Rapid Correction of Turbidity and CDOM Interference on Three-Dimensional Fluorescence Spectra of Live Algae Based on Deep Learning
Abstract
1. Introduction
2. Materials and Methods
2.1. Algal Cultivation
2.2. Sample Preparation
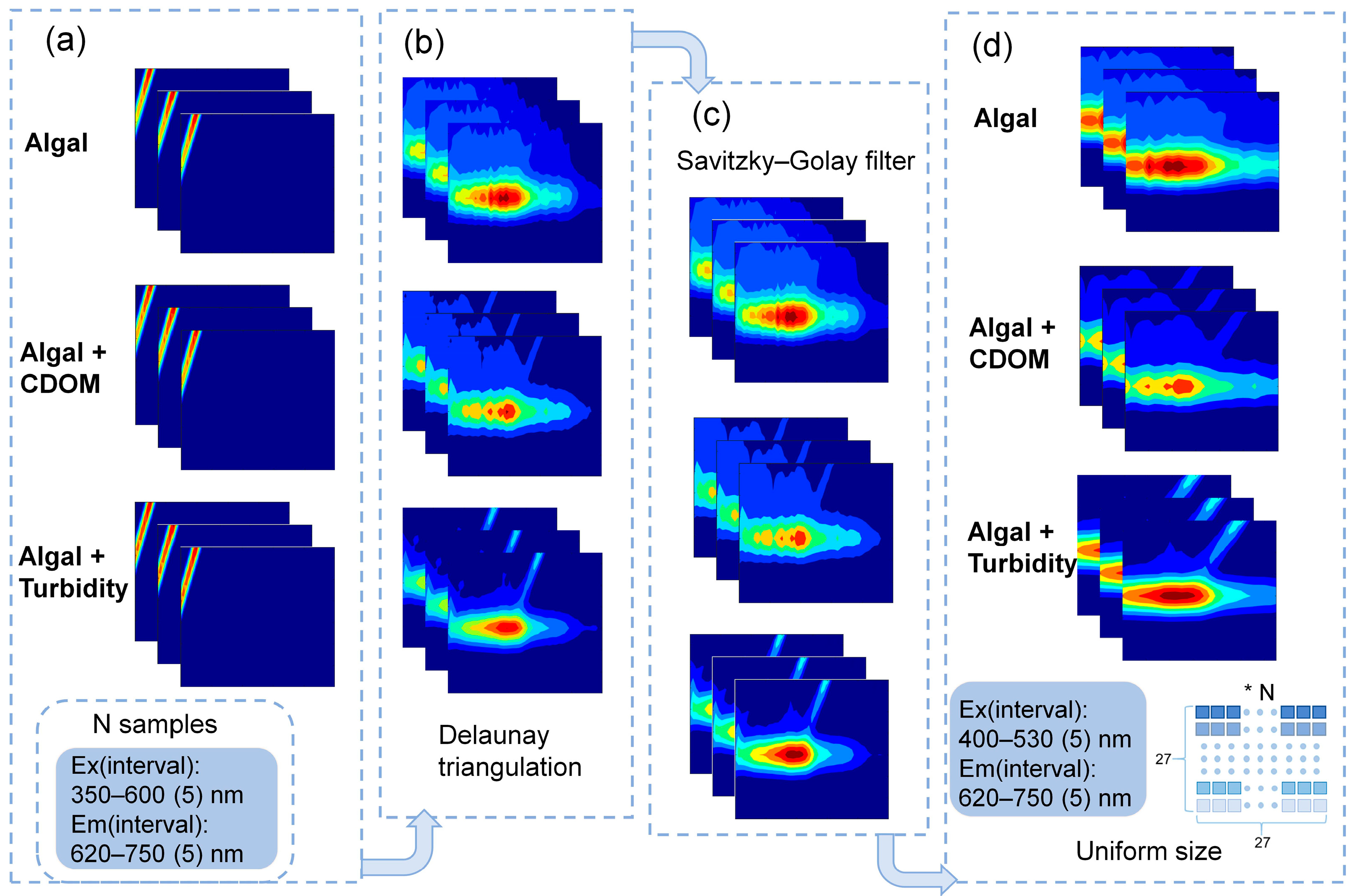
2.3. Fluorescence Measurement and Data Pre-Processing
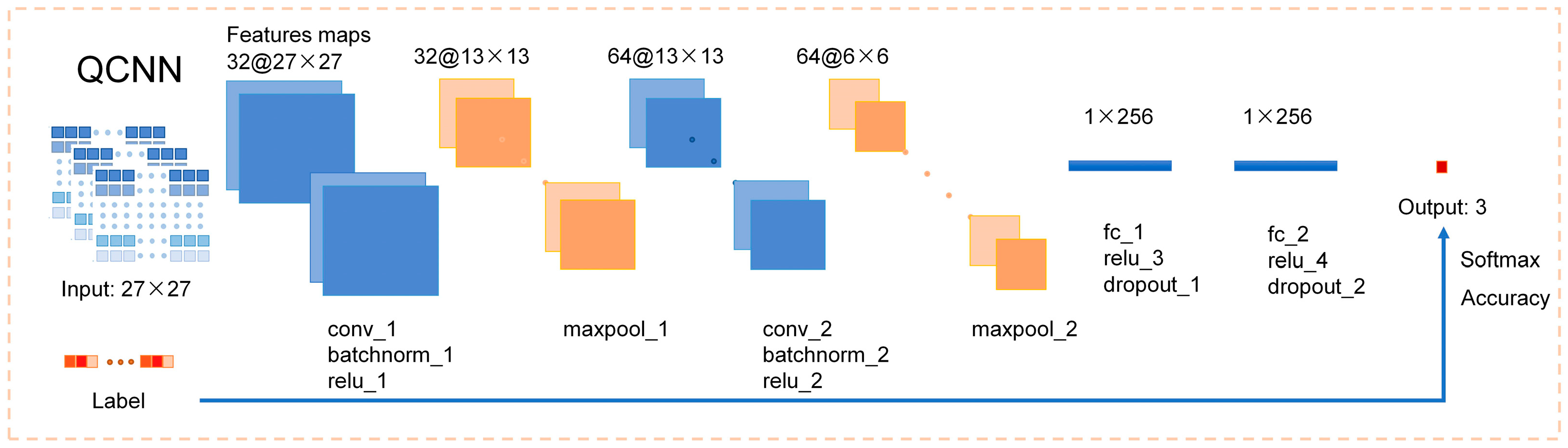
2.4. AFAI-Net Model Establishment Based on Deep Convolutional Neural Network
3. Results and Discussion
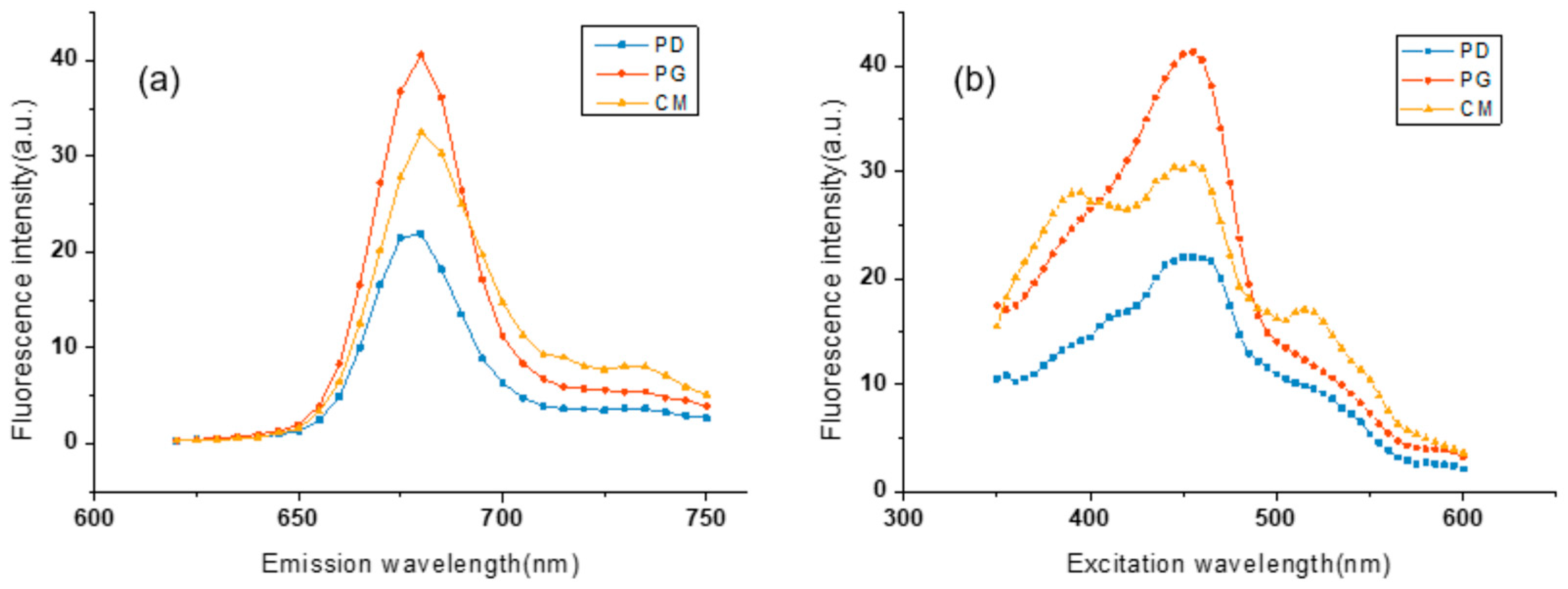
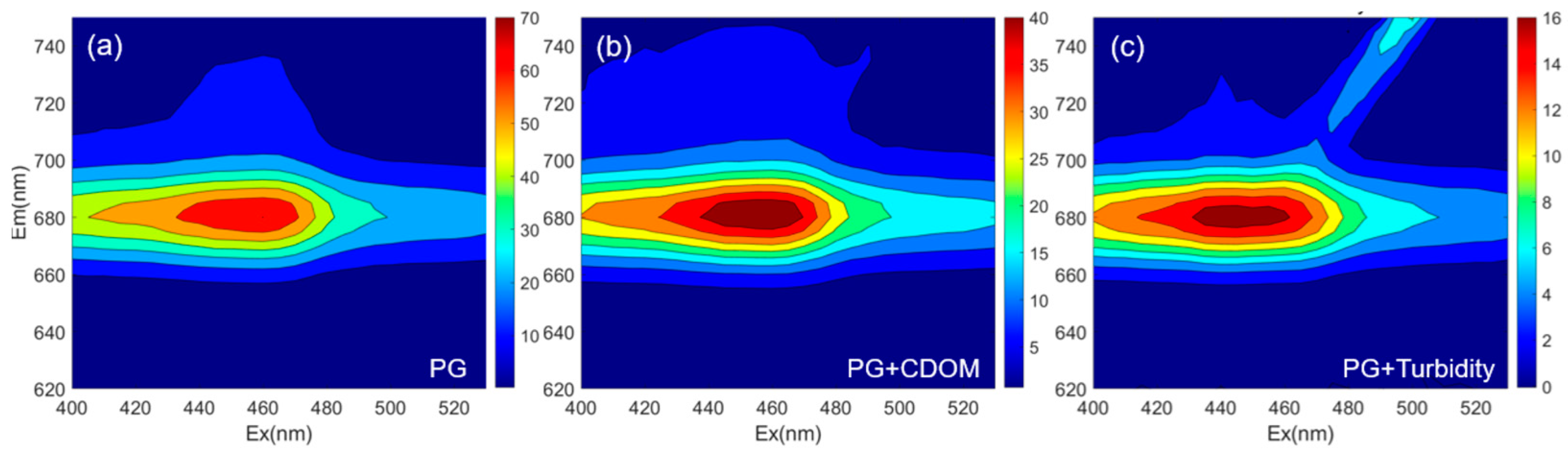
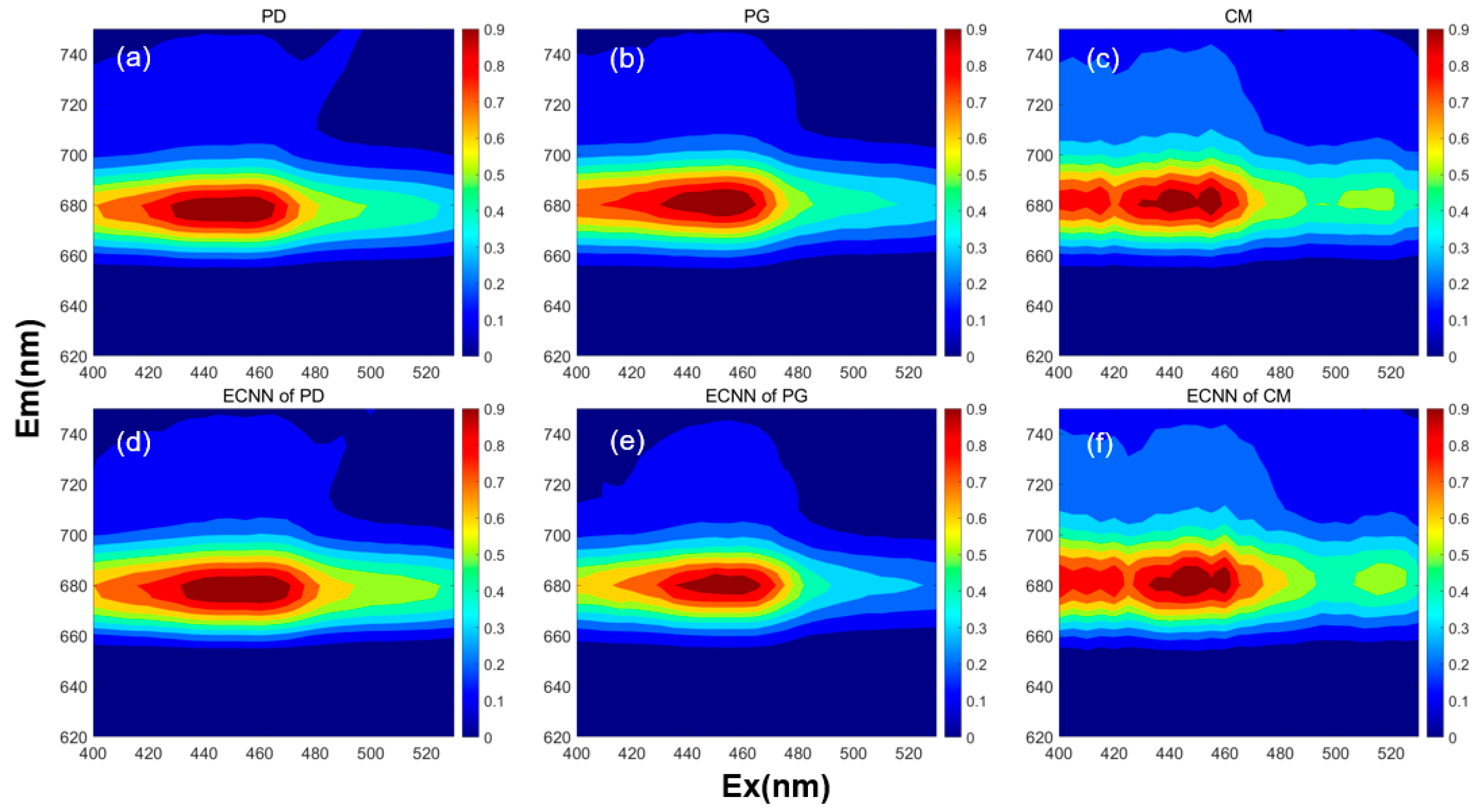
3.1. Characteristics of the 3D-EEMs Data
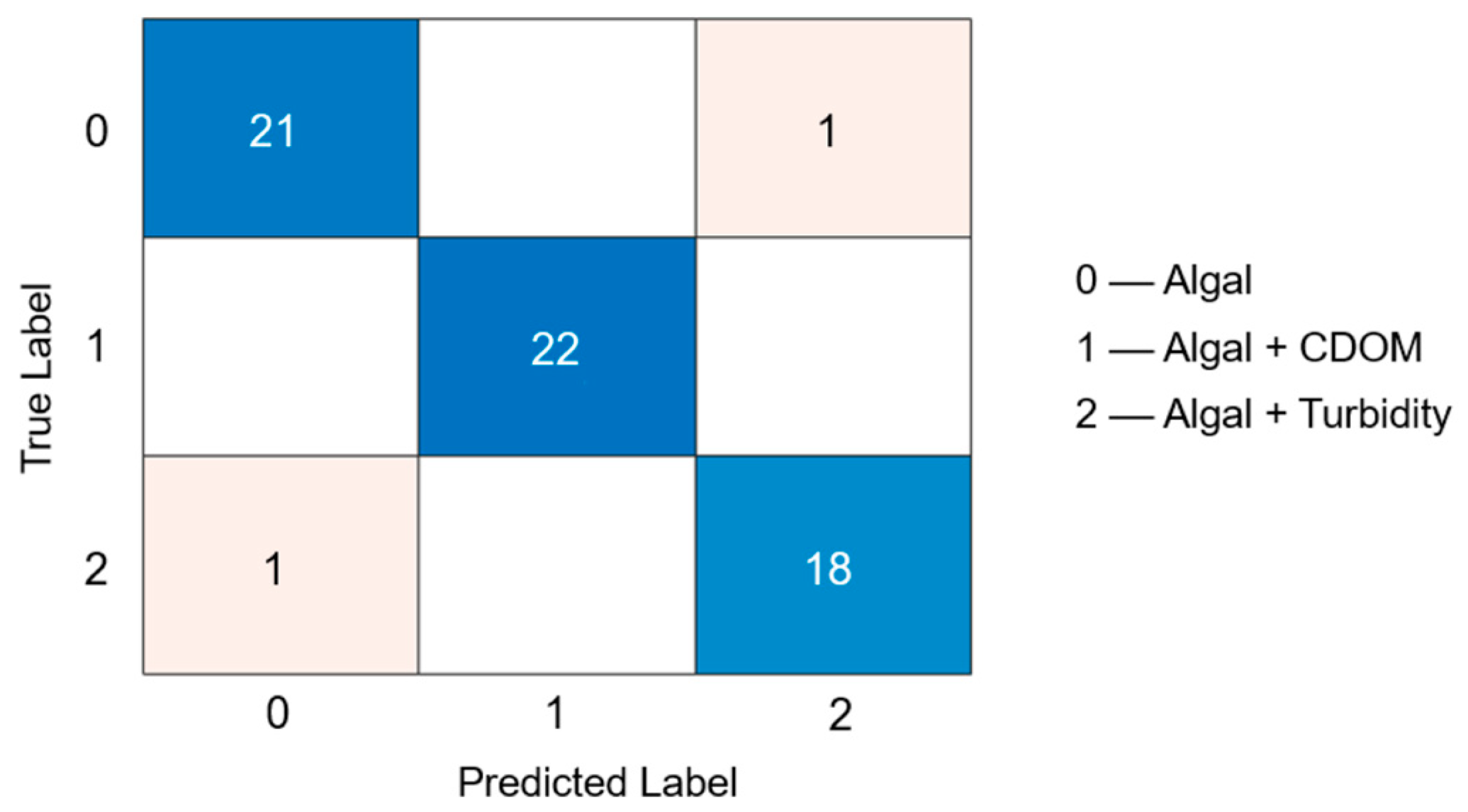
3.2. Training and Evaluation of the AFAI-Net Model
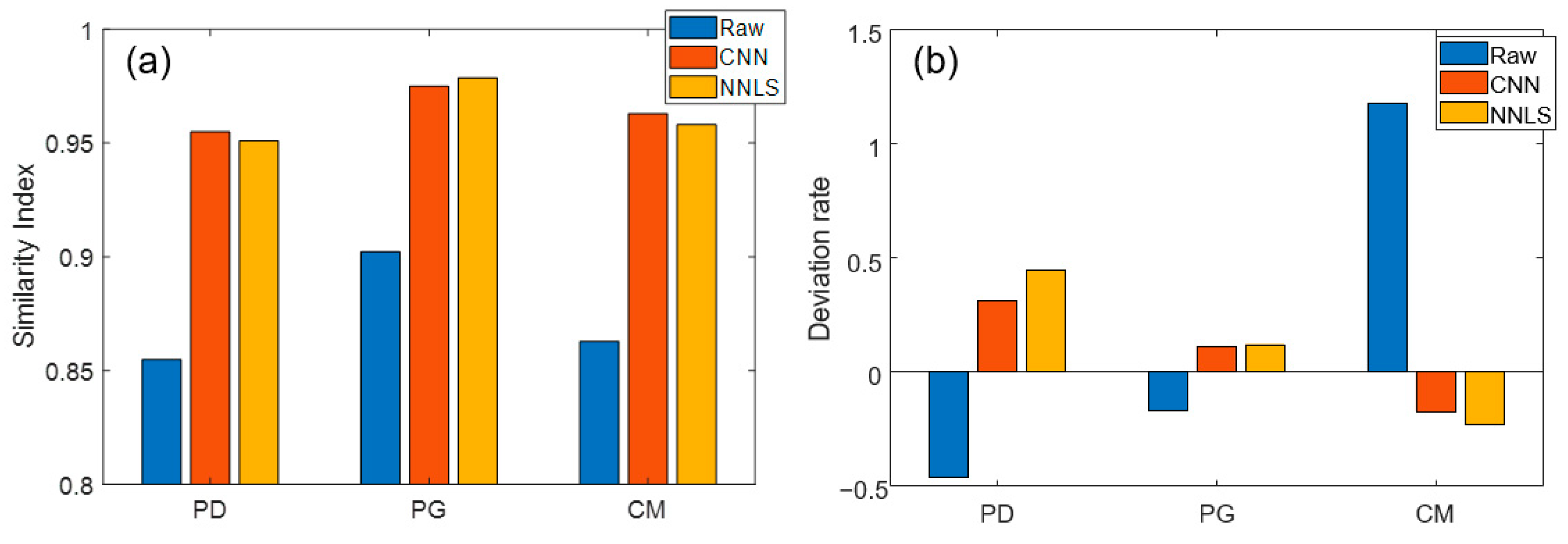
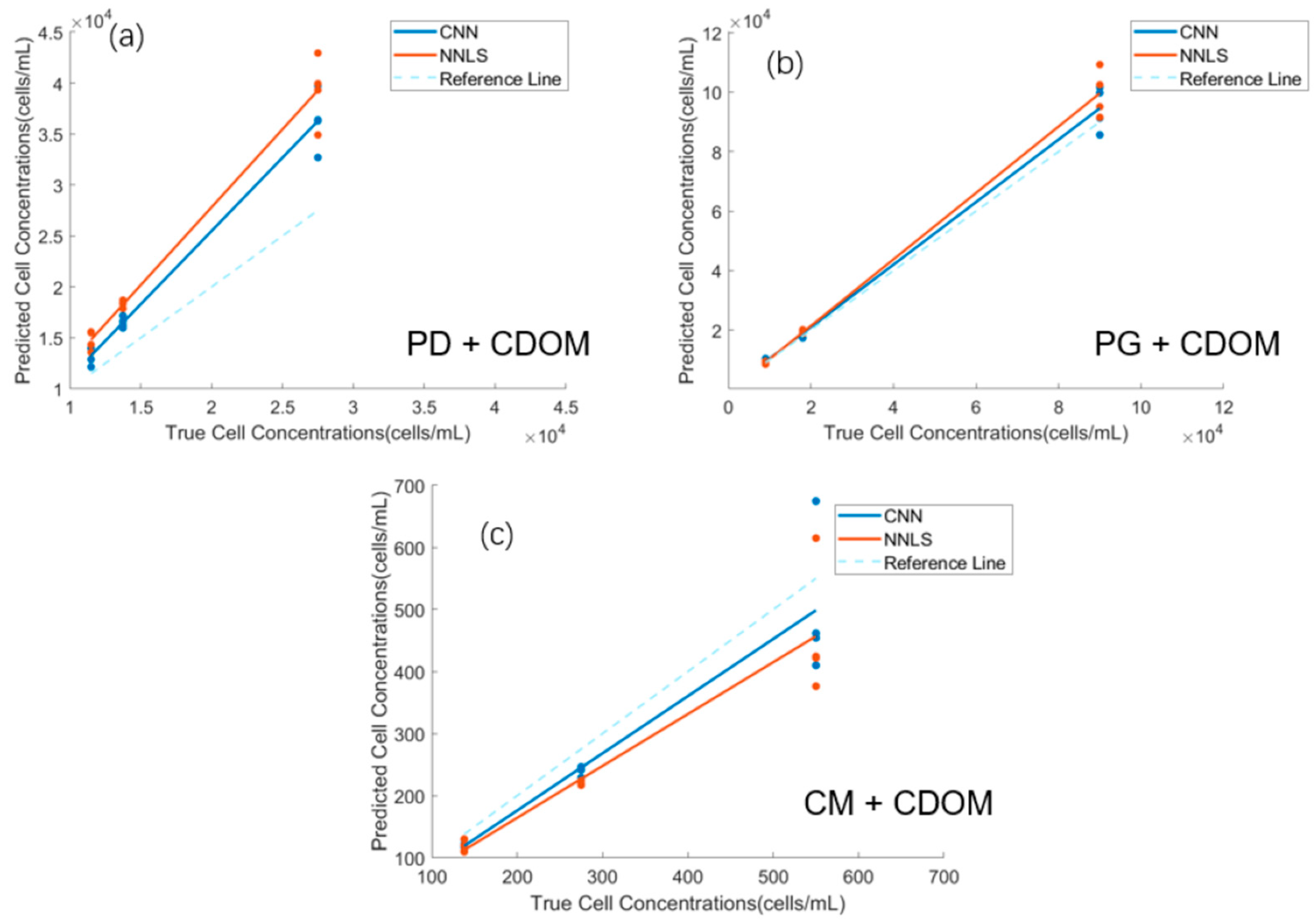
3.3. Comparison of Correction Methods
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Peters, F.; Marrasé, C. Effects of Turbulence on Plankton: An Overview of Experimental Evidence and Some Theoretical Considerations. Mar. Ecol. Prog. Ser. 2000, 205, 291–306. [Google Scholar] [CrossRef]
- Katsiapi, M.; Moustaka-Gouni, M.; Sommer, U. Assessing Ecological Water Quality of Freshwaters: PhyCoI—A New Phytoplankton Community Index. Ecol. Inform. 2016, 31, 22–29. [Google Scholar] [CrossRef]
- Berdalet, E.; Kudela, R.; Urban, E.; Enevoldsen, H.; Banas, N.; Bresnan, E.; Burford, M.; Davidson, K.; Gobler, C.; Karlson, B.; et al. GlobalHAB: A New Program to Promote International Research, Observations, and Modeling of Harmful Algal Blooms in Aquatic Systems. Oceanography 2017, 30, 70–81. [Google Scholar] [CrossRef]
- Goldman, E.A.; Smith, E.M.; Richardson, T.L. Estimation of Chromophoric Dissolved Organic Matter (CDOM) and Photosynthetic Activity of Estuarine Phytoplankton Using a Multiple-Fixed-Wavelength Spectral Fluorometer. Water Res. 2013, 47, 1616–1630. [Google Scholar] [CrossRef]
- Mackey, M.; Mackey, D.; Higgins, H.; Wright, S. CHEMTAX—A Program for Estimating Class Abundances from Chemical Markers:Application to HPLC Measurements of Phytoplankton. Mar. Ecol. Prog. Ser. 1996, 144, 265–283. [Google Scholar] [CrossRef]
- Shin, Y.H.; Barnett, J.Z.; Gutierrez-Wing, M.T.; Rusch, K.A.; Choi, J.W. A Hand-Held Fluorescent Sensor Platform for Selectively Estimating Green Algae and Cyanobacteria Biomass. Sens. Actuators B Chem. 2018, 262, 938–946. [Google Scholar] [CrossRef]
- Sá, M.; Ferrer-Ledo, N.; Gao, F.; Bertinetto, C.G.; Jansen, J.; Crespo, J.G.; Wijffels, R.H.; Barbosa, M.; Galinha, C.F. Perspectives of Fluorescence Spectroscopy for Online Monitoring in Microalgae Industry. Microb. Biotechnol. 2022, 15, 1824–1838. [Google Scholar] [CrossRef]
- Yentsch, C.S.; Phinney, D.A. Spectral Fluorescence: An Ataxonomic Tool for Studying the Structure of Phytoplankton Populations. J. Plankton Res. 1985, 7, 617–632. [Google Scholar] [CrossRef]
- Cadondon, J.G.; Ong, P.M.B.; Vallar, E.A.; Shiina, T.; Galvez, M.C.D. Chlorophyll-a Pigment Measurement of Spirulina in Algal Growth Monitoring Using Portable Pulsed LED Fluorescence Lidar System. Sensors 2022, 22, 2940. [Google Scholar] [CrossRef]
- Chegoonian, A.M.; Zolfaghari, K.; Leavitt, P.R.; Baulch, H.M.; Duguay, C.R. Improvement of Field Fluorometry Estimates of Chlorophyll a Concentration in a Cyanobacteria-Rich Eutrophic Lake. Limnol. Oceanogr. Methods 2022, 20, 193–209. [Google Scholar] [CrossRef]
- Brient, L.; Lengronne, M.; Bertrand, E.; Rolland, D.; Sipel, A.; Steinmann, D.; Baudin, I.; Legeas, M.; Rouzic, B.L.; Bormans, M. A Phycocyanin Probe as a Tool for Monitoring Cyanobacteria in Freshwater Bodies. J. Environ. Monit. 2008, 10, 248–255. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, M.; Fu, X.; Sun, J.; Xie, H. Chromophoric Dissolved Organic Matter (CDOM) Release by Dictyocha Fibula in the Central Bohai Sea. Mar. Chem. 2022, 241, 104107. [Google Scholar] [CrossRef]
- Zhang, Y.; van Dijk, M.A.; Liu, M.; Zhu, G.; Qin, B. The Contribution of Phytoplankton Degradation to Chromophoric Dissolved Organic Matter (CDOM) in Eutrophic Shallow Lakes: Field and Experimental Evidence. Water Res. 2009, 43, 4685–4697. [Google Scholar] [CrossRef]
- Trubetskaya, O.E.; Richard, C.; Patsaeva, S.V.; Trubetskoj, O.A. Evaluation of Aliphatic/Aromatic Compounds and Fluorophores in Dissolved Organic Matter of Contrasting Natural Waters by SEC-HPLC with Multi-Wavelength Absorbance and Fluorescence Detections. Spectrochim. Acta. A Mol. Biomol. Spectrosc. 2020, 238, 118450. [Google Scholar] [CrossRef]
- Del Vecchio, R.; Blough, N.V. Photobleaching of Chromophoric Dissolved Organic Matter in Natural Waters: Kinetics and Modeling. Mar. Chem. 2002, 78, 231–253. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, Y.; Shi, K.; Zhu, G.; Xu, H.; Zhu, M. Absorption and Fluorescence Properties of Chromophoric Dissolved Organic Matter: Implications for the Monitoring of Water Quality in a Large Subtropical Reservoir. Environ. Sci. Pollut. Res. 2014, 21, 14078–14090. [Google Scholar] [CrossRef]
- Choo, F.; Zamyadi, A.; Stuetz, R.M.; Newcombe, G.; Newton, K.; Henderson, R.K. Enhanced Real-Time Cyanobacterial Fluorescence Monitoring through Chlorophyll-a Interference Compensation Corrections. Water Res. 2019, 148, 86–96. [Google Scholar] [CrossRef]
- Xing, X.; Claustre, H.; Boss, E.; Roesler, C.; Organelli, E.; Poteau, A.; Barbieux, M.; D’Ortenzio, F. Correction of Profiles of In-Situ Chlorophyll Fluorometry for the Contribution of Fluorescence Originating from Non-Algal Matter. Limnol. Oceanogr. Methods 2017, 15, 80–93. [Google Scholar] [CrossRef]
- Azil, K.; Altuncu, A.; Ferria, K.; Bouzid, S.; Sadık, Ş.A.; Durak, F.E. A Faster and Accurate Optical Water Turbidity Measurement System Using a CCD Line Sensor. Optik 2021, 231, 166412. [Google Scholar] [CrossRef]
- Ng, C.-L.; Ng, Y.-J.; Chen, Q.-Q.; Hemond, H.F. Corrections for Matrix Effects on Fluorescence Measurement of a Multi-Platform Optical Sensor. Water Pract. Technol. 2016, 11, 644–660. [Google Scholar] [CrossRef]
- Sarmanova, O.E.; Laptinskiy, K.A.; Khmeleva, M.Y.; Burikov, S.A.; Dolenko, S.A.; Tomskaya, A.E.; Dolenko, T.A. Development of the Fluorescent Carbon Nanosensor for PH and Temperature of Liquid Media with Artificial Neural Networks. Spectrochim. Acta. A Mol. Biomol. Spectrosc. 2021, 258, 119861. [Google Scholar] [CrossRef] [PubMed]
- Shin, Y.; Barnett, J.Z.; Song, E.; Gutierrez-Wing, M.T.; Rusch, K.A.; Choi, J.W. A Portable Fluorescent Sensor for On-Site Detection of Microalgae. Microelectron. Eng. 2015, 144, 6–11. [Google Scholar] [CrossRef]
- Zhang, X.; Yin, G.; Zhao, N.; Yang, R.; Liu, J.; Liu, W. Chromophoric Dissolved Organic Matter Influence Correction of Algal Concentration Measurements Using Three-Dimensional Fluorescence Spectra. Spectrochim. Acta. A Mol. Biomol. Spectrosc. 2019, 210, 405–411. [Google Scholar] [CrossRef]
- Li, W.; Liu, T.; Fu, Y.; Huang, M. High Sensitivity and Wide Range Chlorophyll-a Determination by Simultaneous Measurement of Absorbance and Fluorescence Using a Linear CCD. Spectrochim. Acta. A Mol. Biomol. Spectrosc. 2022, 270, 120831. [Google Scholar] [CrossRef]
- Li, Y.; Jia, Y.; Cai, X.; Xie, M.; Zhang, Z. Oil Pollutant Identification Based on Excitation-Emission Matrix of UV-Induced Fluorescence and Deep Convolutional Neural Network. Environ. Sci. Pollut. Res. 2022, 29, 68152–68160. [Google Scholar] [CrossRef]
- Ruan, K.; Zhao, S.; Jiang, X.; Li, Y.; Fei, J.; Ou, D.; Tang, Q.; Lu, Z.; Liu, T.; Xia, J. A 3D Fluorescence Classification and Component Prediction Method Based on VGG Convolutional Neural Network and PARAFAC Analysis Method. Appl. Sci. 2022, 12, 4886. [Google Scholar] [CrossRef]
- Wu, X.; Zhao, Z.; Tian, R.; Shang, Z.; Liu, H. Identification and Quantification of Counterfeit Sesame Oil by 3D Fluorescence Spectroscopy and Convolutional Neural Network. Food Chem. 2020, 311, 125882. [Google Scholar] [CrossRef]
- Xu, R.; Cao, J.; Feng, G.; Luo, J.; Feng, Q.; Ni, B.; Fang, F. Fast Identification of Fluorescent Components in Three-Dimensional Excitation-Emission Matrix Fluorescence Spectra via Deep Learning. Chem. Eng. J. 2022, 430, 132893. [Google Scholar] [CrossRef]
- Hu, Y.; Wen, Y.; Wang, X. Novel Method of Turbidity Compensation for Chemical Oxygen Demand Measurements by Using UV–Vis Spectrometry. Sens. Actuators B Chem. 2016, 227, 393–398. [Google Scholar] [CrossRef]
- Zepp, R.G.; Sheldon, W.M.; Moran, M.A. Dissolved Organic Fluorophores in Southeastern US Coastal Waters: Correction Method for Eliminating Rayleigh and Raman Scattering Peaks in Excitation–Emission Matrices. Mar. Chem. 2004, 89, 15–36. [Google Scholar] [CrossRef]
- Schafer, R.W. What Is a Savitzky-Golay Filter? [Lecture Notes]. IEEE Signal Process. Mag. 2011, 28, 111–117. [Google Scholar] [CrossRef]
- Wan, K.X.; Vidavsky, I.; Gross, M.L. Comparing Similar Spectra: From Similarity Index to Spectral Contrast Angle. J. Am. Soc. Mass Spectrom. 2002, 13, 85–88. [Google Scholar] [CrossRef]
- Beutler, M.; Wiltshire, K.H.; Meyer, B.; Moldaenke, C.; Lüring, C.; Meyerhöfer, M.; Hansen, U.-P.; Dau, H. A Fluorometric Method for the Differentiation of Algal Populations in Vivo and in Situ. Photosynth. Res. 2002, 72, 39–53. [Google Scholar] [CrossRef]
- Shan, S.; Xu, L.; Chen, K.; Tong, M.; Wang, X. A Rapid Fluorescence Approach on Differentiation of Typical Dinoflagellate of East China Sea. Spectrochim. Acta. A Mol. Biomol. Spectrosc. 2022, 276, 121216. [Google Scholar] [CrossRef]
- Liu, J.; Zeng, L.; Ren, Z.; Du, T.; Liu, X. Rapid in Situ Measurements of Algal Cell Concentrations Using an Artificial Neural Network and Single-Excitation Fluorescence Spectrometry. Algal Res. 2020, 45, 101739. [Google Scholar] [CrossRef]


| Phyla | Species | Cultures Media |
|---|---|---|
| Dinophyta | Prorocentrum donghaiense | F/2 |
| Haptophyta | Phaeocystis globosa | F/2 |
| Raphidophyta | Chattonella marinacm | F/2 |
| Type of Samples | Number of Samples | |
|---|---|---|
| Pure | PD | 60 |
| PG | 96 | |
| CM | 36 | |
| Mixed | PD + Turbidity | 42 |
| PG + Turbidity | 108 | |
| CM + Turbidity | 36 | |
| PD + CDOM | 60 | |
| PG + CDOM | 144 | |
| CM + CDOM | 48 | |
| Total | 630 |
| ECNN | RMSE | SI |
|---|---|---|
| ECNN-Tur | 0.3423 | 0.9630 |
| ECNN-CDOM | 0.2274 | 0.9715 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, M.; Chen, T.; Wang, X. Rapid Correction of Turbidity and CDOM Interference on Three-Dimensional Fluorescence Spectra of Live Algae Based on Deep Learning. Photonics 2023, 10, 627. https://doi.org/10.3390/photonics10060627
Wang M, Chen T, Wang X. Rapid Correction of Turbidity and CDOM Interference on Three-Dimensional Fluorescence Spectra of Live Algae Based on Deep Learning. Photonics. 2023; 10(6):627. https://doi.org/10.3390/photonics10060627
Chicago/Turabian StyleWang, Mengwei, Tiantian Chen, and Xiaoping Wang. 2023. "Rapid Correction of Turbidity and CDOM Interference on Three-Dimensional Fluorescence Spectra of Live Algae Based on Deep Learning" Photonics 10, no. 6: 627. https://doi.org/10.3390/photonics10060627
APA StyleWang, M., Chen, T., & Wang, X. (2023). Rapid Correction of Turbidity and CDOM Interference on Three-Dimensional Fluorescence Spectra of Live Algae Based on Deep Learning. Photonics, 10(6), 627. https://doi.org/10.3390/photonics10060627




