Automatic Choroid Vascularity Index Calculation in Optical Coherence Tomography Images with Low-Contrast Sclerochoroidal Junction Using Deep Learning
Abstract
1. Introduction
- A fully automated method with freely available code was proposed for the first time to calculate the CVI value in diabetic retinopathy and pachychoroid spectrum using deep learning methods.
- The proposed modified U-Net segmented the choroid and BM boundaries in challenging cases such as low-contrast images with thickened choroidal areas.
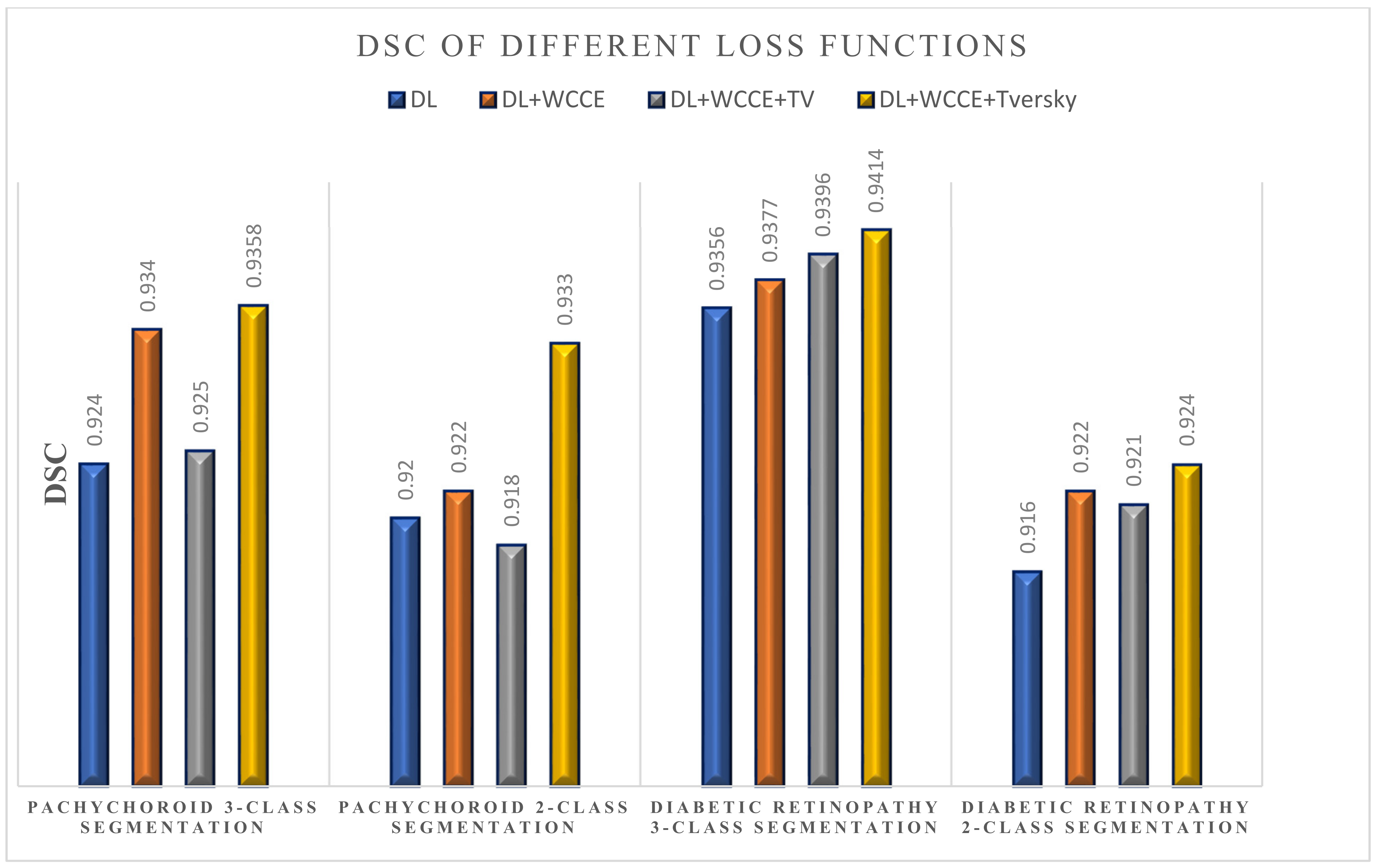
- The proposed loss function (weighted sum of Dice loss (DL), weighted categorical cross entropy (WCCE), and Tversky loss) was shown to overcome the imbalanced data (small foreground vs. big background).
2. Materials and Methods
2.1. OCT Data and Manual Annotation
2.2. Manual Calculation of CVI
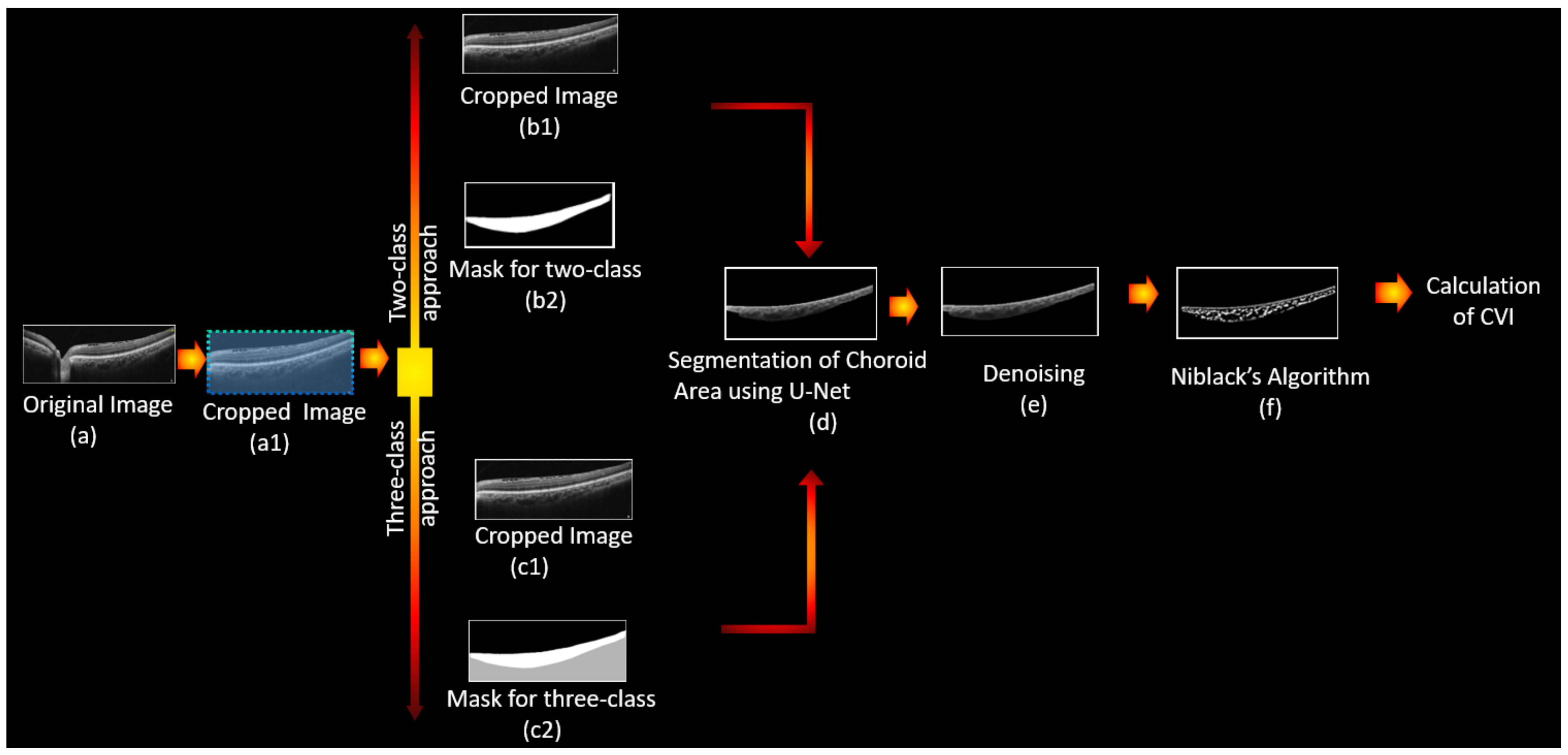
2.3. Fully Automated Calculation of CVI Using Deep Learning
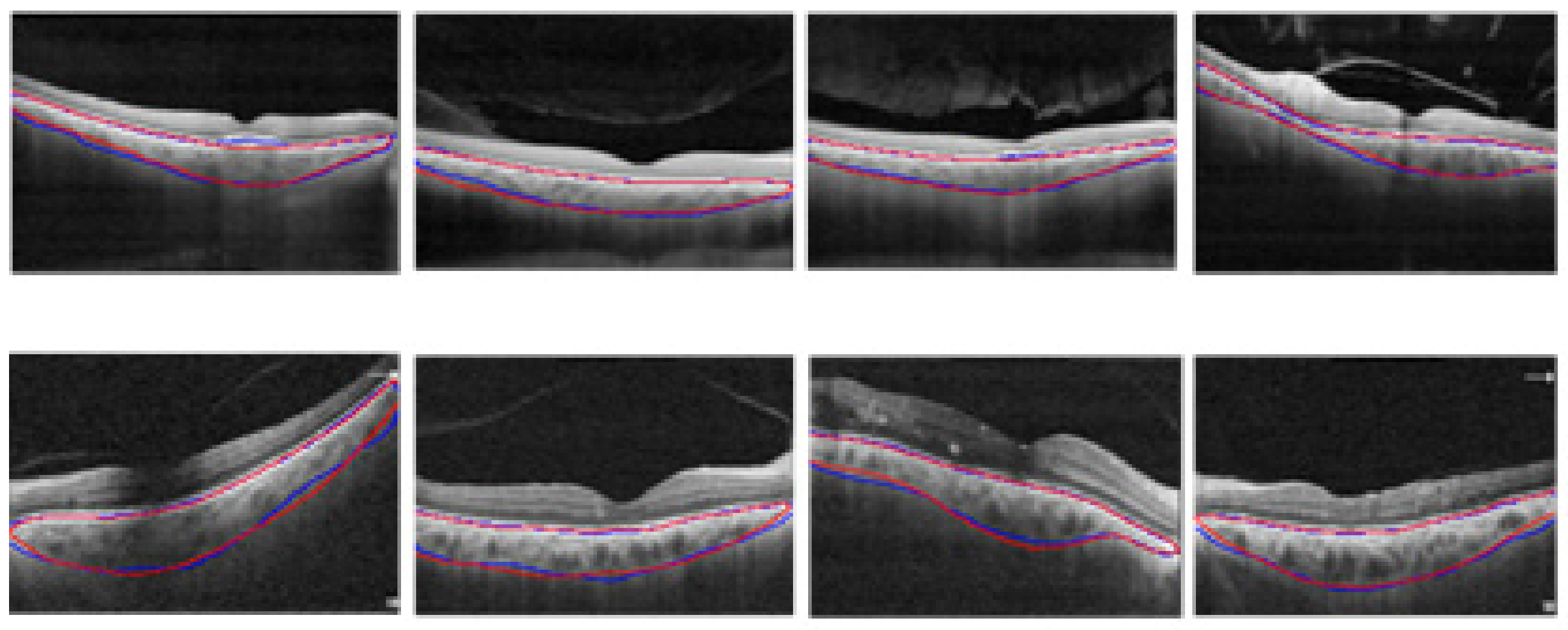
2.3.1. Automatic Segmentation of the Choroidal Layer
Network Architectures
Modified Loss Function
- Loss Function #1: Dice loss
- Loss Function #2: Dice loss + weighted categorical cross entropy (WCCE) [33]
- Loss Function #3: Dice loss + WCCE+ total variation (TV)
- Loss Function #4: Dice loss + WCCE+ Tversky
Train/Test Split and Metrics of the Segmentation Model
2.3.2. Detection of Choroidal Luminal Vessels and Computation of the CVI
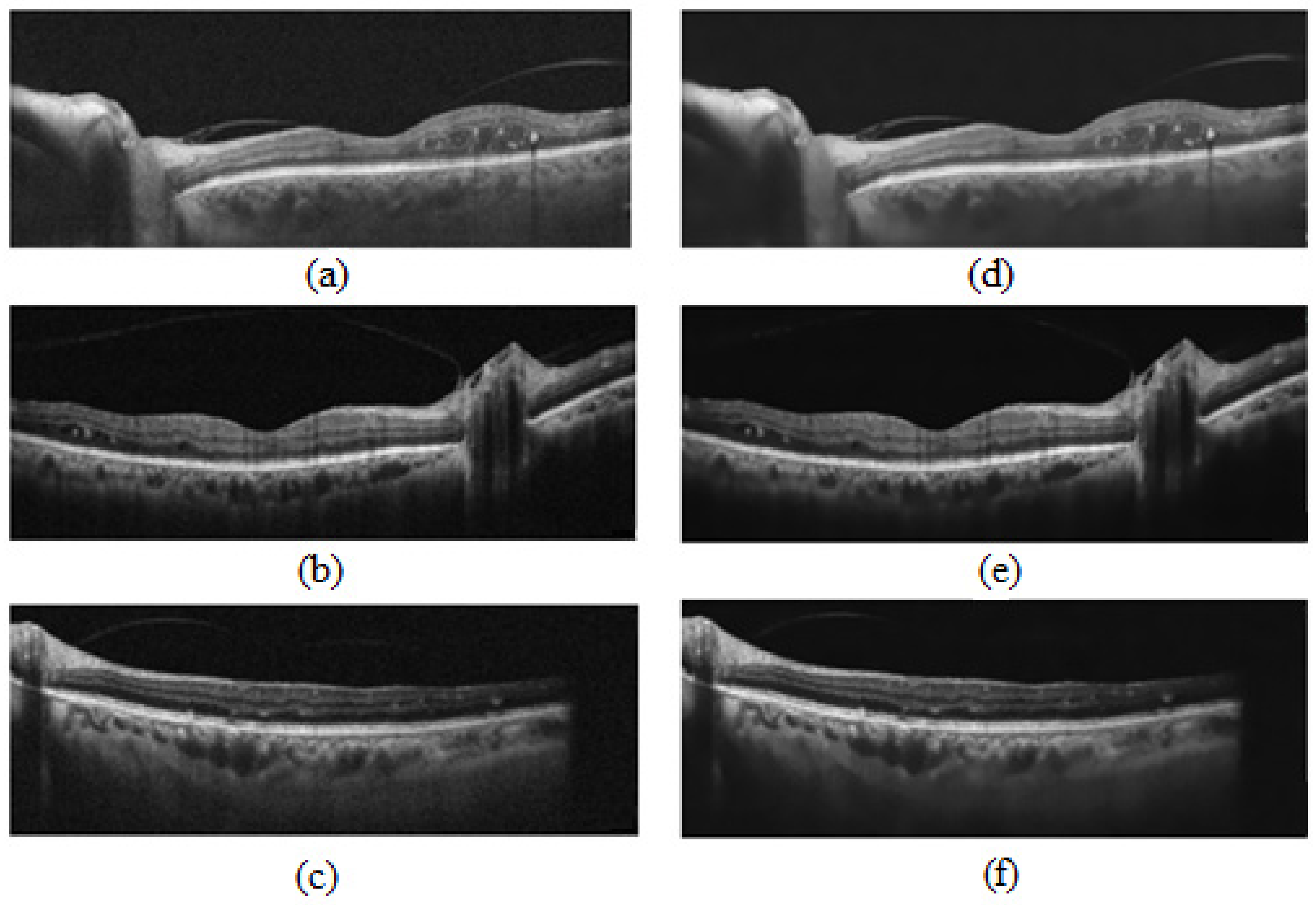
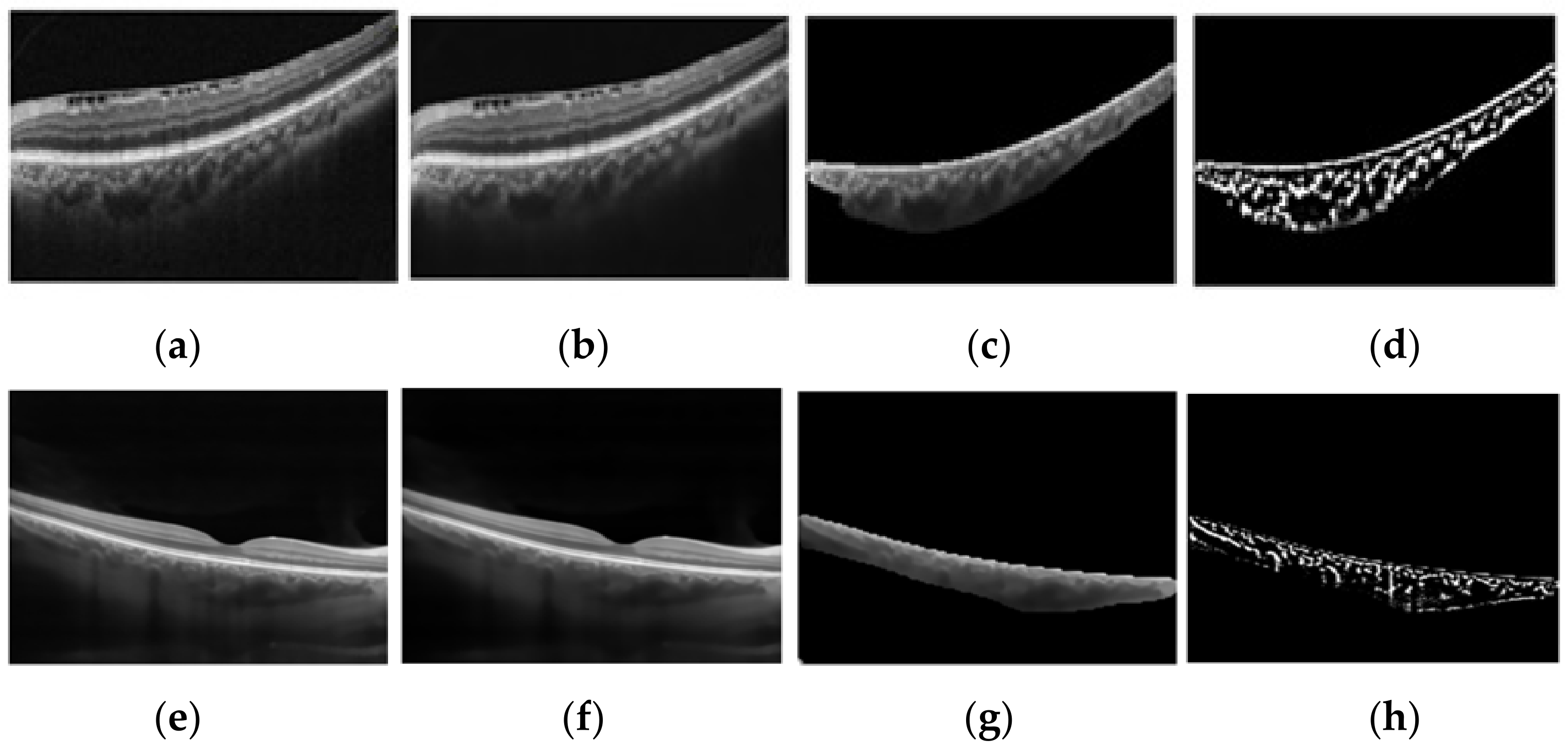
- Noise reduction using a non-local means algorithm with a deciding filter strength of 10 [34]. As shown in Figure 3, noise reduction reduced the errors in finding vascular components by omitting and noisy and disturbing pixels. Moreover, as was mentioned in Section 2.2, in order to calculate the CVI ground truth after finding the choroidal area, noise reduction was performed manually (before applying the Niblack algorithm). Therefore, in the proposed automatic method, we attempted to mimic what is already done in the manual process and achieve the most reliable performance.
- Vascular LA detection using Niblack’s auto local threshold method using Python software. The selected parameters for Niblack’s method are summarized in Table 4 to make the provided code reproducible. The parameters were empirically adjusted to resemble the gold standard values as much as possible in the training dataset.
- Calculation of CVI using:
3. Results
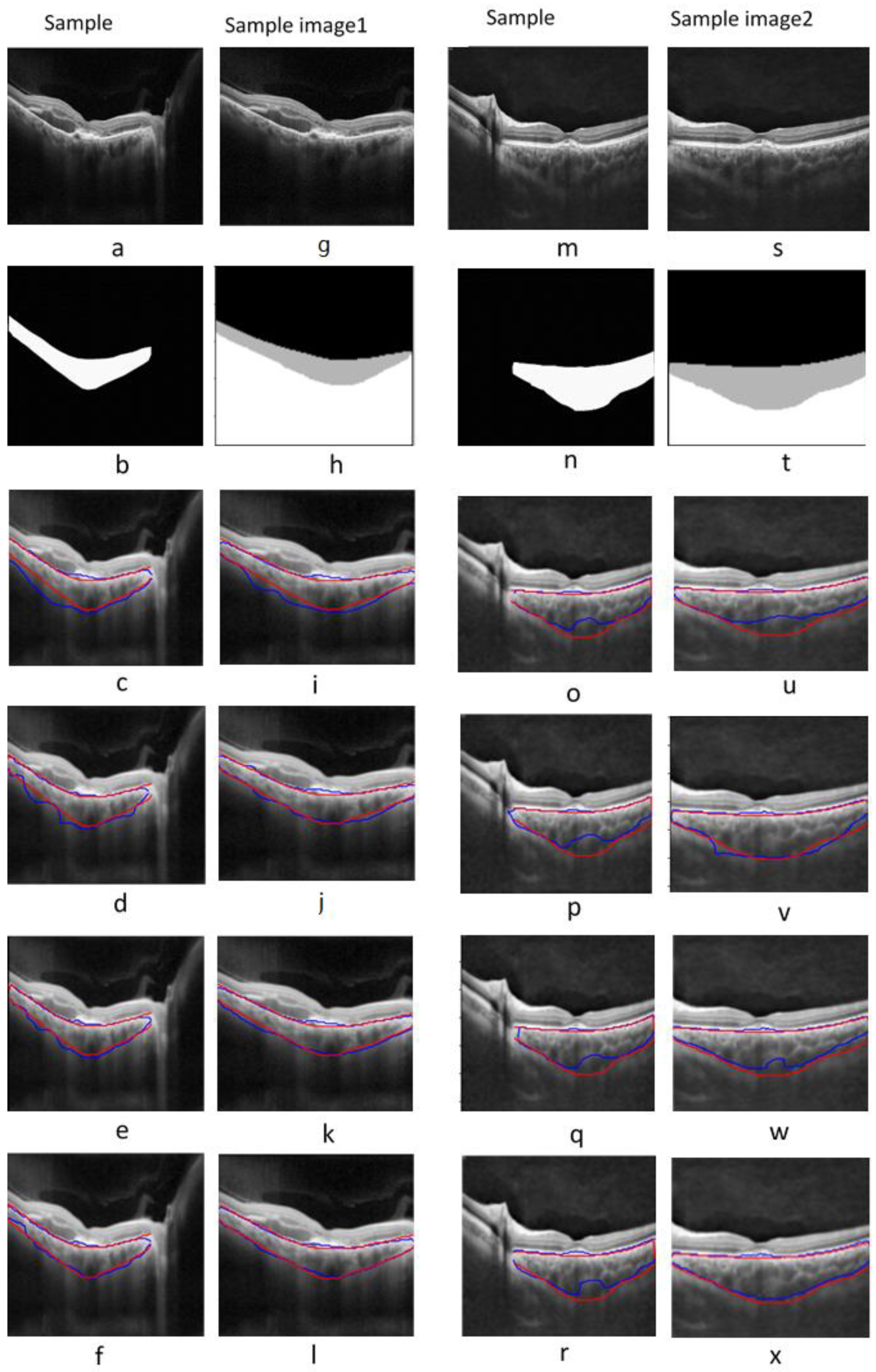
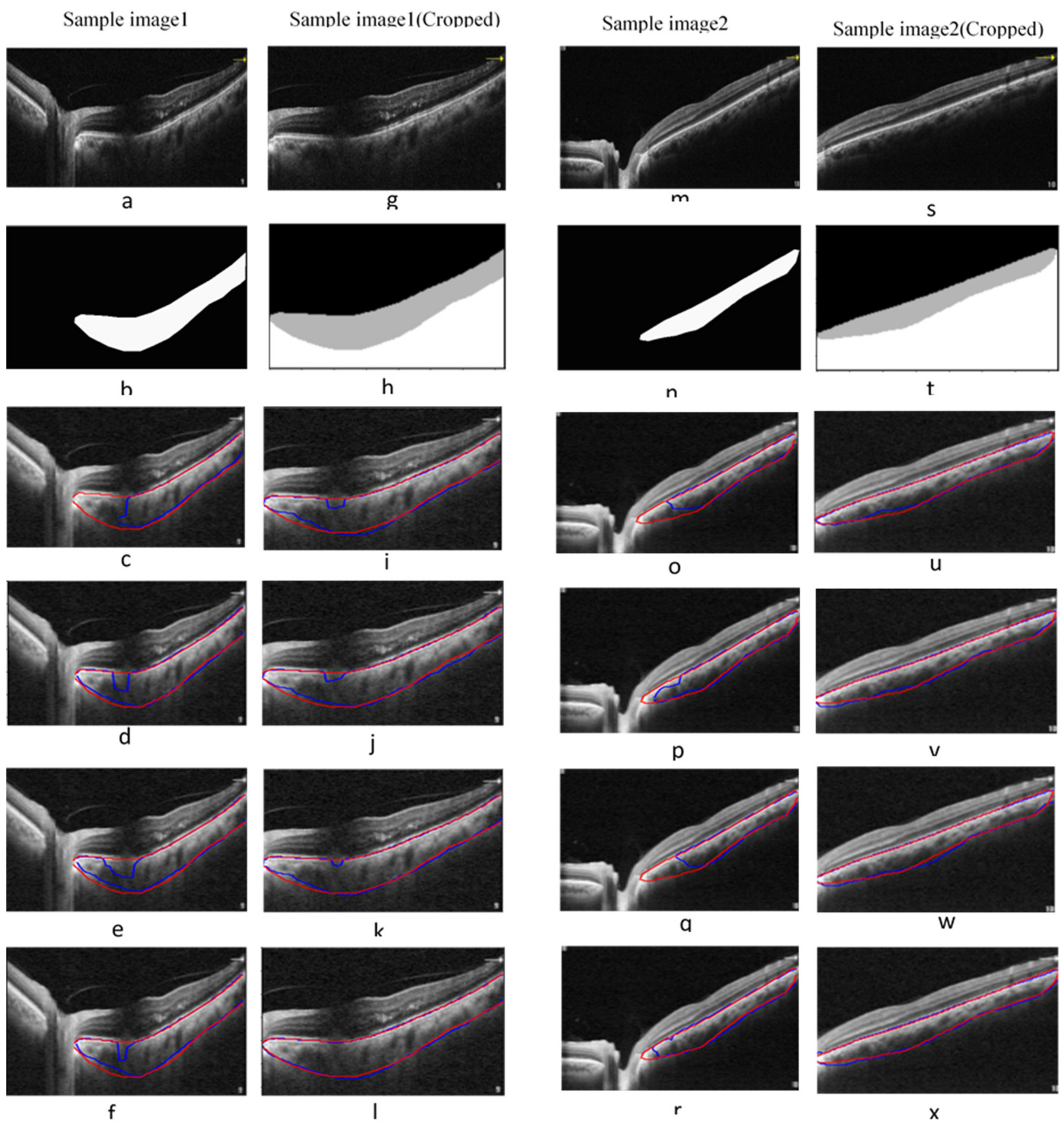
3.1. Choroidal Boundary Segmentation in Pachychoroid Spectrum Dataset
3.2. Choroidal Boundary Segmentation in the Diabetic Retinopathy Dataset
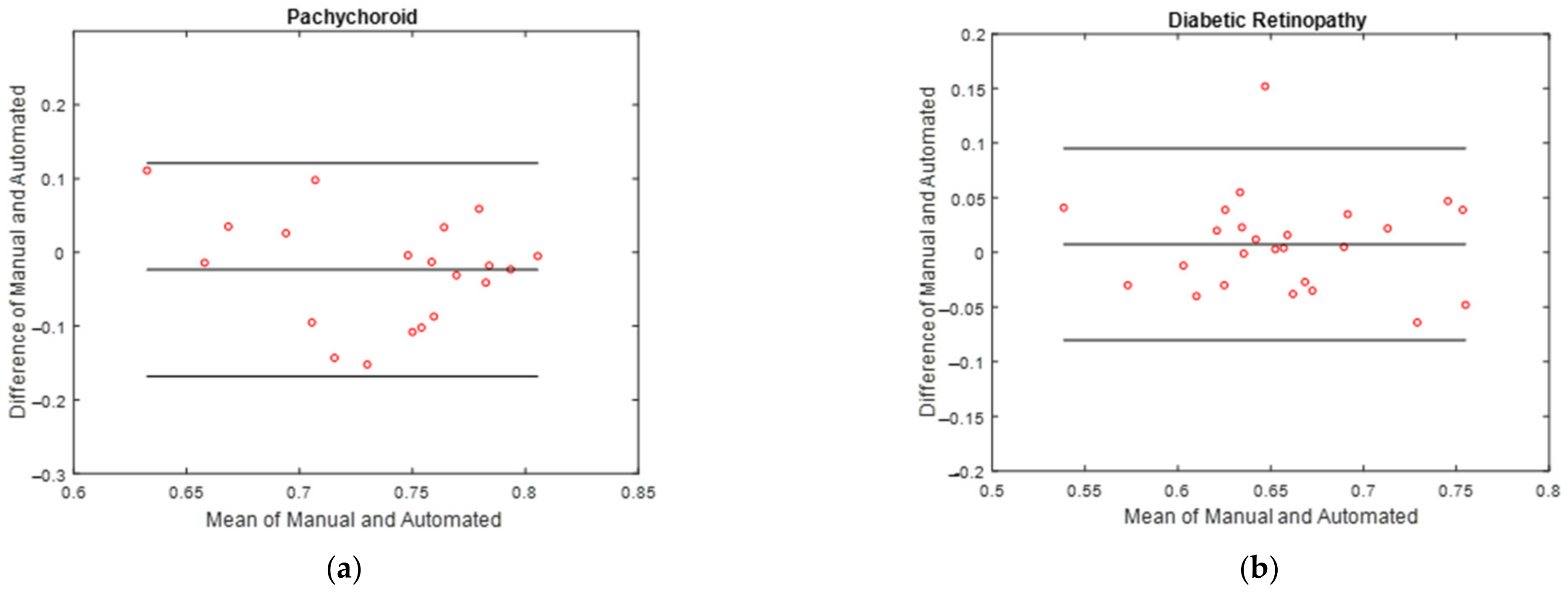
3.3. Vascular LA Segmentation CVI Measurement
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Nickla, D.L.; Wallman, J. The multifunctional choroid. Prog. Retin. Eye Res. 2010, 29, 144–168. [Google Scholar] [CrossRef] [PubMed]
- Tan, K.-A.; Gupta, P.; Agarwal, A.; Chhablani, J.; Cheng, C.-Y.; Keane, P.A.; Agrawal, R. State of science: Choroidal thickness and systemic health. Surv. Ophthalmol. 2016, 61, 566–581. [Google Scholar] [CrossRef]
- Singh, S.R.; Vupparaboina, K.K.; Goud, A.; Dansingani, K.K.; Chhablani, J. Choroidal imaging biomarkers. Surv. Ophthalmol. 2019, 64, 312–333. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, R.; Ding, J.; Sen, P.; Rousselot, A.; Chan, A.; Nivison-Smith, L.; Wei, X.; Mahajan, S.; Kim, R.; Mishra, C.; et al. Exploring choroidal angioarchitecture in health and disease using choroidal vascularity index. Prog. Retin. Eye Res. 2020, 77, 100829. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, R.; Salman, M.; Tan, K.-A.; Karampelas, M.; Sim, D.A.; Keane, P.A.; Pavesio, C. Choroidal Vascularity Index (CVI)—A Novel Optical Coherence Tomography Parameter for Monitoring Patients with Panuveitis? PLoS ONE 2016, 11, e0146344. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, R.; Gupta, P.; Tan, K.-A.; Cheung, C.M.G.; Wong, T.-Y.; Cheng, C.-Y. Choroidal vascularity index as a measure of vascular status of the choroid: Measurements in healthy eyes from a population-based study. Sci. Rep. 2016, 6, 21090. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Medrano, J.; Ruiz-Moreno, J.M.; Goud, A.; Vupparaboina, K.K.; Jana, S.; Chhablani, J. Age-related changes in choroidal vascular density of healthy subjects based on image binarization of swept-source optical coherence tomography. Retina 2018, 38, 508–515. [Google Scholar] [CrossRef]
- Agrawal, R.; Chhablani, J.; Tan, K.A.; Shah, S.; Sarvaiya, C.; Banker, A. CHOROIDAL VASCULARITY INDEX IN CENTRAL SEROUS CHORIORETINOPATHY. Retina 2016, 36, 1646–1651. [Google Scholar] [CrossRef]
- Sezer, T.; Altınışık, M.; Koytak, I.A.; Özdemir, M.H. The Choroid and Optical Coherence Tomography. Turk. J. Ophthalmol. 2016, 46, 30–37. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Ting, D.S.W.; Ng, W.Y.; Khandelwal, N.; Agrawal, R.; Cheung, C.M.G. Choroidal vascularity index—A novel optical coherence tomography based parameter in patients with exudative age-related macular degeneration. Retina 2016, 37, 1120–1125. [Google Scholar] [CrossRef]
- Chhablani, J.; Barteselli, G. Clinical applications of choroidal imaging technologies. Indian J. Ophthalmol. 2015, 63, 384–390. [Google Scholar] [CrossRef]
- Danesh, H.; Kafieh, R.; Rabbani, H.; Hajizadeh, F. Segmentation of Choroidal Boundary in Enhanced Depth Imaging OCTs Using a Multiresolution Texture Based Modeling in Graph Cuts. Comput. Math. Methods Med. 2014, 2014, 479268. [Google Scholar] [CrossRef]
- Li, K.; Wu, X.; Chen, D.; Sonka, M. Optimal Surface Segmentation in Volumetric Images-A Graph-Theoretic Approach. IEEE Trans. Pattern Anal. Mach. Intell. 2005, 28, 119–134. [Google Scholar] [CrossRef]
- Salafian, B.; Kafieh, R.; Rashno, A.; Pourazizi, M.; Sadri, S. Automatic segmentation of choroid layer in edi oct images using graph theory in neutrosophic space. arXiv 2018, arXiv:181201989. [Google Scholar]
- Yazdanpanah, A.; Hamarneh, G.; Smith, B.; Sarunic, M. Intra-retinal layer segmentation in optical coherence tomography using an active contour approach. In Proceedings of the 2009 International Conference on Medical Image Computing and Computer-Assisted Intervention, London, UK, 20–24 September 2009; Springer: Berlin/Heidelber, Germany, 2009. [Google Scholar]
- Haque, I.R.I.; Neubert, J. Deep learning approaches to biomedical image segmentation. Inform. Med. Unlocked 2020, 18, 100297. [Google Scholar] [CrossRef]
- Mahmoudi, T.; Kouzahkanan, Z.M.; Radmard, A.R.; Kafieh, R.; Salehnia, A.; Davarpanah, A.H.; Arabalibeik, H.; Ahmadian, A. Segmentation of pancreatic ductal adenocarcinoma (PDAC) and surrounding vessels in CT images using deep convolutional neural networks and texture descriptors. Sci. Rep. 2022, 12, 3092. [Google Scholar] [CrossRef]
- Pasyar, P.; Mahmoudi, T.; Kouzehkanan, S.-Z.M.; Ahmadian, A.; Arabalibeik, H.; Soltanian, N.; Radmard, A.R. Hybrid classification of diffuse liver diseases in ultrasound images using deep convolutional neural networks. Inform. Med. Unlocked 2020, 22, 100496. [Google Scholar] [CrossRef]
- Sobhaninia, Z.; Danesh, H.; Kafieh, R.; Balaji, J.J.; Lakshminarayanan, V. Determination of foveal avascular zone parameters using a new location-aware deep-learning method. Proc. SPIE 2021, 11843, 1184311. [Google Scholar] [CrossRef]
- Kugelman, J.; Alonso-Caneiro, D.; Read, S.A.; Hamwood, J.; Vincent, S.J.; Chen, F.K.; Collins, M.J. Automatic choroidal segmentation in OCT images using supervised deep learning methods. Sci. Rep. 2019, 9, 13298. [Google Scholar] [CrossRef]
- Masood, S.; Fang, R.; Li, P.; Li, H.; Sheng, B.; Mathavan, A.; Wang, X.; Yang, P.; Wu, Q.; Qin, J.; et al. Automatic Choroid Layer Segmentation from Optical Coherence Tomography Images Using Deep Learning. Sci. Rep. 2019, 9, 3058. [Google Scholar] [CrossRef] [PubMed]
- He, F.; Chun, R.K.M.; Qiu, Z.; Yu, S.; Shi, Y.; To, C.H.; Chen, X. Choroid Segmentation of Retinal OCT Images Based on CNN Classifier and l2-Lq Fitter. Comput. Math. Methods Med. 2021, 2021, 8882801. [Google Scholar] [CrossRef]
- Sui, X.; Zheng, Y.; Wei, B.; Bi, H.; Wu, J.; Pan, X.; Yin, Y.; Zhang, S. Choroid segmentation from Optical Coherence Tomography with graph-edge weights learned from deep convolutional neural networks. Neurocomputing 2017, 237, 332–341. [Google Scholar] [CrossRef]
- Tsuji, S.; Sekiryu, T.; Sugano, Y.; Ojima, A.; Kasai, A.; Okamoto, M.; Eifuku, S. Semantic Segmentation of the Choroid in Swept Source Optical Coherence Tomography Images for Volumetrics. Sci. Rep. 2020, 10, 1088. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.; Zhao, Y.; Chen, B.; Ma, Y.; Gu, Z.; Gu, S.; Yang, J.; Cheng, J.; Liu, J. Deep Learning with Skip Connection Attention for Choroid Layer Segmentation in OCT Images. In Proceedings of the 2020 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Montreal, QC, Canada, 20–24 July 2020. [Google Scholar]
- Xu, Y.; Yan, K.; Kim, J.; Wang, X.; Li, C.; Su, L.; Yu, S.; Xu, X.; Feng, D.D. Dual-stage deep learning framework for pigment epithelium detachment seg-mentation in polypoidal choroidal vasculopathy. Biomed. Opt. Express 2017, 8, 4061–4076. [Google Scholar] [CrossRef]
- Zheng, G.; Jiang, Y.; Shi, C.; Miao, H.; Yu, X.; Wang, Y.; Chen, S.; Lin, Z.; Wang, W.; Lu, F.; et al. Deep learning algorithms to segment and quantify the choroidal thickness and vasculature in swept-source optical coherence tomography images. J. Innov. Opt. Health Sci. 2020, 14, 2140002. [Google Scholar] [CrossRef]
- Vupparaboina, K.K.; Dansingani, K.K.; Goud, A.; Rasheed, M.A.; Jawed, F.; Jana, S.; Richhariya, A.; Freund, K.B.; Chhablani, J. Quantitative shadow compensated optical coherence tomography of choroidal vasculature. Sci. Rep. 2018, 8, 6461. [Google Scholar] [CrossRef]
- Betzler, B.K.; Ding, J.; Wei, X.; Lee, J.M.; Grewal, D.S.; Fekrat, S.; Sadda, S.R.; Zarbin, M.A.; Agarwal, A.; Gupta, V.; et al. Choroidal vascularity index: A step towards software as a medical device. Br. J. Ophthalmol. 2021, 106, 149–155. [Google Scholar] [CrossRef] [PubMed]
- Sonoda, S.; Sakamoto, T.; Yamashita, T.; Uchino, E.; Kawano, H.; Yoshihara, N.; Terasaki, H.; Shirasawa, M.; Tomita, M.; Ishibashi, T. Luminal and stromal areas of choroid deter-mined by binarization method of optical coherence tomographic images. Am. J. Ophthalmol. 2015, 159, 1123–1131. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; Springer: Berlin/Heidelberg, Germany, 2015. [Google Scholar]
- Salehi, S.S.M.; Erdogmus, D.; Gholipour, A. Tversky loss function for image segmentation using 3D fully convolutional deep networks. In Proceedings of the International Workshop on Machine Learning in Medical Imaging, Quebec City, QC, Canada, 10 September 2017; pp. 379–387. [Google Scholar]
- Phan, T.H.; Yamamoto, K. Resolving class imbalance in object detection with weighted cross entropy losses. arXiv 2020, arXiv:200601413. [Google Scholar]
- Buades, A.; Coll, B.; Morel, J.-M. Non-local means denoising. Image Process. Line 2011, 1, 208–212. [Google Scholar] [CrossRef]
- Stokes, S. Available online: https://github.com/sarastokes/OCT-tools (accessed on 1 February 2019).
- Agrawal, R.; Seen, S.; Vaishnavi, S.; Vupparaboina, K.K.; Goud, A.; Rasheed, M.A.; Chhablani, J. Choroidal Vascularity Index Using Swept-Source and Spectral-Domain Optical Coherence Tomography: A Comparative Study. Ophthalmic Surg. Lasers Imaging Retin. 2019, 50, e26–e32. [Google Scholar] [CrossRef] [PubMed]

| Paper | Data | Device Name | Method | Loss Function | Metrics | Performance | |
|---|---|---|---|---|---|---|---|
| Mao et al. [25] | 20 normal human subjects | Topcon DRI-OCT-1 | SCA-CENet | Not reported | Sensitivity | 0.918 | |
| F1-score | 0.952 | ||||||
| Dice coefficient | 0.951 | ||||||
| IoU | 0.909 | ||||||
| Mean absolute error (MAE) (BM) (in pixels) | 1.945 | ||||||
| MAE (SCI) (in pixels) | 8.946 | ||||||
| Kugelman et al. [20] | 99 children, 594 B-scans | SD-OCT | Patch-based classification (CNN-RNN) | Tverksy loss | Mean error (ME) (in pixels) | ILM | 0.01 |
| BM | 0.03 | ||||||
| CSI | −0.02 | ||||||
| MAE (in pixels) | ILM | 0.45 | |||||
| BM | 0.46 | ||||||
| CSI | 3.22 | ||||||
| Masood et al. [21] | 11 normal, 4 Shortsightedness, 4 glaucoma, 3 DME | Swept-source OCT | Morphological processing and CNN | Cross entropy loss | ME (in pixels) | BM | 0.43 ± 1.01 |
| Choroid | 2.8 ± 1.50 | ||||||
| MAE (in pixels) | BM | 1.39 ± 0.25 | |||||
| Choroid | 2.89 ± 1.05 | ||||||
| Sui et al. [23] | 912 B-scans (618 B-scans normal, 294 macular edema) | EDI-OCT | Graph-based and CNN | MSE | MAE (in pixels) | BM | 4.6 ± 4.8 |
| CSI | 11.4 ± 11.0 | ||||||
| Xu et al. [26] | 50 PCV patients (1800 B-scans) | SD-OCT | Dual-stage DNN | Log-loss | MAE (in µm) | BM | 5.71 ± 3.53 |
| Tsuji et al. [24] | 43 eyes from 34 healthy individual | SS-OCT | SegNet and graph cut | Not reported | Dice coefficient | 0.909 ± 0.505 | |
| He et al. [22] | 146 OCT images | SD-OCT | Patch-based CNN classifier | Focal loss | Dice coefficient | 0.904 ± 0.055 | |
| Zheng et al. [27] | 450 images from 12 healthy individual | SS-OCT | Residual U-Net | Not reported | Failure ratio less than 0.02 mm | 68.84% | |
| Data | Pachychoroid Spectrum | Diabetic Retinopathy |
|---|---|---|
| No. of patients | 44 | 112 |
| No. of B-scan Images | 98 | 439 |
| Mean age (mean ± SD) | 50.6 ± 11.2 (range: 29–74 years) | 61 ± 8 (range:47–78 years) |
| Gender | 37 (84.1%) male | 59 (52.6%) male |
| Best corrected visual acuity (BCVA) (mean ± SD) | 0.50 ± 0.38 | 0.57 ± 0.25 |
| Subfoveal choroidal thickness (range in µm) | (265–510 µm) | (135–370 µm) |
| Resolution | 3000 × 12,000 µm | 1500 × 12,000 µm |
| Interclass Correlation Coefficient (ICC) | 95% Confidence Interval (CI) | |
|---|---|---|
| CVI | 0.969 | 0.918–0.988 |
| Parameter of Niblack’s Algorithm | Diabetic Retinopathy Images | Pachychoroid Spectrum Images |
|---|---|---|
| Window Size | 15 | 13 |
| K | 0.001 | 0.01 |
| Threshold Coefficient | 1.03 | 1.02 |
| Unit | Loss Function | Two-Class Segmentation | Three-Class Segmentation | ||||||
|---|---|---|---|---|---|---|---|---|---|
| BM Boundary | Choroid Boundary | BM Boundary | Choroid Boundary | ||||||
| U_E | S_E | U_E | S_E | U_E | S_E | U_E | S_E | ||
| µm | Loss#1 | 44.7 | −9.6 | 95.7 | −37.8 | 22.2 | −16.5 | 91.5 | 16.8 |
| Loss#2 | 33.9 | −31.8 | 92.7 | 22.5 | 21.9 | −19.8 | 78 | 12.6 | |
| Loss#3 | 39 | 3.9 | 87.3 | 11.7 | 22.5 | −11.1 | 78.3 | 15.3 | |
| Loss#4 | 30.3 | −25.5 | 84.3 | −5.7 | 21.6 | −14.4 | 76.2 | −25.5 | |
| Unit | Loss Function | Two-Class Segmentation | Three-Class Segmentation | ||||||
|---|---|---|---|---|---|---|---|---|---|
| BM Boundary | Sclerochoroidal Boundary | BM Boundary | Sclerochoroidal Boundary | ||||||
| U_E | S_E | U_E | S_E | U_E | S_E | U_E | S_E | ||
| µm | Loss#1 | 30.15 | 6.75 | 45.15 | 19.35 | 4.95 | −4.35 | 21.45 | −3.3 |
| Loss#2 | 22.5 | 3.45 | 37.2 | 14.55 | 3.15 | −1.5 | 20.85 | 0.45 | |
| Loss#3 | 22.8 | 4.8 | 37.65 | 16.5 | 3.15 | −1.05 | 22.2 | −7.35 | |
| Loss#4 | 21.15 | 1.65 | 31.35 | −0.9 | 3 | 0.15 | 20.7 | 0.15 | |
| Unit | Method | Pachychoroid | Diabetic Retinopathy | ||||||
|---|---|---|---|---|---|---|---|---|---|
| BM Boundary | Choroid Boundary | BM Boundary | Choroid Boundary | ||||||
| U_E | S_E | U_E | S_E | U_E | S_E | U_E | S_E | ||
| µm | Proposed model | 21.6 | −14.4 | 76.2 | −25.5 | 3 | 0.15 | 20.7 | 0.15 |
| Semantic segmentation [20] | 27.1 | −5.76 | 88.5 | 45.7 | 4.5 | 1.56 | 29.3 | −4.67 | |
| Patch-based [20] | 32.2 | 25.3 | 96.5 | 4.32 | 6.1 | −3.4 | 40.1 | 0.26 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arian, R.; Mahmoudi, T.; Riazi-Esfahani, H.; Faghihi, H.; Mirshahi, A.; Ghassemi, F.; Khodabande, A.; Kafieh, R.; Khalili Pour, E. Automatic Choroid Vascularity Index Calculation in Optical Coherence Tomography Images with Low-Contrast Sclerochoroidal Junction Using Deep Learning. Photonics 2023, 10, 234. https://doi.org/10.3390/photonics10030234
Arian R, Mahmoudi T, Riazi-Esfahani H, Faghihi H, Mirshahi A, Ghassemi F, Khodabande A, Kafieh R, Khalili Pour E. Automatic Choroid Vascularity Index Calculation in Optical Coherence Tomography Images with Low-Contrast Sclerochoroidal Junction Using Deep Learning. Photonics. 2023; 10(3):234. https://doi.org/10.3390/photonics10030234
Chicago/Turabian StyleArian, Roya, Tahereh Mahmoudi, Hamid Riazi-Esfahani, Hooshang Faghihi, Ahmad Mirshahi, Fariba Ghassemi, Alireza Khodabande, Raheleh Kafieh, and Elias Khalili Pour. 2023. "Automatic Choroid Vascularity Index Calculation in Optical Coherence Tomography Images with Low-Contrast Sclerochoroidal Junction Using Deep Learning" Photonics 10, no. 3: 234. https://doi.org/10.3390/photonics10030234
APA StyleArian, R., Mahmoudi, T., Riazi-Esfahani, H., Faghihi, H., Mirshahi, A., Ghassemi, F., Khodabande, A., Kafieh, R., & Khalili Pour, E. (2023). Automatic Choroid Vascularity Index Calculation in Optical Coherence Tomography Images with Low-Contrast Sclerochoroidal Junction Using Deep Learning. Photonics, 10(3), 234. https://doi.org/10.3390/photonics10030234






