef][Green Version]
- Cheng, Y.; Zhang, Y.; Gong, Z.; Zhang, X.; Li, Y.; Shi, X.; Pei, Y.; You, H. High Mechanical Stability and Slow Unfolding Rates Are Prevalent in Parallel-Stranded DNA G-Quadruplexes. J. Phys. Chem. Lett. 2020, 11, 7966–7971. [Google Scholar] [CrossRef] [PubMed]
- You, H.; Wu, J.; Shao, F.; Yan, J. Stability and Kinetics of C-MYC Promoter G-Quadruplexes Studied by Single-Molecule Manipulation. J. Am. Chem. Soc. 2015, 137, 2424–2427. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Tang, Q.; Li, Y.; Zhang, Y.; Zhao, C.; Yan, J.; You, H. Folding/Unfolding Kinetics of G-Quadruplexes Upstream of the P1 Promoter of the Human BCL-2 Oncogene. J. Biol. Chem. 2019, 294, 5890–5895. [Google Scholar] [CrossRef] [PubMed]
- Buglione, E.; Salerno, D.; Marrano, C.A.; Cassina, V.; Vesco, G.; Nardo, L.; Dacasto, M.; Rigo, R.; Sissi, C.; Mantegazza, F. Nanomechanics of G-Quadruplexes within the Promoter of the KIT Oncogene. Nucleic Acids Res. 2021, 49, 4564–4573. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Hou, X.-M.; Wang, P.-Y.; Xi, X.-G.; Li, M. Direct Measurement of Sequential Folding Pathway and Energy Landscape of Human Telomeric G-Quadruplex Structures. J. Am. Chem. Soc. 2013, 135, 6423–6426. [Google Scholar] [CrossRef]
- You, H.; Guo, S.; Le, S.; Tang, Q.; Yao, M.; Zhao, X.; Yan, J. Two-State Folding Energy Determination Based on Transition Points in Nonequilibrium Single-Molecule Experiments. J. Phys. Chem. Lett. 2018, 9, 811–816. [Google Scholar] [CrossRef]
- Zhang, Y.; Cheng, Y.; Chen, J.; Zheng, K.; You, H. Mechanical Diversity and Folding Intermediates of Parallel-Stranded G-Quadruplexes with a Bulge. Nucleic Acids Res. 2021, 49, 7179–7188. [Google Scholar] [CrossRef]
- You, H.; Lattmann, S.; Rhodes, D.; Yan, J. RHAU Helicase Stabilizes G4 in Its Nucleotide-Free State and Destabilizes G4 upon ATP Hydrolysis. Nucleic Acids Res. 2017, 45, 206–214. [Google Scholar] [CrossRef]
- You, H.; Zhou, Y.; Yan, J. Using Magnetic Tweezers to Unravel the Mechanism of the G-Quadruplex Binding and Unwinding Activities of DHX36 Helicase. In RNA Remodeling Proteins: Methods and Protocols; Boudvillain, M., Ed.; Methods in Molecular Biology; Springer: New York, NY, USA, 2021; pp. 175–191. ISBN 978-1-07-160935-4. [Google Scholar]
- Ju, H.-P.; Wang, Y.-Z.; You, J.; Hou, X.-M.; Xi, X.-G.; Dou, S.-X.; Li, W.; Wang, P.-Y. Folding Kinetics of Single Human Telomeric G-Quadruplex Affected by Cisplatin. ACS Omega 2016, 1, 244–250. [Google Scholar] [CrossRef]
- Wang, Y.-Z.; Hou, X.-M.; Ju, H.-P.; Xiao, X.; Xi, X.-G.; Dou, S.-X.; Wang, P.-Y.; Li, W. Interaction between Human Telomeric G-Quadruplexes Characterized by Single Molecule Magnetic Tweezers. Chin. Phys. B 2018, 27, 068701. [Google Scholar] [CrossRef]
- Tran, P.L.T.; Rieu, M.; Hodeib, S.; Joubert, A.; Ouellet, J.; Alberti, P.; Bugaut, A.; Allemand, J.-F.; Boulé, J.-B.; Croquette, V. Folding and Persistence Times of Intramolecular G-Quadruplexes Transiently Embedded in a DNA Duplex. Nucleic Acids Res. 2021, 49, 5189–5201. [Google Scholar] [CrossRef] [PubMed]
- Long, X.; Parks, J.W.; Bagshaw, C.R.; Stone, M.D. Mechanical Unfolding of Human Telomere G-Quadruplex DNA Probed by Integrated Fluorescence and Magnetic Tweezers Spectroscopy. Nucleic Acids Res. 2013, 41, 2746–2755. [Google Scholar] [CrossRef] [PubMed]
- Mitra, J.; Ha, T. Streamlining Effects of Extra Telomeric Repeat on Telomeric DNA Folding Revealed by Fluorescence-Force Spectroscopy. Nucleic Acids Res. 2019, 47, 11044–11056. [Google Scholar] [CrossRef] [PubMed]
- Mitra, J.; Makurath, M.A.; Ngo, T.T.M.; Troitskaia, A.; Chemla, Y.R.; Ha, T. Extreme Mechanical Diversity of Human Telomeric DNA Revealed by Fluorescence-Force Spectroscopy. Proc. Natl. Acad. Sci. USA 2019, 116, 8350–8359. [Google Scholar] [CrossRef]
- Kar, A.; Jones, N.; Arat, N.Ö.; Fishel, R.; Griffith, J.D. Long Repeating (TTAGGG)n Single-Stranded DNA Self-Condenses into Compact Beaded Filaments Stabilized by G-Quadruplex Formation. J. Biol. Chem. 2018, 293, 9473–9485. [Google Scholar] [CrossRef]
- Li, S.; Wang, X.; Li, Z.; Huang, Z.; Lin, S.; Hu, J.; Tu, Y. Research Progress of Single Molecule Force Spectroscopy Technology Based on Atomic Force Microscopy in Polymer Materials: Structure, Design Strategy and Probe Modification. Nano Sel. 2021, 2, 909–931. [Google Scholar] [CrossRef]
- Haynes, P.J.; Main, K.H.S.; Pyne, A. Atomic Force Microscopy of DNA and DNA-Protein Interactions. In Chromosome Architecture: Methods and Protocols; Springer: New York, NY, USA, 2020. [Google Scholar]
- Pyne, A.L.B.; Noy, A.; Main, K.H.S.; Velasco-Berrelleza, V.; Piperakis, M.M.; Mitchenall, L.A.; Cugliandolo, F.M.; Beton, J.G.; Stevenson, C.E.M.; Hoogenboom, B.W.; et al. Base-Pair Resolution Analysis of the Effect of Supercoiling on DNA Flexibility and Major Groove Recognition by Triplex-Forming Oligonucleotides. Nat. Commun. 2021, 12, 1053. [Google Scholar] [CrossRef]
- Funayama, R.; Nakahara, Y.; Kado, S.; Tanaka, M.; Kimura, K. A Single-Molecule Force-Spectroscopic Study on Stabilization of G-Quadruplex DNA by a Telomerase Inhibitor. Analyst 2014, 139, 4037–4043. [Google Scholar] [CrossRef]
- Müller, D.J.; Dumitru, A.C.; Lo Giudice, C.; Gaub, H.E.; Hinterdorfer, P.; Hummer, G.; De Yoreo, J.J.; Dufrêne, Y.F.; Alsteens, D. Atomic Force Microscopy-Based Force Spectroscopy and Multiparametric Imaging of Biomolecular and Cellular Systems. Chem. Rev. 2021, 121, 11701–11725. [Google Scholar] [CrossRef]
- Lynch, S.; Baker, H.; Byker, S.G.; Zhou, D.; Sinniah, K. Single molecule force spectroscopy on G-quadruplex DNA. Chemistry 2009, 15, 8113. Available online: https://chemistry-europe.onlinelibrary.wiley.com/doi/full/10.1002/chem.200901390 (accessed on 25 October 2023). [CrossRef]
- Zhang, X.; Zhang, Y.; Zhang, W. Dynamic Topology of Double-Stranded Telomeric DNA Studied by Single-Molecule Manipulation in Vitro. Nucleic Acids Res. 2020, 48, 6458–6470. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.-Q.; Wu, J.; Liang, J.-H.; Yan, J.-W.; Zhu, Z.; Yang, C.J.; Mao, B.-W. Single-Molecule Force Spectroscopic Studies on Intra- and Intermolecular Interactions of G-Quadruplex Aptamer with Target Shp2 Protein. J. Phys. Chem. B 2012, 116, 11397–11404. [Google Scholar] [CrossRef] [PubMed]
- Costa, L.T.; Kerkmann, M.; Hartmann, G.; Endres, S.; Bisch, P.M.; Heckl, W.M.; Thalhammer, S. Structural Studies of Oligonucleotides Containing G-Quadruplex Motifs Using AFM. Biochem. Biophys. Res. Commun. 2004, 313, 1065–1072. [Google Scholar] [CrossRef]
- Chiorcea-Paquim, A.-M.; Viegas Santos, P.; Eritja, R.; Maria Oliveira-Brett, A. Self-Assembled G-Quadruplex Nanostructures: AFM and Voltammetric Characterization. Phys. Chem. Chem. Phys. 2013, 15, 9117–9124. [Google Scholar] [CrossRef]
- Neaves, K.J.; Huppert, J.L.; Henderson, R.M.; Edwardson, J.M. Direct Visualization of G-Quadruplexes in DNA Using Atomic Force Microscopy. Nucleic Acids Res. 2009, 37, 6269–6275. [Google Scholar] [CrossRef]
- Mela, I.; Kranaster, R.; Henderson, R.M.; Balasubramanian, S.; Edwardson, J.M. Demonstration of Ligand Decoration, and Ligand-Induced Perturbation, of G-Quadruplexes in a Plasmid Using Atomic Force Microscopy. Biochemistry 2012, 51, 578–585. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Nora, G.J.; Ghodke, H.; Opresko, P.L. Single Molecule Studies of Physiologically Relevant Telomeric Tails Reveal POT1 Mechanism for Promoting G-Quadruplex Unfolding. J. Biol. Chem. 2011, 286, 7479–7489. [Google Scholar] [CrossRef]
- Sannohe, Y.; Endo, M.; Katsuda, Y.; Hidaka, K.; Sugiyama, H. Visualization of Dynamic Conformational Switching of the G-Quadruplex in a DNA Nanostructure. J. Am. Chem. Soc. 2010, 132, 16311–16313. [Google Scholar] [CrossRef]
- Endo, M.; Sugiyama, H. Single-Molecule Imaging of Dynamic Motions of Biomolecules in DNA Origami Nanostructures Using High-Speed Atomic Force Microscopy. Acc. Chem. Res. 2014, 47, 1645–1653. [Google Scholar] [CrossRef]
- Endo, M.; Xing, X.; Zhou, X.; Emura, T.; Hidaka, K.; Tuesuwan, B.; Sugiyama, H. Single-Molecule Manipulation of the Duplex Formation and Dissociation at the G-Quadruplex/i-Motif Site in the DNA Nanostructure. ACS Nano 2015, 9, 9922–9929. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lamperti, M.; Rigo, R.; Sissi, C.; Nardo, L. Probing G-Quadruplexes Conformational Dynamics and Nano-Mechanical Interactions at the Single Molecule Level: Techniques and Perspectives. Photonics 2024, 11, 1061. https://doi.org/10.3390/photonics11111061
Lamperti M, Rigo R, Sissi C, Nardo L. Probing G-Quadruplexes Conformational Dynamics and Nano-Mechanical Interactions at the Single Molecule Level: Techniques and Perspectives. Photonics. 2024; 11(11):1061. https://doi.org/10.3390/photonics11111061
Chicago/Turabian StyleLamperti, Marco, Riccardo Rigo, Claudia Sissi, and Luca Nardo. 2024. "Probing G-Quadruplexes Conformational Dynamics and Nano-Mechanical Interactions at the Single Molecule Level: Techniques and Perspectives" Photonics 11, no. 11: 1061. https://doi.org/10.3390/photonics11111061
APA StyleLamperti, M., Rigo, R., Sissi, C., & Nardo, L. (2024). Probing G-Quadruplexes Conformational Dynamics and Nano-Mechanical Interactions at the Single Molecule Level: Techniques and Perspectives. Photonics, 11(11), 1061. https://doi.org/10.3390/photonics11111061






