Abstract
Artificial intelligence (AI) represents a growing and promising branch of computer science that is expanding the horizon of prediction, screening, and disease monitoring. The use of multimodal imaging in retinal diseases is particularly advantageous to valorize the integration of machine learning and deep learning for early diagnosis, prediction, and management of retinal disorders. In age-related macular degeneration (AMD) beyond its diagnosis and characterization, the prediction of AMD high-risk phenotypes evolving into late forms remains a critical point. The main multimodal imaging modalities adopted included color fundus photography, fundus autofluorescence, and optical coherence tomography (OCT), which represents undoubtful advantages over other methods. OCT features identified as predictors of late AMD include the morphometric evaluation of retinal layers, drusen volume and topographic distribution, reticular pseudodrusen, and hyperreflective foci quantification. The present narrative review proposes to analyze the current evidence on AI models and biomarkers identified to predict disease progression with particular attention to OCT-based features and to highlight potential perspectives for future research.
1. Introduction
Artificial Intelligence (AI) is a new branch of computer science that has significantly revolutionized the field of medicine [1,2,3]. The development of AI has allowed applications in several fields of ophthalmology, particularly in retinal disorders. The most significant advances were performed in age-related macular degeneration (AMD), covering different clinical aspects including screening, diagnosis, prediction, and monitoring [4]. The most critical element in managing AMD is still represented by the identification of predictive models allowing prompt identification of patients at risk on a large scale. It can be advantageous to refer individuals who deserve further testing, treatment, and more strict follow-up examinations [5]. The early detection of high-risk AMD phenotypes is particularly useful to predict future exudation that may benefit from timely management with anti-vascular endothelial growth factor (anti-VEGF), leading to better clinical outcomes [6]. Although no current treatment is available for GA, promising therapies are on the horizon, highlighting the need to refine disease activity and the prognostic implications and potential clinical endpoints to be incorporated in future clinical trials [7,8,9,10,11,12,13].
Despite the extensive literature available on screening and diagnosis with high performance in detecting AMD at any stage [14,15,16,17,18], one of the main challenges remains the assessment of the risk of conversion and disease progression [19]. Thus, the need to expand the knowledge on potential predictors and models that can help predict AMD phenotypes evolving into late stages. Regarding imaging biomarkers, the use of optical coherence tomography (OCT)-based features seems to present several technical and practical advantages. Beyond its use on a large scale in routine clinical practice for the diagnosis and management of AMD and in trials [20], the high resolution of both spectral-domain and swept-source technologies offers the opportunity to identify preceding alterations and early stages of the disease before the lesions appear clinically evident. Therefore, recently, OCT-defined classification of atrophy and neovascular features in the setting of AMD has been precisely defined by a consensus group of leading experts [21,22,23]. Furthermore, several OCT qualitative features have been identified, enabling the evaluation of disease progression and treatment response and corroborating the importance of OCT biomarker identification to achieve optimal AMD management [24].
AI based on deep learning (DL) offered tremendous advantages in ophthalmology, providing the opportunity to be applied to medical imaging analysis. DL techniques found the best application in retinal pathologies, where ocular imaging is routinely used for diagnosis and management, including fundus photographs, fundus autofluorescence, and OCT [14,25,26,27,28]. Understanding the existing DL models and biomarkers explored so far may further expand the development of more accurate predictive models in AMD.
This narrative review proposes to analyze the strengths and weaknesses of the existing AI predictive models. Imaging technologies, biomarkers, and AI models are analyzed and discussed to improve the understanding of the key elements considered in the models. These aspects would help delineate future research on biomarker identification, imaging modalities, and refining predictive AI models.
2. Historical Background and Principles of Artificial Intelligence in Ophthalmology
The term AI was first coined by John McCarthy in 1956 and referred to “hardware or software that exhibits behavior which appears intelligent”, capable of resembling human intelligence in useful tasks such as learning, identifying images, and problem solving [29]. AI has proved to be particularly suitable to imaging-centric specialties, and thus exceptionally useful in ophthalmology, showing efficacy comparable to that of the specialist mainly in identifying diseases with a high incidence, such as glaucoma, retinopathy of prematurity, diabetic retinopathy, and AMD [30,31,32,33].
More specifically, the integration of AI machine learning (ML) and DL in ophthalmic settings for the early diagnosis, prediction, and timely treatment of the most common sight-threatening eye diseases is an urgent need given the lifespan extension and, consequently, a large amount of medical data. Within this framework, AI technologies have the promising potential to revolutionize ophthalmic health care services creating a significative clinical impact and minimizing doctor burden [34,35]. ML is a subfield of AI technology that was introduced in the 1980s and includes DL and conventional machine learning (CML). ML allows the computer system to learn and improve how to complete a task on its own without being explicitly programmed [36]. Among CML algorithms used in AI, random forests (RF) [37] and support vector machines (SVM) [38] are the most commonly used in the field of ophthalmology.
DL was introduced in the 2000s as a subset of ML that learns features in data using an artificial neural network (ANN) structure inspired by the human brain structure and function. DL is composed of multiple stimulus inputs into the so-called hidden layers of neurons, each of which can learn different features from the offered stimuli. This allows this machine model to complete complex tasks resulting in the output being recognized [39].
Among the various DL methods, deep convolutional neural network (DCNN) has proved to be particularly suitable for medical image recognition [29]. DCNNs have evolved from traditional artificial neural networks, using a three-dimensional neural pattern and employing a special mathematical filtering operation called convolution [40]. Full training of DCNN implies a large amount of training data already labeled from medical experts, extensive computational and memory resources, and complicated by overfitting and convergence issues that require repetitive and time-consuming adjustments [41]. A valid alternative for CNN training from scratch involves fine-tuning a CNN using transfer learning, which involves transferring the learned features from a pre-trained CNN to initialize another task [41,42]. CNN-based transfer learning models demonstrated a good performance in classifying OCT-based features in AMD [43]. Several state-of-the-art DL techniques have been applied for retinal image segmentation, such as FCNN, U-Net, Seg-net, Deeplabv3, and AlexNet [14,44,45,46,47].
3. Model Analysis in Age-Related Macular Degeneration
DL models have been implemented using a combination of imaging and non-imaging features. Several imaging biomarkers have been identified and characterized using different imaging modalities. Most of the models were trained using large databases, such as the Age-Related Eye Disease Study (AREDS) study database [27,48,49,50], which mainly used color fundus photographs (CFP). More recently, with the multiplication of clinical trials in AMD [51,52,53,54], different imaging techniques were adopted, expanding the spectrum of the predictors. The present chapter summarizes the DL models developed using different multimodal imaging modalities.
3.1. Imaged-Based Features
3.1.1. Fundus Photographs
CNN models represented the state-of-the-art image classification in retinal diseases using CFP. Several DL models were trained to predict AMD stage from the AREDS study database with variable levels of accuracy ranging from 63.3% to 92.1% [5,14,35,55]. The images were classified according to the AREDS 9-steps plus three scales to identify ungradable images. The accuracies increased by restricting the analysis of fundus images from individuals 55 years or older, with 82.2% of sensitivity and 97.1% specificity in categorizing intermediate AMD (AREDS classes 4–9) features. The algorithm performed better for late AMD (AREDS classes 10–12) with a sensitivity of 100% and specificity of 96.5% [56]. Burlina et al. [28] used AREDS data and employed DCNN (ResNet-50 network) to stratify images in 4-step and 9-step severity scales. The five-year progression risk was then estimated by creating three DCNNs to produce three different predictions. The 9-step AMD severity scale was based on the data from AREDS report 17 [56], as summarized in Table 1.

Table 1.
The 9-step age-related macular degeneration (AMD) severity scale modified from the AREDS 17 report.
A step forward was performed by drusen quantification, which contemplated assessing the total area and number of large drusen (125 μm), and the recognition of reticular pseudodrusen (RPD), achieving an accuracy of 96% [5]. The CNN DenseNet model demonstrated superior performance to other CNNs (VGG version 16, VGG version 19, Inception V3, and ResNet version 101) in detecting RPD on both CFP and FAF images [57].
DeepSeeNet is a DL model designed as a CNN with an Inception-v3 architecture, developed to determine patient-based AREDS Simplified Severity Scale scores using bilateral CFP. This model consisted of three main subnetworks: (a) Drusen-Net (D-Net), which classifies drusen according to dimensions (small/none, medium, and large); (b) Pigment-Net (P-Net), which identifies the presence of pigmentary abnormalities; and (c) Late AMD-Net (LA-Net), which detects the presence of late complications including neovascular AMD and/or geographic atrophy. The performance of DeepSeeNet was superior to retinal specialists in classifying AMD according to AREDS scores. When considering the D-Net and P-net subnetworks, the model was superior in assessing large drusen and pigmentary abnormalities. However, the recognition of late AMD through the LA-net was similar to that of retinal specialists. Noteworthy, DeepSeeNet is publicly available on https://github.com/ncbi-nlp/DeepSeeNet (accessed on 1 November 2022) [35].
Using longitudinal images on a dataset of 4903 eyes with AMD selected from the AREDS, a three-step model comprised of a pre-trained SNN (Inception V3) was developed by reducing each image into a single feature vector; the feature vectors were then combined by applying an interval scaling to account for the uneven time intervals. Finally, a recurrent neural network classified the images according to the progression [58]. The authors compared their results with a model that used a CNN of retinal fundus images combined with single nucleotide polymorphisms (SNPs) and AMD severity to predict late AMD progression. The severity scale was assessed on a centralized grading based on the AREDS AMD scale of fundus images at each semi-annual or annual follow-up visit. The model was simplified into sub-models. The fundus image taken at the current visit was used alone or combined with the SNPs to predict whether the progression time to late AMD exceeded the inquired year [59]. By comparing the two methodological approaches, the longitudinal prognostic model, taking into account two- and three-times points, performed better over the single time point method. However, the three-time points model had a non-significant increase over the two-time points, suggesting that using more than two-time points does not affect the predictive value of the model [58].
3.1.2. Fundus Autofluorescence
Fundus autofluorescence (FAF) has a limited application in DL models for AMD, mainly restricted to geographic atrophy (GA) assessment and, more recently, to its prediction [26,56,60,61].
A recent DL model used data from the study of eyes of patients with bilateral GA enrolled in lampalizumab phase 3 clinical trials (Chroma [NCT02247479]; Spectri [NCT02247531]) or in an observational study (Proxima A [NCT02479386]) to predict GA progression. The model was designed as a regression task, with three multi-task CNN trained using FAF alone, OCT alone, and a multimodal approach (combined FAF/OCT) at the same visit. The GA growth rate prediction performance was high for both the FAF alone and the multimodal model, with r2 of 0.48 and 0.47, respectively [51].
DenseNet achieved the highest performance at 0.939 over the other 4 CNN models tested in detecting RPD on FAF. The FAF image analysis using DenseNet demonstrated the highest κ value for the primary performance metric (0.789) compared to human graders, with higher specificity and precision levels of two retinal fellows [56]. FAF-based algorithms can segment the GA lesions 6-times more quickly than human evaluation with very high performance, validation, and testing scores [59].
3.1.3. Optical Coherence Tomography
The CNN-based models were used to transfer learning for classifying OCT images. Transfer learning employed pre-trained models as the starting points to process other tasks reducing the computation time, resources, and the need to develop neuronal network models ex novo. Using pre-trained CNN with transfer learning is considered faster and easier than building a new CNN. The model can be fine-tuned to learn specific features from a new data set of OCT images [43,62].
OCT imaging modality applied in AI prediction models is gaining consideration as it provides both a qualitative assessment of the drusen and other features such as hyperreflective foci (HRF) and a quantitative estimation of the morphometric changes in retinal layers [63,64,65,66,67,68,69]. Different strategies have been developed to create predictive models using retinal morphological features. To extract and measure OCT features, fully automated image analysis is essential to identify specific biomarkers of interest, such as the status of outer retinal layers, drusen, RPD, and HRF [70,71]. The main biomarkers considered for AMD progression included the automated drusen segmentation on OCT volumetric cube extrapolating different quantitative drusen features such as number, mean volume, topographic distribution, maximum height, slope, reflectivity, and drusen area. Furthermore, the presence and distribution of HRF, choroidal thickness, presence of RPD, and photoreceptor outer segment loss were also estimated for an accurate prediction [72,73,74].
3.2. Non-Imaging Features
3.2.1. Demographic Features
Demographic features are confounding factors for AMD progression that should be incorporated to implement the risk prediction algorithm [75]. Implementing a hybrid sequential prediction model permitted to incorporate longitudinal OCT images and demographic information in a recursive neural network (RNN) model. The demographic factors included age, gender, race, smoking status, and visual acuity. The model performance in predicting AMD progression was high in both the short term, with an AUC of 0.96 within 3 months, and long term, reaching an AUC of 0.97 within 21 months, supporting the importance of combining imaging and demographic factors [70]. A 5-year predictive model revealed an AUROC of 0.92, obtained by incorporating baseline demographic features with both qualitative and quantitative OCT features [76].
An ML algorithm predicting the progression from early into late AMD at 1 or 2 years was built using a logistic model tree (LMT), combining socio-demographic characteristics with color fundus images. The demographic parameters included gender, age, smoking status, diabetes, body mass index, blood pressure, sunlight exposure, visual acuity, and AMD in the fellow eye. By stratifying the patients according to gender, smoking, and age, the authors noticed that all the models performed better on females and nonsmokers, while the 1-year models performed worse in subjects less than 60 years old [5].
Another model obtained by the bootstrap least absolute shrinkage and selection operator (LASSO) considered a combination of retinal phenotypes, demographic characteristics, and genetic features. The model found that the most relevant non-imaging predictors were represented by age, smoking status, pulse pressure, education, and Mediterranean diet score, with AUCs comprised between 0.88 and 0.92 [77].
3.2.2. Genetic Factors
Genetic features integrated into predictive models incorporated risk alleles of single-nucleotide polymorphisms (SNPs) at different AMD-associated loci [69]. Despite the strong evidence of a genetic association in advanced AMD, no univocal associations were found to be directly linked to GA progression [78]. Likewise, DL models interpolating demographic, genetic, and imaging features failed to identify specific AMD risk-associated SNPs with a predictive value for neovascularization and GA [69]. Yan et al. [58] combined 52 AMD genetic variants previously identified (Appendix A, Table A1) [79] with fundus images and AMD severity, demonstrating that the addition of genotypes improved only modestly the accuracy of the predictive model. Moreover, Peng et al. [80] confirmed that the 52-SNPs AMD genetic risk score did not improve the prediction of the DL model at 5 years, while the accuracy only slightly improved when considering two SNPs (ARMS2 rs10490924, CFH rs1061170).
A bootstrap LASSO model indicated that the genetic risk score was one of the four most relevant predictors, together with the presence of intermediate drusen, the AREDS simplified scale, and age. The prediction model had high performances with AUC between 0.91 and 0.92, but when subtracting the genetic score the model was similar with AUCs between 0.88 and 0.93. The authors specified that in a minority of cases the genetic evaluation helped identify high-risk patients, especially when the simplified AREDS score [76] was 0 in patients with a high genetic score instead [62]. Taken together, the results from different studies suggested that including genotype information may have a marginal role in the predictive models, with a negligible contribution to the predictive power further complicated by the scarce availability of genetic analysis in clinical practice.
4. Automated Analysis of OCT Biomarkers and Morphometric Parameters
The use of DL models allowed the automatic quantification of imaging biomarkers and morphometrics parameters, revolutionizing the clinical approach to predicting disease progression [25]. The leading automated imaging analyses included the evaluation of retinal layers thickness, drusen volume and topographic distribution, reticular pseudodrusen estimation, and hyperreflective foci distribution. These features have an important role in predicting AMD progression, as summarized below.
4.1. Retinal Layers Morphometric Analysis
The morphometric changes in retinal layers were deemed important quantitative biomarkers of AMD progression. Studies have demonstrated a progressive thinning of the outer retinal layers reflecting photoreceptor degeneration, but also indicating involvement of rod, bipolar, and horizontal cells [81,82,83]. Structure–function correlations further corroborated the role of outer nuclear layer (ONL) thickness as a surrogate of cone-rod dysfunction [65,66,84]. The progressive ONL thinning was considered a robust predictor of conversion with a faster thinning in eyes developing GA [68].
Ellipsoid zone (EZ) retinal pigment epithelium (RPE) thickness was included as an OCT surrogate of photoreceptor outer segment length and EZ integrity, while the RPE-Bruch’s membrane (BrM) thickness was included as a morphometric parameter for drusen load and topographical distribution [75,85]. The segmentation of RPE with photoreceptors was previously labeled as RPE+ inner segment/outer segment (IS/OS) [69]. OCT B-scans segmentation also included the retinal nerve fiber layer, ganglion cell layer, inner plexiform layer, and the choroid performed through the CNN model using the raw B-scan with normalized intensity as input. Retinal layers segmentation can be better visualized using the en face thickness and drusen maps [86].
Retinal thickness maps obtained through a fine-tuned DCNN (VGG16) were compared to a CNN model trained from scratch (AMDnet), demonstrating that the layer segmentation-based preprocessing showed the strongest predictive power for progression into advanced AMD, reaching AUC of 0.89 for the B-scan and 0.91 for the volumes [87].
4.2. Drusen Volumetric Evaluation and Reticular Pseudodrusen Estimation
Automated drusen segmentation algorithms have been developed that allow easy drusen growth and regression detection over time [65,84,88]. Considering drusen as RPE elevations, the delineation of the RPE with respect to the BrM plane into every single B-scan resulted in a three-dimensional segmentation and a topographical map of the drusen in the OCT volume [70]. Drusen thickness is often reported with an en face topographical representation with quantitative correlate displayed in a colorimetric scale [65,67,69,84]. The quantification of total drusen area, number of large drusen (≥125 μm), and RPD were used for late AMD prediction increasing the predictive model performance [5,67,88]. The most remarkable drusen features involved in the prediction included the mean drusen thickness, maximum drusen height, and the mean drusen attenuation considering the attenuation within drusen in comparison with the overlying outer retinal bands and the ONL [89].
The recognition of RPD represents a crucial element in AMD progression [90,91,92,93,94,95,96,97,98] that was largely overlooked in AREDS studies for using CFP as the only imaging modality. The proper identification of RPD needs a multimodal imaging approach, where the use of FAF, near-infrared reflectance, and OCT allows the correctly identification of the stage and distribution of such subretinal lesions representing the histopathological correlate of subretinal drusenoid deposits (SDD) [93,99,100,101]. The deep learning detection of RPD was possible by utilizing a large dataset provided by the AREDS2 ancillary study of FAF imaging, using a standard protocol for their recognition and grading [102]. The DenseNet model could discern the presence of RPD on FAF with high accuracy (AUC 0.94). Label transfer was applied between FAF and CFP images and classified the corresponding CFP image according to the graded FAF for model training. The model then identified RPD on CFP with an AUC of 0.83 and a high specificity of 0.90. Therefore, the DenseNet DL model was considered highly accurate in identifying RPD presence from FAF images, suggesting that the model can exceed the performance of nonspecialized ophthalmologists in routine clinical practice [56]. Agrón et al. [103] used a simplified severity scale for AMD [48] (Figure 1), assigning 1 point for each feature in both eyes, but in the absence of large drusen 1 point was assigned if both eyes had medium-sized drusen. Moreover, the original scale modification consisted of including any GA (including noncentral GA) as late AMD. The authors found that the presence of RPD was associated with a higher risk for progression (Hazard ratio, HR of 4.7, 95%CI, 3.9–5.8) independently from the severity scale. When stratifying the patients according to the severity levels, the RPD risk was less prominent and non-significant for the 3–4 levels [103].
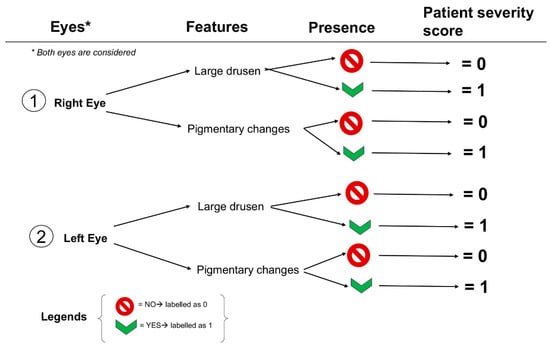
Figure 1.
A schematization of the 4-steps simplified severity scale from AREDS report No. 18.
4.3. Hyperreflective Foci
Hyperreflective foci (HRF) on OCT were identified as well-circumscribed hyperreflective roundish formations above drusen [75]. The identification of HRF is of leading importance in neovascular conversion, representing a distinctive hallmark alone and a more robust indicator when overlaying drusen [64,69]. HRF are also important predictors of GA development, likely representing the histopathological correlates of migrating RPE cells and disaggregated photoreceptors. The increasing number of HRF, often accompanying a reduced retinal thickness and ONL thinning, was found to be associated with RPE atrophy increase [62,63,69,104,105].
An approach to defining the presence of HRF consisted of identifying a set of connected HRF voxels greater or equal to four, corresponding to approximately 5.570 μm3, through a connected components algorithm [106]. A random forest classifier was also trained to provide a probability that a given pixel on B-scan corresponded to HRF, refining the results through an iterative approach considering previous classifications. A total of 150 annotated B-scans by certified readers were used as a dataset for the classifier training [88].
A component filtering algorithm was applied, and all the HRF colocalizing with retinal vessels were removed to account for the potential interference of retinal vessels leading to a false estimation of HRF. In this study, HRF voxels detected in a three-dimensional OCT volume were identified through a U-NET semantic segmentation architecture trained on a pre-existing dataset. Moreover, to assess the HRF dynamics, en face HRF thickness maps were compared between consecutive examinations [107]. The automated identification of HRF allowed evaluation of not only the presence but also load and dynamics, which were best obtained using a ResUNet+ model tested on Cirrus and Spectralis OCTs. The authors suggested preferring the utilization of cross entropy training loss over Dice-based training to obtain a higher performance in HRF automated identification [67,108].
5. Predictive Models for Disease Progression in Age-Related Macular Degeneration
5.1. Geographic Atrophy Prediction
A predictive model of GA conversion indicated outer retinal (RPE+ inner segment/outer segment -IS/OS) and ONL thinning and increasing HRF at the ONL layer as morphometric factors predominantly involved; the only non-imaging factor was represented by age. SNPs did not contribute significantly to predicting GA conversion [69]. Eyes progressing to macular atrophy presented the greatest drusen height and HRF localized at 0.5-mm eccentricity, and the distribution of HRF was not concentrated in areas overlying drusen [67]. Schmidt-Erfurth et al. [69] also noted that the amount of HRF and the alterations seen in GA progressors did not colocalize with drusen but tended to spread more into the retina [69]. HRF distribution was found to be associated with the GA border, with the majority of HRF (65%) situated within a 1-mm junctional zone; at this level, increasing 2D HRF concentrations and counts were associated with GA progression [106].
Another feature associated with a higher risk for GA over neovascular conversion is represented by the presence of RPD in isolation, not considering the AMD severity scale [102]. Other quantitative outer retinal and sub-RPE features identified through ML demonstrated a role in identifying GA converters, including EZ-RPE thickness, EZ total and partial attenuation, RPE-BrM thickness, and RPE total attenuation [75]. More recently, a DL model quantified multimodal imaging biomarkers involved in GA progression. The best fit model associated with GA progression included FAF patterns alone, the interaction with the HRF concentration (HRF*FAF patterns), and the presence of RPD. In contrast, the interaction of age*sqrt GA area was negatively associated with GA growth, indicating that larger lesions in older patients were less prone to enlarge [108].
Using a variation in the U-Net architecture, where the model outputs a likelihood estimate for a given feature for every input image pixel, RPE loss, photoreceptor degeneration, and hypertransmission were identified. A fourth model segmented RPE and outer retina atrophy (RORA) as regions of overlapping RPE loss, photoreceptor degeneration, and hypertransmission. The presence of RORA (30.5%), followed by hypertransmission (21.5%), contributed the most to the prediction of visual acuity in GA, whilst the low-luminance (VA) was predicted by importance from photoreceptor degeneration (38.9%) followed by hypertransmission (26%) [109].
5.2. Predictive Factors of Neovascular Conversion
The prediction towards neovascular conversion included thickening of the RPE-drusen complex, an increase in the drusen area, and drusen-centric HRF associated with a thickening of the ONL where HRF tended to migrate. In this predictive model, demographic and genetic factors did not contribute to the neovascular conversion (AUC = 0.68) [69]. The relationship between the drusen topographical distribution and HRF was confirmed to be relevant when differentiating neovascular and GA conversion. In eyes progressing to macular neovascularization (MNV), the greatest drusen volume was observable within the foveal center, with a slight increase in mean HRF volume over time [67]. A choroidal thinning was also observable in MNV progressors, but the difference was not significant after adjusting for the false discovery rate. However, longitudinal evaluation confirmed a faster thinning of the ONL and outer retinal bands, as well as choroidal thinning towards the point of MNV conversion [68].
A greater score in the modified severity scale in conjunction with RPD presence was associated with a higher risk of neovascular AMD [102]. Nevertheless, most of the literature currently available investigates prognostic indicators or biomarkers of treatment response and clinical outcomes after anti-VEGF treatment [110,111,112,113,114,115,116,117,118,119].
6. Discussion
The biomarkers and artificial intelligence prediction models in AMD analyzed in this narrative review are summarized in Table 2. The DL models created using CFP included three DCNNs estimating the 5-year conversion risk (%) according to clinical features, including total drusen area, increased pigmentation, and depigmentation [55]. A pre-trained CNN (Inception V3) model of multiple longitudinal images with two and three time points [42]. Using FAF alone or combined with OCT was dedicated to the GA growth prediction using multi-task CNNs [51].

Table 2.
Biomarkers considered in artificial intelligence prediction models in age-related macular degeneration.
The most crucial multimodal imaging technique is represented by OCT, which allows an exhaustive evaluation of several biomarkers and morphometric parameters within a single volumetric acquisition. In our opinion, the importance of OCT should also be interpreted in light of its wide use in clinical practice, the readiness of execution, and the numerous preliminary studies investigating potential predictors to be included in DL models. Through the automated segmentation of the OCT volumetric cube it is possible to extrapolate different quantitative drusen features, which include drusen number, mean volume, drusen area, topographic distribution, maximum height, slope, and reflectivity [57]. The most remarkable drusen features involved in the prediction included the mean drusen thickness, maximum drusen height, and the mean drusen attenuation considering the attenuation within drusen in comparison with the overlying outer retinal bands and ONL [88]. An automated grading system can evaluate the presence and distribution of HRF, choroidal thickness, the presence of RPD, and photoreceptor outer segment loss [60]. The identification of HRF is of leading importance in neovascular conversion, representing a distinctive hallmark alone and a more robust indicator when overlying drusen HRF, also important predictors of GA development [64,69].
Several studies also analyzed the influence of demographic factors alone or in combination with clinical or OCT features, as summarized in Table 2. However, the specific AMD risk associated with SNPs did not improve the prediction of the DL models for both GA and neovascularization [62,69,79].
Quantitative outer retinal and sub-RPE features identified through ML that demonstrated a role in determining GA converters included EZ-RPE thickness, EZ total and partial attenuation, RPE-BrM thickness, and RPE total attenuation [75]. A DL model quantified multimodal imaging biomarkers involved in GA progression. The best-fit model associated with GA progression included FAF patterns alone, the interaction with the HRF, and the presence of RPD [108]. Furthermore, the progressive ONL thinning visible on OCT was found to be a robust predictor of conversion with a faster thinning in eyes developing GA [68]. Using a variation in the U-Net architecture, the presence of RORA (30.5%), followed by hypertransmission (21.5%), contributed the most to the prediction of visual acuity in GA [109].
Factors contributing to the prediction towards neovascular conversion included the thickening of the RPE-drusen complex, an increase in the drusen area, and drusen-centric HRF associated with a thickening of the ONL where HRF tended to migrate [69]. Moreover, the most significant drusen volume was observable within the foveal center in eyes progressing to MNV, with a slight increase in mean HRF volume over time [67].
7. Conclusions and Future Perspectives
With recent advances in DL, this field is significantly expanding the spectrum of AMD prediction through multimodal imaging. DCNN represented the most relevant and suitable DL methodology for recognizing imaging output data. However, training a DCNN model from scratch is particularly time-consuming and expensive. A reasonable option is represented by fine-tuning CNN, consisting of transferring pre-learned features from a pre-trained CNN to initialize a new model.
When approaching AMD prediction, it is essential to know and apply the AMD severity scales that allow an improvement in disease prediction along with other imaging and non-imaging parameters. Although the imaging biomarkers represented the most relevant predictive features, some DL models performed better by including non-imaging parameters, such as demographic baseline characteristics including age, gender, race, smoking status, fellow eye status, body mass index, blood pressure, sunlight exposure, education, and Mediterranean diet score. Contrariwise, the incorporation of genetic features such as SNPs modified the predictive power only slightly, suggesting a marginal role of genotypic characterization in identifying patients at high risk of AMD evolution into late stages.
Different multimodal imaging modalities were used in the predictive algorithms, including CFP, FAF, and OCT. Among those, OCT represented the most informative modality, allowing the qualitative identification of specific biomarkers and the quantitative estimation of morphometric changes in retinal layers, drusen volume, and choroidal thickness. The most relevant predictors are represented by ONL and outer retinal layer thickness changes, drusen and volume morphometric modifications, quantitative and qualitative changes in HRF, and the presence of RPD. For GA conversion, the most important factors identified were outer retina and ONL thinning, increasing HRF and their topographic distribution, as well as age as the unique non-imaging factor. The prediction towards neovascular conversion was less investigated, but the main factors identified were represented by an increasing drusen area and volume within the foveal center with overlying HRF and outer retinal, ONL, and choroidal thinning. Identifying OCT predictors presented significant advantages, including the routinary use of this imaging modality in clinical practice, the large number of qualitative and quantitative features that can be extrapolated at each examination, and the repeatability of acquisition and metrics. In AMD, the interpolation of imaging biomarkers and risk factors allowed a good risk stratification that can be estimated through DL models with high diagnostic performance. The understanding of the strengths and limitation of different DL systems is crucial to identify future methodologies. Moreover, further studies are encouraged to assess more accurate metrics and predictors and to build even more accurate predictive models that can be available in clinical practice.
Author Contributions
Conceptualization, S.A. and S.F.; methodology, S.F. and F.G.; data curation, F.G.; writing—original draft preparation, S.F. and F.G.; writing—review and editing, S.A.; supervision, S.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Appendix A

Table A1.
A summary of the 52 independent AMD risk variants identified in 34 loci.
Table A1.
A summary of the 52 independent AMD risk variants identified in 34 loci.
| # | Locus Name | Variants |
|---|---|---|
| 1 | CFH | rs10922109; rs570618; rs121913059; rs148553336; rs187328863; rs61818925; rs35292876; rs191281603 |
| 2 | COL4A3 | rs11884770 |
| 3 | ADAMTS9-AS2 | rs62247658 |
| 4 | COL8A1 | rs140647181; rs55975637 |
| 5 | CFI | rs10033900; rs141853578 |
| 6 | C9 | rs62358361 |
| 7 | PRLR/SPEF2 | rs114092250 |
| 8 | C2/CFB/SKIV2L | rs116503776; rs144629244; rs114254831; rs181705462 |
| 9 | VEGFA | rs943080 |
| 10 | KMT2E/SRPK2 | rs1142 |
| 11 | PILRB/PILRA | rs7803454 |
| 12 | TNFRSF10A | rs79037040 |
| 13 | MIR6130/RORB | rs10781182 |
| 14 | TRPM3 | rs71507014 |
| 15 | TGFBR1 | rs1626340 |
| 16 | ABCA1 | rs2740488 |
| 17 | ARHGAP21 | rs12357257 |
| 18 | ARMS2/HTRA1 | rs3750846 |
| 19 | RDH5/CD63 | rs3138141 |
| 20 | ACAD10 | rs61941274 |
| 21 | B3GALTL | rs9564692 |
| 22 | RAD51B | rs61985136; rs2842339 |
| 23 | LIPC | rs2043085; rs2070895 |
| 24 | CETP | rs5817082; rs17231506 |
| 25 | CTRB2/CTRB1 | rs72802342 |
| 26 | TMEM97/VTN | rs11080055 |
| 27 | NPLOC4/TSPAN10 | rs6565597 |
| 28 | C3 | rs2230199; rs147859257; rs12019136 |
| 29 | CNN2 | rs67538026 |
| 30 | APOE | rs429358; rs73036519 |
| 31 | MMP9 | rs142450006 |
| 32 | C20orf85 | rs201459901 |
| 33 | SYN3/TIMP3 | rs5754227 |
| 34 | SLC16A8 | rs8135665 |
References
- Doi, K. Diagnostic imaging over the last 50 years: Research and development in medical imaging science and technology. Phys. Med. Biol. 2006, 51, R5–R27. [Google Scholar] [CrossRef] [PubMed]
- Doi, K. Computer-aided diagnosis in medical imaging: Historical review, current status and future potential. Comput. Med. Imaging Graph. 2007, 31, 198–211. [Google Scholar] [CrossRef]
- Kermany, D.S.; Goldbaum, M.; Cai, W.; Valentim, C.C.S.; Liang, H.; Baxter, S.L.; McKeown, A.; Yang, G.; Wu, X.; Yan, F.; et al. Identifying Medical Diagnoses and Treatable Diseases by Image-Based Deep Learning. Cell 2018, 172, 1122–1131.e1129. [Google Scholar] [CrossRef] [PubMed]
- Dow, E.R.; Keenan, T.D.L.; Lad, E.M.; Lee, A.Y.; Lee, C.S.; Loewenstein, A.; Eydelman, M.B.; Chew, E.Y.; Keane, P.A.; Lim, J.I.; et al. From Data to Deployment: The Collaborative Community on Ophthalmic Imaging Roadmap for Artificial Intelligence in Age-Related Macular Degeneration. Ophthalmology 2022, 129, e43–e59. [Google Scholar] [CrossRef] [PubMed]
- Bhuiyan, A.; Wong, T.Y.; Ting, D.S.W.; Govindaiah, A.; Souied, E.H.; Smith, R.T. Artificial Intelligence to Stratify Severity of Age-Related Macular Degeneration (AMD) and Predict Risk of Progression to Late AMD. Transl. Vis. Sci. Technol. 2020, 9, 25. [Google Scholar] [CrossRef]
- Sacconi, R.; Fragiotta, S.; Sarraf, D.; Sadda, S.R.; Freund, K.B.; Parravano, M.; Corradetti, G.; Cabral, D.; Capuano, V.; Miere, A.; et al. Towards a better understanding of non-exudative choroidal and macular neovascularization. Prog. Retin. Eye Res. 2022, 92, 101113. [Google Scholar] [CrossRef] [PubMed]
- Liao, D.S.; Metapally, R.; Joshi, P. Pegcetacoplan treatment for geographic atrophy due to age-related macular degeneration: A plain language summary of the FILLY study. Immunotherapy 2022, 14, 995–1006. [Google Scholar] [CrossRef] [PubMed]
- Nittala, M.G.; Metlapally, R.; Ip, M.; Chakravarthy, U.; Holz, F.G.; Staurenghi, G.; Waheed, N.; Velaga, S.B.; Lindenberg, S.; Karamat, A.; et al. Association of Pegcetacoplan With Progression of Incomplete Retinal Pigment Epithelium and Outer Retinal Atrophy in Age-Related Macular Degeneration: A Post Hoc Analysis of the FILLY Randomized Clinical Trial. JAMA Ophthalmol. 2022, 140, 243–249. [Google Scholar] [CrossRef] [PubMed]
- Wykoff, C.C.; Rosenfeld, P.J.; Waheed, N.K.; Singh, R.P.; Ronca, N.; Slakter, J.S.; Staurenghi, G.; Monés, J.; Baumal, C.R.; Saroj, N.; et al. Characterizing New-Onset Exudation in the Randomized Phase 2 FILLY Trial of Complement Inhibitor Pegcetacoplan for Geographic Atrophy. Ophthalmology 2021, 128, 1325–1336. [Google Scholar] [CrossRef] [PubMed]
- Steinle, N.C.; Pearce, I.; Monés, J.; Metlapally, R.; Saroj, N.; Hamdani, M.; Ribeiro, R.; Rosenfeld, P.J.; Lad, E.M. Impact of Baseline Characteristics on Geographic Atrophy Progression in the FILLY Trial Evaluating the Complement C3 Inhibitor Pegcetacoplan. Am. J. Ophthalmol. 2021, 227, 116–124. [Google Scholar] [CrossRef] [PubMed]
- Tolentino, M.J.; Tolentino, A.J. Investigational drugs in clinical trials for macular degeneration. Expert Opin. Investig. Drugs 2022, 31, 1067–1085. [Google Scholar] [CrossRef] [PubMed]
- Jaffe, G.J.; Westby, K.; Csaky, K.G.; Monés, J.; Pearlman, J.A.; Patel, S.S.; Joondeph, B.C.; Randolph, J.; Masonson, H.; Rezaei, K.A. C5 Inhibitor Avacincaptad Pegol for Geographic Atrophy Due to Age-Related Macular Degeneration: A Randomized Pivotal Phase 2/3 Trial. Ophthalmology 2021, 128, 576–586. [Google Scholar] [CrossRef] [PubMed]
- Kuppermann, B.D.; Patel, S.S.; Boyer, D.S.; Augustin, A.J.; Freeman, W.R.; Kerr, K.J.; Guo, Q.; Schneider, S.; López, F.J. Phase 2 study of the safety and efficacy of brimonidine drug delivery system (brimo dds) generation 1 in patients with geographic atrophy secondary to age-related macular degeneration. Retina 2021, 41, 144–155. [Google Scholar] [CrossRef] [PubMed]
- Burlina, P.M.; Joshi, N.; Pekala, M.; Pacheco, K.D.; Freund, D.E.; Bressler, N.M. Automated Grading of Age-Related Macular Degeneration From Color Fundus Images Using Deep Convolutional Neural Networks. JAMA Ophthalmol. 2017, 135, 1170–1176. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, R.; Pramanik, A. DCNN-based prediction model for detection of age-related macular degeneration from color fundus images. Med. Biol. Eng. Comput. 2022, 60, 1431–1448. [Google Scholar] [CrossRef]
- Sotoudeh-Paima, S.; Jodeiri, A.; Hajizadeh, F.; Soltanian-Zadeh, H. Multi-scale convolutional neural network for automated AMD classification using retinal OCT images. Comput. Biol. Med. 2022, 144, 105368. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Jin, K.; Yan, Y.; Liu, X.; Huang, X.; Gao, Z.; Wang, Y.; Wang, S.; Ye, J. Automated diagnosis of age-related macular degeneration using multi-modal vertical plane feature fusion via deep learning. Med. Phys. 2022, 49, 2324–2333. [Google Scholar] [CrossRef] [PubMed]
- Yellapragada, B.; Hornauer, S.; Snyder, K.; Yu, S.; Yiu, G. Self-Supervised Feature Learning and Phenotyping for Assessing Age-Related Macular Degeneration Using Retinal Fundus Images. Ophthalmol. Retina 2022, 6, 116–129. [Google Scholar] [CrossRef] [PubMed]
- Romond, K.; Alam, M.; Kravets, S.; Sisternes, L.; Leng, T.; Lim, J.I.; Rubin, D.; Hallak, J.A. Imaging and artificial intelligence for progression of age-related macular degeneration. Exp. Biol. Med. 2021, 246, 2159–2169. [Google Scholar] [CrossRef]
- Arevalo, J.F.; Lasave, A.F.; Arias, J.D.; Serrano, M.A.; Arevalo, F.A. Clinical applications of optical coherence tomography in the posterior pole: The 2011 Jose Manuel Espino Lecture–Part II. Clin. Ophthalmol. 2013, 7, 2181–2206. [Google Scholar] [CrossRef] [PubMed]
- Guymer, R.H.; Rosenfeld, P.J.; Curcio, C.A.; Holz, F.G.; Staurenghi, G.; Freund, K.B.; Schmitz-Valckenberg, S.; Sparrow, J.; Spaide, R.F.; Tufail, A.; et al. Incomplete Retinal Pigment Epithelial and Outer Retinal Atrophy in Age-Related Macular Degeneration: Classification of Atrophy Meeting Report 4. Ophthalmology 2020, 127, 394–409. [Google Scholar] [CrossRef] [PubMed]
- Sadda, S.R.; Guymer, R.; Holz, F.G.; Schmitz-Valckenberg, S.; Curcio, C.A.; Bird, A.C.; Blodi, B.A.; Bottoni, F.; Chakravarthy, U.; Chew, E.Y.; et al. Consensus definition for atrophy associated with age-related macular degeneration on OCT: Classification of atrophy report 3. Ophthalmology 2018, 125, 537–548. [Google Scholar] [CrossRef] [PubMed]
- Spaide, R.F.; Jaffe, G.J.; Sarraf, D.; Freund, K.B.; Sadda, S.R.; Staurenghi, G.; Waheed, N.K.; Chakravarthy, U.; Rosenfeld, P.J.; Holz, F.G.; et al. Consensus Nomenclature for Reporting Neovascular Age-Related Macular Degeneration Data: Consensus on Neovascular Age-Related Macular Degeneration Nomenclature Study Group. Ophthalmology 2020, 127, 616–636. [Google Scholar] [CrossRef]
- Metrangolo, C.; Donati, S.; Mazzola, M.; Fontanel, L.; Messina, W.; D’Alterio, G.; Rubino, M.; Radice, P.; Premi, E.; Azzolini, C. OCT Biomarkers in Neovascular Age-Related Macular Degeneration: A Narrative Review. J. Ophthalmol. 2021, 2021, 9994098. [Google Scholar] [CrossRef] [PubMed]
- Ting, D.S.W.; Pasquale, L.R.; Peng, L.; Campbell, J.P.; Lee, A.Y.; Raman, R.; Tan, G.S.W.; Schmetterer, L.; Keane, P.A.; Wong, T.Y. Artificial intelligence and deep learning in ophthalmology. Br. J. Ophthalmol. 2019, 103, 167–175. [Google Scholar] [CrossRef]
- Fujinami-Yokokawa, Y.; Ninomiya, H.; Liu, X.; Yang, L.; Pontikos, N.; Yoshitake, K.; Iwata, T.; Sato, Y.; Hashimoto, T.; Tsunoda, K.; et al. Prediction of causative genes in inherited retinal disorder from fundus photography and autofluorescence imaging using deep learning techniques. Br. J. Ophthalmol. 2021, 105, 1272–1279. [Google Scholar] [CrossRef]
- Burlina, P.; Pacheco, K.D.; Joshi, N.; Freund, D.E.; Bressler, N.M. Comparing humans and deep learning performance for grading AMD: A study in using universal deep features and transfer learning for automated AMD analysis. Comput. Biol. Med. 2017, 82, 80–86. [Google Scholar] [CrossRef]
- Burlina, P.M.; Joshi, N.; Pacheco, K.D.; Freund, D.E.; Kong, J.; Bressler, N.M. Use of Deep Learning for Detailed Severity Characterization and Estimation of 5-Year Risk Among Patients With Age-Related Macular Degeneration. JAMA Ophthalmol. 2018, 136, 1359–1366. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Yousefi, S.; Goldbaum, M.H.; Balasubramanian, M.; Jung, T.-P.; Weinreb, R.N.; Medeiros, F.A.; Zangwill, L.M.; Liebmann, J.M.; Girkin, C.A.; Bowd, C. Glaucoma progression detection using structural retinal nerve fiber layer measurements and functional visual field points. IEEE Trans. Biomed. Eng. 2014, 61, 1143–1154. [Google Scholar] [CrossRef]
- Gelman, R.; Jiang, L.; Du, Y.E.; Martinez-Perez, M.E.; Flynn, J.T.; Chiang, M.F. Plus disease in retinopathy of prematurity: Pilot study of computer-based and expert diagnosis. J. AAPOS 2007, 11, 532–540. [Google Scholar] [CrossRef]
- Gulshan, V.; Peng, L.; Coram, M.; Stumpe, M.C.; Wu, D.; Narayanaswamy, A.; Venugopalan, S.; Widner, K.; Madams, T.; Cuadros, J.; et al. Development and Validation of a Deep Learning Algorithm for Detection of Diabetic Retinopathy in Retinal Fundus Photographs. JAMA 2016, 316, 2402. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.S.; Baughman, D.M.; Lee, A.Y. Deep learning is effective for the classification of OCT images of normal versus Age-related Macular Degeneration. Ophthalmol. Retina 2017, 1, 322–327. [Google Scholar] [CrossRef] [PubMed]
- Lu, W.; Tong, Y.; Yu, Y.; Xing, Y.; Chen, C.; Shen, Y. Applications of Artificial Intelligence in Ophthalmology: General Overview. J. Ophthalmol. 2018, 2018, 5278196. [Google Scholar] [CrossRef]
- Peng, Y.; Dharssi, S.; Chen, Q.; Keenan, T.D.; Agrón, E.; Wong, W.T.; Chew, E.Y.; Lu, Z. DeepSeeNet: A Deep Learning Model for Automated Classification of Patient-based Age-related Macular Degeneration Severity from Color Fundus Photographs. Ophthalmology 2019, 126, 565–575. [Google Scholar] [CrossRef]
- Samuel, A.L. Some Studies in Machine Learning Using the Game of Checkers. I. In Computer Games I; Springer: New York, NY, USA, 1988; pp. 335–365. [Google Scholar]
- Breiman, L. Machine Learning; Springer: Berlin/Heidelberg, Germany, 2001; Volume 45, pp. 5–32. [Google Scholar] [CrossRef]
- Chang, C.-C.; Lin, C.-J. LIBSVM. ACM Trans. Intell. Syst. Technol. 2011, 2, 1–27. [Google Scholar] [CrossRef]
- Rahimy, E. Deep learning applications in ophthalmology. Curr. Opin. Ophthalmol. 2018, 29, 254–260. [Google Scholar] [CrossRef] [PubMed]
- Schmidt-Erfurth, U.; Sadeghipour, A.; Gerendas, B.S.; Waldstein, S.M.; Bogunović, H. Artificial intelligence in retina. Prog. Retin. Eye Res. 2018, 67, 1–29. [Google Scholar] [CrossRef]
- Tajbakhsh, N.; Shin, J.Y.; Gurudu, S.R.; Hurst, R.T.; Kendall, C.B.; Gotway, M.B.; Jianming, L. Convolutional Neural Networks for Medical Image Analysis: Full Training or Fine Tuning? IEEE Trans. Med. Imaging 2016, 35, 1299–1312. [Google Scholar] [CrossRef]
- Rattani, A.; Derakhshani, R. On fine-tuning convolutional neural networks for smartphone based ocular recognition. In Proceedings of the 2017 IEEE International Joint Conference on Biometrics (IJCB), Denver, CO, USA, 1–4 October 2017; pp. 762–767. [Google Scholar]
- Chen, Y.M.; Huang, W.T.; Ho, W.H.; Tsai, J.T. Classification of age-related macular degeneration using convolutional-neural-network-based transfer learning. BMC Bioinform. 2021, 22, 99. [Google Scholar] [CrossRef]
- Long, J.; Shelhamer, E.; Darrell, T. Fully convolutional networks for semantic segmentation. In Proceedings of the 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Lecture Notes in Computer Science; Springer International Publishing: New York, NY, USA, 2015; pp. 234–241. [Google Scholar]
- Badrinarayanan, V.; Kendall, A.; Cipolla, R. SegNet: A Deep Convolutional Encoder-Decoder Architecture for Image Segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 2481–2495. [Google Scholar] [CrossRef]
- Chen, L.-C.; Zhu, Y.; Papandreou, G.; Schroff, F.; Adam, H. Encoder-Decoder with Atrous Separable Convolution for Semantic Image Segmentation. In Computer Vision–ECCV 2018; Springer International Publishing: New York, NY, USA, 2018; pp. 833–851. [Google Scholar]
- Ferris, F.L.; Davis, M.D.; Clemons, T.E.; Lee, L.Y.; Chew, E.Y.; Lindblad, A.S.; Milton, R.C.; Bressler, S.B.; Klein, R. A simplified severity scale for age-related macular degeneration: AREDS Report No. 18. Arch Ophthalmol. 2005, 123, 1570–1574. [Google Scholar] [CrossRef]
- Ghahramani, G.; Brendel, M.; Lin, M.; Chen, Q.; Keenan, T.; Chen, K.; Chew, E.; Lu, Z.; Peng, Y.; Wang, F. Multi-task deep learning-based survival analysis on the prognosis of late AMD using the longitudinal data in AREDS. AMIA Annu. Symp. Proc. 2021, 2021, 506–515. [Google Scholar]
- Busbee, B.G.; Ho, A.C.; Brown, D.M.; Heier, J.S.; Suner, I.J.; Li, Z.; Rubio, R.G.; Lai, P.; Group, H.S. Twelve-month efficacy and safety of 0.5 mg or 2.0 mg ranibizumab in patients with subfoveal neovascular age-related macular degeneration. Ophthalmology 2013, 120, 1046–1056. [Google Scholar] [CrossRef]
- Anegondi, N.; Gao, S.S.; Steffen, V.; Spaide, R.F.; Sadda, S.R.; Holz, F.G.; Rabe, C.; Honigberg, L.; Newton, E.M.; Cluceru, J.; et al. Deep Learning to Predict Geographic Atrophy Area and Growth Rate from Multi-modal Imaging (90 characters; max 135 characters). Ophthalmol. Retina 2022, in press. [Google Scholar] [CrossRef]
- Holz, F.G.; Sadda, S.R.; Busbee, B.; Chew, E.Y.; Mitchell, P.; Tufail, A.; Brittain, C.; Ferrara, D.; Gray, S.; Honigberg, L.; et al. Efficacy and Safety of Lampalizumab for Geographic Atrophy Due to Age-Related Macular Degeneration: Chroma and Spectri Phase 3 Randomized Clinical Trials. JAMA Ophthalmol. 2018, 136, 666–677. [Google Scholar] [CrossRef] [PubMed]
- Holekamp, N.; Wykoff, C.C.; Schmitz-Valckenberg, S.; Mones, J.; Souied, E.H.; Lin, H.; Rabena, M.D.; Cantrell, R.A.; Henry, E.C.; Tang, F.; et al. Natural History of Geographic Atrophy Secondary to Age-Related Macular Degeneration: Results from the Prospective Proxima A and B Clinical Trials. Ophthalmology 2020, 127, 769–783. [Google Scholar] [CrossRef] [PubMed]
- Grassmann, F.; Mengelkamp, J.; Brandl, C.; Harsch, S.; Zimmermann, M.E.; Linkohr, B.; Peters, A.; Heid, I.M.; Palm, C.; Weber, B.H.F. A Deep Learning Algorithm for Prediction of Age-Related Eye Disease Study Severity Scale for Age-Related Macular Degeneration from Color Fundus Photography. Ophthalmology 2018, 125, 1410–1420. [Google Scholar] [CrossRef]
- Davis, M.D.; Gangnon, R.E.; Lee, L.Y.; Hubbard, L.D.; Klein, B.E.; Klein, R.; Ferris, F.L.; Bressler, S.B.; Milton, R.C.; Age-Related Eye Disease Study, G. The Age-Related Eye Disease Study severity scale for age-related macular degeneration: AREDS Report No. 17. Arch. Ophthalmol. 2005, 123, 1484–1498. [Google Scholar] [CrossRef]
- Keenan, T.D.L.; Chen, Q.; Peng, Y.; Domalpally, A.; Agrón, E.; Hwang, C.K.; Thavikulwat, A.T.; Lee, D.H.; Li, D.; Wong, W.T.; et al. Deep Learning Automated Detection of Reticular Pseudodrusen from Fundus Autofluorescence Images or Color Fundus Photographs in AREDS2. Ophthalmology 2020, 127, 1674–1687. [Google Scholar] [CrossRef] [PubMed]
- Bridge, J.; Harding, S.; Zheng, Y. Development and validation of a novel prognostic model for predicting AMD progression using longitudinal fundus images. BMJ Open Ophthalmol. 2020, 5, e000569. [Google Scholar] [CrossRef]
- Yan, Q.; Weeks, D.E.; Xin, H.; Swaroop, A.; Chew, E.Y.; Huang, H.; Ding, Y.; Chen, W. Deep-learning-based Prediction of Late Age-Related Macular Degeneration Progression. Nat. Mach. Intell. 2020, 2, 141–150. [Google Scholar] [CrossRef]
- Arslan, J.; Samarasinghe, G.; Sowmya, A.; Benke, K.K.; Hodgson, L.A.B.; Guymer, R.H.; Baird, P.N. Deep Learning Applied to Automated Segmentation of Geographic Atrophy in Fundus Autofluorescence Images. Transl. Vis. Sci. Technol. 2021, 10, 2. [Google Scholar] [CrossRef] [PubMed]
- Miere, A.; Capuano, V.; Kessler, A.; Zambrowski, O.; Jung, C.; Colantuono, D.; Pallone, C.; Semoun, O.; Petit, E.; Souied, E. Deep learning-based classification of retinal atrophy using fundus autofluorescence imaging. Comput. Biol. Med. 2021, 130, 104198. [Google Scholar] [CrossRef] [PubMed]
- Pan, S.J.; Yang, Q. A survey on transfer learning. IEEE Trans. Knowl. Data Eng. 2009, 22, 1345–1359. [Google Scholar] [CrossRef]
- Christenbury, J.G.; Folgar, F.A.; O’Connell, R.V.; Chiu, S.J.; Farsiu, S.; Toth, C.A. Progression of intermediate age-related macular degeneration with proliferation and inner retinal migration of hyperreflective foci. Ophthalmology 2013, 120, 1038–1045. [Google Scholar] [CrossRef]
- Curcio, C.A.; Zanzottera, E.C.; Ach, T.; Balaratnasingam, C.; Freund, K.B. Activated retinal pigment epithelium, an optical coherence tomography biomarker for progression in age-related macular degeneration. Investig. Ophthalmol. Vis. Sci. 2017, 58, BIO211–BIO226. [Google Scholar] [CrossRef]
- Fragiotta, S.; Rossi, T.; Cutini, A.; Grenga, P.L.; Vingolo, E.M. Predictive factors for development of neovascular age-related macular degeneration: A Spectral-Domain Optical Coherence Tomography Study. Retina 2018, 38, 245–252. [Google Scholar] [CrossRef]
- Fragiotta, S.; Costanzo, E.; Viggiano, P.; De Geronimo, D.; Scuderi, G.; Varano, M.; Parravano, M. Functional Correlates of Outer Retina Remodeling in Intermediate Age-Related Macular Degeneration Using Microperimetry. Invest. Ophthalmol. Vis. Sci. 2022, 63, 16. [Google Scholar] [CrossRef]
- Sassmannshausen, M.; Zhou, J.; Pfau, M.; Thiele, S.; Steinberg, J.; Fleckenstein, M.; Holz, F.G.; Schmitz-Valckenberg, S. Longitudinal Analysis of Retinal Thickness and Retinal Function in Eyes with Large Drusen Secondary to Intermediate Age-Related Macular Degeneration. Ophthalmol. Retina 2021, 5, 241–250. [Google Scholar] [CrossRef]
- Waldstein, S.M.; Vogl, W.D.; Bogunovic, H.; Sadeghipour, A.; Riedl, S.; Schmidt-Erfurth, U. Characterization of Drusen and Hyperreflective Foci as Biomarkers for Disease Progression in Age-Related Macular Degeneration Using Artificial Intelligence in Optical Coherence Tomography. JAMA Ophthalmol. 2020, 138, 740–747. [Google Scholar] [CrossRef]
- Vogl, W.D.; Bogunovic, H.; Waldstein, S.M.; Riedl, S.; Schmidt-Erfurth, U. Spatio-temporal alterations in retinal and choroidal layers in the progression of age-related macular degeneration (AMD) in optical coherence tomography. Sci. Rep. 2021, 11, 5743. [Google Scholar] [CrossRef]
- Schmidt-Erfurth, U.; Waldstein, S.M.; Klimscha, S.; Sadeghipour, A.; Hu, X.; Gerendas, B.S.; Osborne, A.; Bogunovic, H. Prediction of Individual Disease Conversion in Early AMD Using Artificial Intelligence. Investig. Ophthalmol. Vis. Sci. 2018, 59, 3199–3208. [Google Scholar] [CrossRef]
- Banerjee, I.; de Sisternes, L.; Hallak, J.A.; Leng, T.; Osborne, A.; Rosenfeld, P.J.; Gregori, G.; Durbin, M.; Rubin, D. Prediction of age-related macular degeneration disease using a sequential deep learning approach on longitudinal SD-OCT imaging biomarkers. Sci. Rep. 2020, 10, 15434. [Google Scholar] [CrossRef]
- de Sisternes, L.; Jonna, G.; Greven, M.A.; Chen, Q.; Leng, T.; Rubin, D.L. Individual Drusen Segmentation and Repeatability and Reproducibility of Their Automated Quantification in Optical Coherence Tomography Images. Transl. Vis. Sci. Technol. 2017, 6, 12. [Google Scholar] [CrossRef] [PubMed]
- Saha, S.; Nassisi, M.; Wang, M.; Lindenberg, S.; Kanagasingam, Y.; Sadda, S.; Hu, Z.J. Automated detection and classification of early AMD biomarkers using deep learning. Sci. Rep. 2019, 9, 10990. [Google Scholar] [CrossRef] [PubMed]
- Kanagasingam, Y.; Bhuiyan, A.; Abramoff, M.D.; Smith, R.T.; Goldschmidt, L.; Wong, T.Y. Progress on retinal image analysis for age related macular degeneration. Prog. Retin. Eye Res. 2014, 38, 20–42. [Google Scholar] [CrossRef] [PubMed]
- Seddon, J.M.; Reynolds, R.; Yu, Y.; Daly, M.J.; Rosner, B. Risk models for progression to advanced age-related macular degeneration using demographic, environmental, genetic, and ocular factors. Ophthalmology 2011, 118, 2203–2211. [Google Scholar] [CrossRef] [PubMed]
- Sarici, K.; Abraham, J.R.; Sevgi, D.D.; Lunasco, L.; Srivastava, S.K.; Whitney, J.; Cetin, H.; Hanumanthu, A.; Bell, J.M.; Reese, J.L.; et al. Risk Classification for Progression to Subfoveal Geographic Atrophy in Dry Age-Related Macular Degeneration Using Machine Learning-Enabled Outer Retinal Feature Extraction. Ophthalmic. Surg. Lasers Imaging Retina 2022, 53, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Ajana, S.; Cougnard-Grégoire, A.; Colijn, J.M.; Merle, B.M.J.; Verzijden, T.; de Jong, P.; Hofman, A.; Vingerling, J.R.; Hejblum, B.P.; Korobelnik, J.F.; et al. Predicting Progression to Advanced Age-Related Macular Degeneration from Clinical, Genetic, and Lifestyle Factors Using Machine Learning. Ophthalmology 2021, 128, 587–597. [Google Scholar] [CrossRef] [PubMed]
- Fleckenstein, M.; Mitchell, P.; Freund, K.B.; Sadda, S.; Holz, F.G.; Brittain, C.; Henry, E.C.; Ferrara, D. The Progression of Geographic Atrophy Secondary to Age-Related Macular Degeneration. Ophthalmology 2018, 125, 369–390. [Google Scholar] [CrossRef]
- Fritsche, L.G.; Igl, W.; Bailey, J.N.; Grassmann, F.; Sengupta, S.; Bragg-Gresham, J.L.; Burdon, K.P.; Hebbring, S.J.; Wen, C.; Gorski, M.; et al. A large genome-wide association study of age-related macular degeneration highlights contributions of rare and common variants. Nat. Genet 2016, 48, 134–143. [Google Scholar] [CrossRef]
- Peng, Y.; Keenan, T.D.; Chen, Q.; Agrón, E.; Allot, A.; Wong, W.T.; Chew, E.Y.; Lu, Z. Predicting risk of late age-related macular degeneration using deep learning. NPJ Digit. Med. 2020, 3, 111. [Google Scholar] [CrossRef]
- Jones, B.W.; Watt, C.B.; Frederick, J.M.; Baehr, W.; Chen, C.K.; Levine, E.M.; Milam, A.H.; Lavail, M.M.; Marc, R.E. Retinal remodeling triggered by photoreceptor degenerations. J. Comp. Neurol. 2003, 464, 1–16. [Google Scholar] [CrossRef]
- Marc, R.E.; Jones, B.W.; Watt, C.B.; Vazquez-Chona, F.; Vaughan, D.K.; Organisciak, D.T. Extreme retinal remodeling triggered by light damage: Implications for age related macular degeneration. Mol. Vis. 2008, 14, 782–806. [Google Scholar]
- Strettoi, E.; Pignatelli, V.; Rossi, C.; Porciatti, V.; Falsini, B. Remodeling of second-order neurons in the retina of rd/rd mutant mice. Vis. Res. 2003, 43, 867–877. [Google Scholar] [CrossRef]
- Pfau, M.; von der Emde, L.; Dysli, C.; Moller, P.T.; Thiele, S.; Lindner, M.; Schmid, M.; Rubin, D.L.; Fleckenstein, M.; Holz, F.G.; et al. Determinants of Cone and Rod Functions in Geographic Atrophy: AI-Based Structure-Function Correlation. Am. J. Ophthalmol. 2020, 217, 162–173. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Shen, M.; Wang, L.; de Sisternes, L.; Durbin, M.K.; Feuer, W.; Rosenfeld, P.J.; Gregori, G. Validation of a Novel Automated Algorithm to Measure Drusen Volume and Area Using Swept Source Optical Coherence Tomography Angiography. Transl. Vis. Sci. Technol. 2021, 10, 11. [Google Scholar] [CrossRef] [PubMed]
- Gigon, A.; Mosinska, A.; Montesel, A.; Derradji, Y.; Apostolopoulos, S.; Ciller, C.; De Zanet, S.; Mantel, I. Personalized Atrophy Risk Mapping in Age-Related Macular Degeneration. Transl. Vis. Sci. Technol. 2021, 10, 18. [Google Scholar] [CrossRef] [PubMed]
- Russakoff, D.B.; Lamin, A.; Oakley, J.D.; Dubis, A.M.; Sivaprasad, S. Deep Learning for Prediction of AMD Progression: A Pilot Study. Investig. Ophthalmol. Vis. Sci. 2019, 60, 712–722. [Google Scholar] [CrossRef]
- Schlanitz, F.G.; Baumann, B.; Kundi, M.; Sacu, S.; Baratsits, M.; Scheschy, U.; Shahlaee, A.; Mittermuller, T.J.; Montuoro, A.; Roberts, P.; et al. Drusen volume development over time and its relevance to the course of age-related macular degeneration. Br. J. Ophthalmol. 2017, 101, 198–203. [Google Scholar] [CrossRef] [PubMed]
- Bogunovic, H.; Montuoro, A.; Baratsits, M.; Karantonis, M.G.; Waldstein, S.M.; Schlanitz, F.; Schmidt-Erfurth, U. Machine Learning of the Progression of Intermediate Age-Related Macular Degeneration Based on OCT Imaging. Investig. Ophthalmol. Vis. Sci. 2017, 58, Bio141–Bio150. [Google Scholar] [CrossRef]
- Fragiotta, S.; Parravano, M.; Sacconi, R.; Costanzo, E.; Viggiano, P.; Prascina, F.; Capuano, V.; Souied, E.H.; Querques, G. A Common Finding in Foveal-Sparing Extensive Macular Atrophy with Pseudodrusen Implicates Basal Laminar Deposits. Retina 2022, 42, 1319–1329. [Google Scholar] [CrossRef]
- Smith, R.T.; Sohrab, M.A.; Busuioc, M.; Barile, G. Reticular macular disease. Am. J. Ophthalmol. 2009, 148, 733–743.e732. [Google Scholar] [CrossRef] [PubMed]
- Spaide, R.F.; Ooto, S.; Curcio, C.A. Subretinal drusenoid deposits AKA pseudodrusen. Surv. Ophthalmol. 2018, 63, 782–815. [Google Scholar] [CrossRef] [PubMed]
- Zweifel, S.A.; Imamura, Y.; Spaide, T.C.; Fujiwara, T.; Spaide, R.F. Prevalence and significance of subretinal drusenoid deposits (reticular pseudodrusen) in age-related macular degeneration. Ophthalmology 2010, 117, 1775–1781. [Google Scholar] [CrossRef] [PubMed]
- Zweifel, S.A.; Spaide, R.F.; Curcio, C.A.; Malek, G.; Imamura, Y. Reticular pseudodrusen are subretinal drusenoid deposits. Ophthalmology 2010, 117, 303–312.e1. [Google Scholar] [CrossRef]
- Wu, Z.; Fletcher, E.L.; Kumar, H.; Greferath, U.; Guymer, R.H. Reticular pseudodrusen: A critical phenotype in age-related macular degeneration. Prog. Retin. Eye Res. 2022, 88, 101017. [Google Scholar] [CrossRef]
- Abdolrahimzadeh, S.; Di Pippo, M.; Sordi, E.; Cusato, M.; Lotery, A.J. Subretinal drusenoid deposits as a biomarker of age-related macular degeneration progression via reduction of the choroidal vascularity index. Eye 2022. [Google Scholar] [CrossRef]
- Abdolrahimzadeh, S.; Parisi, F.; Marcelli, M.; Giustolisi, R.; Gharbiya, M. Optical coherence tomography evidence of macular ganglion cell-inner plexiform layer thinning in eyes with subretinal drusenoid deposits. Eye 2019, 33, 1290–1296. [Google Scholar] [CrossRef]
- Abdolrahimzadeh, S.; Di Pippo, M.; Sordi, E.; Zweifel, S.A. Inner Retinal Layer Thickness Alterations in Early Age Related Macular Degeneration in Eyes with Subretinal Drusenoid Deposits or Conventional Drusen. J. Clin. Med. 2021, 10, 5136. [Google Scholar] [CrossRef] [PubMed]
- Curcio, C.A.; Messinger, J.D.; Sloan, K.R.; McGwin, G.; Medeiros, N.E.; Spaide, R.F. Subretinal drusenoid deposits in non-neovascular age-related macular degeneration: Morphology, prevalence, topography, and biogenesis model. Retina 2013, 33, 265–276. [Google Scholar] [CrossRef]
- Querques, G.; Canoui-Poitrine, F.; Coscas, F.; Massamba, N.; Querques, L.; Mimoun, G.; Bandello, F.; Souied, E.H. Analysis of progression of reticular pseudodrusen by spectral domain-optical coherence tomography. Investig. Ophthalmol. Vis. Sci. 2012, 53, 1264–1270. [Google Scholar] [CrossRef]
- Querques, G.; Querques, L.; Martinelli, D.; Massamba, N.; Coscas, G.; Soubrane, G.; Souied, E.H. Pathologic insights from integrated imaging of reticular pseudodrusen in age-related macular degeneration. Retina 2011, 31, 518–526. [Google Scholar] [CrossRef] [PubMed]
- Domalpally, A.; Agron, E.; Pak, J.W.; Keenan, T.D.; Ferris, F.L., 3rd; Clemons, T.E.; Chew, E.Y. Prevalence, Risk, and Genetic Association of Reticular Pseudodrusen in Age-related Macular Degeneration: Age-Related Eye Disease Study 2 Report 21. Ophthalmology 2019, 126, 1659–1666. [Google Scholar] [CrossRef]
- Agrón, E.; Domalpally, A.; Cukras, C.A.; Clemons, T.E.; Chen, Q.; Lu, Z.; Chew, E.Y.; Keenan, T.D.L. Reticular Pseudodrusen: The Third Macular Risk Feature for Progression to Late Age-related Macular Degeneration: Age-Related Eye Disease Study 2 Report 30. Ophthalmology 2022, 129, 1107–1119. [Google Scholar] [CrossRef] [PubMed]
- Fragiotta, S.; Abdolrahimzadeh, S.; Dolz-Marco, R.; Sakurada, Y.; Gal-Or, O.; Scuderi, G. Significance of Hyperreflective Foci as an Optical Coherence Tomography Biomarker in Retinal Diseases: Characterization and Clinical Implications. J. Ophthalmol. 2021, 2021, 6096017. [Google Scholar] [CrossRef] [PubMed]
- Balaratnasingam, C.; Yannuzzi, L.A.; Curcio, C.A.; Morgan, W.H.; Querques, G.; Capuano, V.; Souied, E.; Jung, J.; Freund, K.B. Associations between retinal pigment epithelium and drusen volume changes during the lifecycle of large drusenoid pigment epithelial detachments. Investig. Ophthalmol. Vis. Sci. 2016, 57, 5479–5489. [Google Scholar] [CrossRef]
- Yim, J.; Chopra, R.; Spitz, T.; Winkens, J.; Obika, A.; Kelly, C.; Askham, H.; Lukic, M.; Huemer, J.; Fasler, K.; et al. Predicting conversion to wet age-related macular degeneration using deep learning. Nat. Med. 2020, 26, 892–899. [Google Scholar] [CrossRef] [PubMed]
- Schmidt-Erfurth, U.; Bogunovic, H.; Grechenig, C.; Bui, P.; Fabianska, M.; Waldstein, S.; Reiter, G.S. Role of Deep Learning-Quantified Hyperreflective Foci for the Prediction of Geographic Atrophy Progression. Am. J. Ophthalmol. 2020, 216, 257–270. [Google Scholar] [CrossRef]
- Schlegl, T.; Bogunovic, H.; Klimscha, S.; Seeböck, P.; Sadeghipour, A.; Gerendas, B.; Waldstein, S.M.; Langs, G.; Schmidt-Erfurth, U. Fully automated segmentation of hyperreflective foci in optical coherence tomography images. arXiv 2018, arXiv:1805.03278. [Google Scholar]
- Bui, P.T.A.; Reiter, G.S.; Fabianska, M.; Waldstein, S.M.; Grechenig, C.; Bogunovic, H.; Arikan, M.; Schmidt-Erfurth, U. Fundus autofluorescence and optical coherence tomography biomarkers associated with the progression of geographic atrophy secondary to age-related macular degeneration. Eye 2022, 36, 2013–2019. [Google Scholar] [CrossRef]
- Balaskas, K.; Glinton, S.; Keenan, T.D.L.; Faes, L.; Liefers, B.; Zhang, G.; Pontikos, N.; Struyven, R.; Wagner, S.K.; McKeown, A.; et al. Prediction of visual function from automatically quantified optical coherence tomography biomarkers in patients with geographic atrophy using machine learning. Sci. Rep. 2022, 12, 15565. [Google Scholar] [CrossRef]
- Zhao, X.; Zhang, X.; Lv, B.; Meng, L.; Zhang, C.; Liu, Y.; Lv, C.; Xie, G.; Chen, Y. Optical coherence tomography-based short-term effect prediction of anti-vascular endothelial growth factor treatment in neovascular age-related macular degeneration using sensitive structure guided network. Graefes Arch. Clin. Exp. Ophthalmol. 2021, 259, 3261–3269. [Google Scholar] [CrossRef]
- Yeh, T.C.; Luo, A.C.; Deng, Y.S.; Lee, Y.H.; Chen, S.J.; Chang, P.H.; Lin, C.J.; Tai, M.C.; Chou, Y.B. Prediction of treatment outcome in neovascular age-related macular degeneration using a novel convolutional neural network. Sci. Rep. 2022, 12, 5871. [Google Scholar] [CrossRef]
- Song, X.; Xu, Q.; Li, H.; Fan, Q.; Zheng, Y.; Zhang, Q.; Chu, C.; Zhang, Z.; Yuan, C.; Ning, M.; et al. Automatic quantification of retinal photoreceptor integrity to predict persistent disease activity in neovascular age-related macular degeneration using deep learning. Front. Neurosci. 2022, 16, 952735. [Google Scholar] [CrossRef]
- Sodhi, S.K.; Pereira, A.; Oakley, J.D.; Golding, J.; Trimboli, C.; Russakoff, D.B.; Choudhry, N. Utilization of deep learning to quantify fluid volume of neovascular age-related macular degeneration patients based on swept-source OCT imaging: The ONTARIO study. PLoS ONE 2022, 17, e0262111. [Google Scholar] [CrossRef] [PubMed]
- Romo-Bucheli, D.; Erfurth, U.S.; Bogunovic, H. End-to-End Deep Learning Model for Predicting Treatment Requirements in Neovascular AMD From Longitudinal Retinal OCT Imaging. IEEE J. Biomed. Health Inform. 2020, 24, 3456–3465. [Google Scholar] [CrossRef] [PubMed]
- Pfau, M.; Sahu, S.; Rupnow, R.A.; Romond, K.; Millet, D.; Holz, F.G.; Schmitz-Valckenberg, S.; Fleckenstein, M.; Lim, J.I.; de Sisternes, L.; et al. Probabilistic Forecasting of Anti-VEGF Treatment Frequency in Neovascular Age-Related Macular Degeneration. Transl. Vis. Sci. Technol. 2021, 10, 30. [Google Scholar] [CrossRef] [PubMed]
- Paul, S.K.; Pan, I.; Sobol, W.M. A Systematic Review Of Deep Learning Applications For Optical Coherence Tomography In Age-Related Macular Degeneration. Retina 2022, 42, 1417–1424. [Google Scholar] [CrossRef]
- Mantel, I.; Mosinska, A.; Bergin, C.; Polito, M.S.; Guidotti, J.; Apostolopoulos, S.; Ciller, C.; De Zanet, S. Automated Quantification of Pathological Fluids in Neovascular Age-Related Macular Degeneration, and Its Repeatability Using Deep Learning. Transl. Vis. Sci. Technol. 2021, 10, 17. [Google Scholar] [CrossRef] [PubMed]
- Gutfleisch, M.; Ester, O.; Aydin, S.; Quassowski, M.; Spital, G.; Lommatzsch, A.; Rothaus, K.; Dubis, A.M.; Pauleikhoff, D. Clinically applicable deep learning-based decision aids for treatment of neovascular AMD. Graefes Arch. Clin. Exp. Ophthalmol. 2022, 260, 2217–2230. [Google Scholar] [CrossRef] [PubMed]
- Fu, D.J.; Faes, L.; Wagner, S.K.; Moraes, G.; Chopra, R.; Patel, P.J.; Balaskas, K.; Keenan, T.D.L.; Bachmann, L.M.; Keane, P.A. Predicting Incremental and Future Visual Change in Neovascular Age-Related Macular Degeneration Using Deep Learning. Ophthalmol. Retina 2021, 5, 1074–1084. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).