Integrating Equation Coding with Residual Networks for Efficient ODE Approximation in Biological Research
Abstract
1. Introduction
2. Materials and Methods
2.1. Model Layout
2.2. Training
2.3. Data Preparation
2.3.1. Synthetic Data Studies
2.3.2. Metabolite Data Study
2.3.3. Single-Cell RNA Sequencing (scRNA-seq)
3. Results
3.1. LazyNet’s Estimation of HIV Strain ODE System
3.2. Adapting to Periodic Relationships in Gene Regulatory Networks (GRNs)
3.3. Real-World Data in Mass Spectrometry and Synthetic Biology
3.4. Integrating LazyNet with scRNA-seq Data for Enhanced Trajectory Analysis
3.5. Summary
4. Discussion
5. Conclusions
Supplementary Materials
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ODE | ordinary differential equations |
| ResNet | Residual Network |
| KAN | Kolmogorov–Arnold networks |
| MS | mass spectrometry |
| GRN | gene regulatory networks |
| SCRNA | single-cell RNA |
| RNN | recurrent neural network |
| TPOT | Tree-based Pipeline Optimization Tool |
References
- Katebi, A.; Ramirez, D.; Lu, M. Computational systems-biology approaches for modeling gene networks driving epithelial-mesenchymal transitions. Comput. Syst. Oncol. 2021, 1, e1021. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, M.; Lipson, H. Distilling Free-Form Natural Laws from Experimental Data. Science 2009, 324, 81–85. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.T.Q.; Rubanova, Y.; Bettencourt, J.; Duvenaud, D. Neural Ordinary Differential Equations. arXiv 2019, arXiv:1806.07366. [Google Scholar]
- Qin, J. Neural Networks for ResNet ODE Fitting. arXiv 2019, arXiv:1806.07366. [Google Scholar]
- Kim, S.; Lu, P.Y.; Mukherjee, S.; Gilbert, M.; Jing, L.; Ceperic, V.; Soljacic, M. Integration of Neural Network-Based Symbolic Regression in Deep Learning for Scientific Discovery. IEEE Trans. Neural Netw. Learn. Syst. 2021, 32, 4166–4177. [Google Scholar] [CrossRef]
- Chen, Z.; Liu, Y.; Sun, H. Physics-informed learning of governing equations from scarce data. Nat. Commun. 2021, 12, 6136. [Google Scholar] [CrossRef]
- Qin, T.; Wu, K.; Xiu, D. Data driven governing equations approximation using deep neural networks. J. Comput. Phys. 2019, 395, 620–635. [Google Scholar] [CrossRef]
- Yulia, R.; Ricky, T.; Chen, Q.; David, D. Latent ODEs for irregularly-sampled time series. In Proceedings of the 33rd International Conference on Neural Information Processing Systems, Vancouver, BC, Canada, 8–14 December 2019; Curran Associates Inc.: Red Hook, NY, USA, 2019; Volume 478, pp. 5320–5330. [Google Scholar]
- Golovanev, Y.; Hvatov, A. On the balance between the training time and interpretability of neural ODE for time series modelling. arXiv 2022, arXiv:2206.03304. [Google Scholar]
- Quax, S.C.; D’asaro, M.; van Gerven, M.A.J. Adaptive time scales in recurrent neural networks. Sci. Rep. 2020, 10, 11360. [Google Scholar] [CrossRef]
- Quaghebeur, W.; Torfs, E.; De Baets, B.; Nopens, I. Hybrid differential equations: Integrating mechanistic and data-driven techniques for modelling of water systems. Water Res. 2022, 213, 118166. [Google Scholar] [CrossRef]
- Ye, H.; Zhang, Y.; Liu, H.; Li, X.; Chang, J.; Zheng, H. Light Recurrent Unit: Towards an Interpretable Recurrent Neural Network for Modeling Long-Range Dependency. Electronics 2024, 13, 3204. [Google Scholar] [CrossRef]
- Wu, H.; Lu, T.; Xue, H.; Liang, H. Sparse Additive ODEs for Dynamic Gene Regulatory Network Modeling. J. Am. Stat. Assoc. 2014, 109, 700–716. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Xue, H.; Kumar, A. Numerical Discretization-Based Estimation Methods for Ordinary Differential Equation Models via Penalized Spline Smoothing with Applications in Biomedical Research. Biometrics 2012, 68, 344–352. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.; Miao, H.; Wu, H. Neural Generalized Ordinary Differential Equations with Layer-Varying Parameters. J. Data Sci. 2024, 22, 10–24. [Google Scholar] [CrossRef]
- Fakhoury, D.; Fakhoury, E.; Speleers, H. ExSpliNet: An interpretable and expressive spline-based neural network. Neural Netw. 2022, 152, 332–346. [Google Scholar] [CrossRef]
- Liu, Z.; Wang, Y.; Vaidya, S.; Ruehle, F.; Halverson, J.; Soljačić, M.; Hou, T.Y.; Tegmark, M. KAN: Kolmogorov-Arnold Networks. arXiv 2024, arXiv:2404.19756. [Google Scholar]
- van Deutekom, H.W.M.; Wijnker, G.; de Boer, R.J. The rate of immune escape vanishes when multiple immune responses control an HIV infection. J. Immunol. 2013, 191, 3277–3286. [Google Scholar] [CrossRef]
- Zhang, Q.; Yu, Y.; Zhang, J.; Liang, H. Using single-index ODEs to study dynamic gene regulatory network. PLoS ONE 2018, 13, e0192833. [Google Scholar] [CrossRef]
- Yi, Z.; Geng, S.; Li, L. Comparative analyses of monocyte memory dynamics from mice to humans. Inflamm. Res. 2023, 72, 1539–1549. [Google Scholar] [CrossRef]
- Costello, Z.; Martin, H.G. A machine learning approach to predict metabolic pathway dynamics from time-series multiomics data. NPJ Syst. Biol. Appl. 2018, 4, 19. [Google Scholar] [CrossRef]
- Trang, T.; Le, W.F.; Jason, H.M. Scaling tree-based automated machine learning to biomedical big data with a feature set selector. Bioinformatics 2020, 36, 250–256. [Google Scholar]
- Randal, S.O.; Ryan, J.U.; Peter, C.A.; Nicole, A.L.; La Creis, K.; Jason, H.M. Automating biomedical data science through tree-based pipeline optimization. Appl. Evol. Comput. 2016, 123–137. [Google Scholar] [CrossRef]
- Randal, S.O.; Nathan, B.; Ryan, J.U.; Jason, H.M. Evaluation of a Tree-based Pipeline Optimization Tool for Automating Data Science. In Proceedings of the GECCO 2016, Denver, CO, USA, 20–24 July 2016; pp. 485–492. [Google Scholar]
- La Manno, G.; Soldatov, R.; Zeisel, A.; Braun, E.; Hochgerner, H.; Petukhov, V.; Lidschreiber, K.; Kastriti, M.E.; Lönnerberg, P.; Furlan, A.; et al. RNA velocity of single cells. Nature 2018, 560, 494–498. [Google Scholar] [CrossRef]
- Hao, Y.; Stuart, T.; Kowalski, M.H.; Choudhary, S.; Hoffman, P.; Hartman, A.; Srivastava, A.; Molla, G.; Madad, S.; Fernandez-Granda, C.; et al. Dictionary learning for integrative, multimodal and scalable single-cell analysis. Nat. Biotechnol. 2024, 42, 293–304. [Google Scholar] [CrossRef]
- Akhter, N.; Hasan, A.; Shenouda, S.; Wilson, A.; Kochumon, S.; Ali, S.; Tuomilehto, J.; Sindhu, S.; Ahmad, R. TLR4/MyD88 -mediated CCL2 production by lipopolysaccharide (endotoxin): Implications for metabolic inflammation. J. Diabetes Metab. Disord. 2018, 17, 77–84. [Google Scholar] [CrossRef]
- Kochumon, S.; Wilson, A.; Chandy, B.; Shenouda, S.; Tuomilehto, J.; Sindhu, S.; Ahmad, R. Palmitate Activates CCL4 Expression in Human Monocytic Cells via TLR4/MyD88 Dependent Activation of NF-κB/MAPK/PI3K Signaling Systems. Cell. Physiol. Biochem. 2018, 46, 953–964. [Google Scholar] [CrossRef]
- Zhou, H.; Zhao, C.; Shao, R.; Xu, Y.; Zhao, W. The functions and regulatory pathways of S100A8/A9 and its receptors in cancers. Front. Pharmacol. 2023, 14, 1187741. [Google Scholar] [CrossRef]
- Tsai, S.-Y.; Segovia, J.A.; Chang, T.-H.; Morris, I.R.; Berton, M.T.; Tessier, P.A.; Tardif, M.R.; Cesaro, A.; Bose, S. DAMP molecule S100A9 acts as a molecular pattern to enhance inflammation during influenza A virus infection: Role of DDX21-TRIF-TLR4-MyD88 pathway. PLoS Pathog. 2014, 10, e1003848. [Google Scholar] [CrossRef]
- Weighardt, H.; Mages, J.; Jusek, G.; Kaiser-Moore, S.; Lang, R.; Holzmann, B. Organ-specific role of MyD88 for gene regulation during polymicrobial peritonitis. Infect. Immun. 2006, 74, 3618–3632. [Google Scholar] [CrossRef]
- Gao, X.; Gao, L.-F.; Zhang, Y.-N.; Kong, X.-Q.; Jia, S.; Meng, C.-Y. Huc-MSCs-derived exosomes attenuate neuropathic pain by inhibiting activation of the TLR2/MyD88/NF-κB signaling pathway in the spinal microglia by targeting Rsad2. Int. Immunopharmacol. 2023, 114, 109505. [Google Scholar] [CrossRef]
- Zhang, T.; Fu, J.-N.; Chen, G.-B.; Zhang, X. Plac8-ERK pathway modulation of monocyte function in sepsis. Cell Death Discov. 2024, 10, 308. [Google Scholar] [CrossRef] [PubMed]
- Pradhan, K.; Yi, Z.; Geng, S.; Li, L. Development of Exhausted Memory Monocytes and Underlying Mechanisms. Front. Immunol. 2021, 12, 778830. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald, K.A.; Rowe, D.C.; Barnes, B.J.; Caffrey, D.R.; Visintin, A.; Latz, E.; Monks, B.; Pitha, P.M.; Golenbock, D.T. LPS-TLR4 signaling to IRF-3/7 and NF-kappaB involves the toll adapters TRAM and TRIF. J. Exp. Med. 2003, 198, 1043–1055. [Google Scholar] [CrossRef] [PubMed]
- Kawai, T.; Takeuchi, O.; Fujita, T.; Inoue, J.-I.; Mühlradt, P.F.; Sato, S.; Hoshino, K.; Akira, S. Lipopolysaccharide stimulates the MyD88-independent pathway and results in activation of IFN-regulatory factor 3 and the expression of a subset of lipopolysaccharide-inducible genes. J. Immunol. 2001, 167, 5887–5894. [Google Scholar] [CrossRef]
- Owen, A.M.; Luan, L.; Burelbach, K.R.; McBride, M.A.; Stothers, C.L.; Boykin, O.A.; Sivanesam, K.; Schaedel, J.F.; Patil, T.K.; Wang, J.; et al. MyD88-dependent signaling drives toll-like receptor-induced trained immunity in macrophages. Front. Immunol. 2022, 13, 1044662. [Google Scholar] [CrossRef]

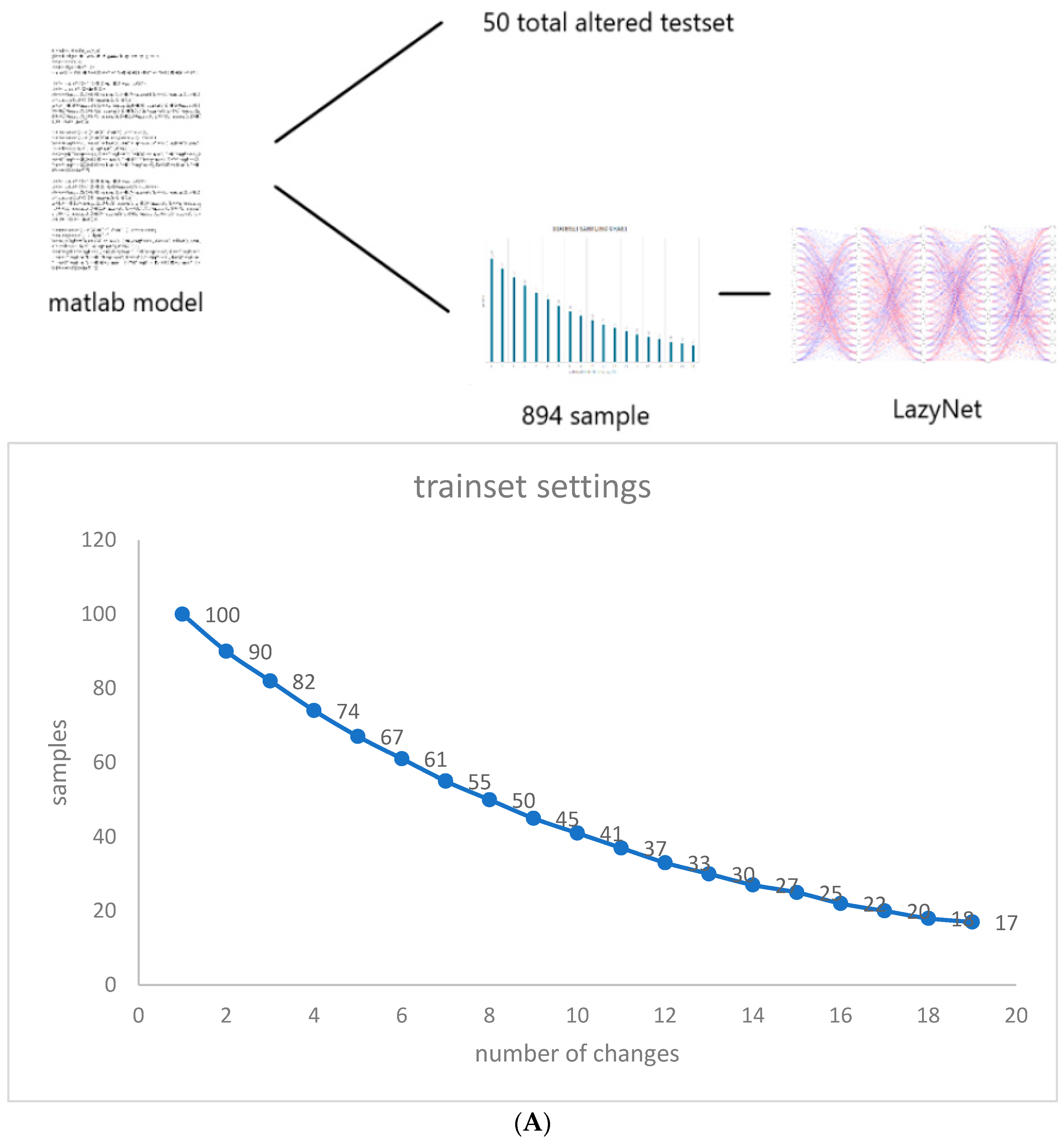


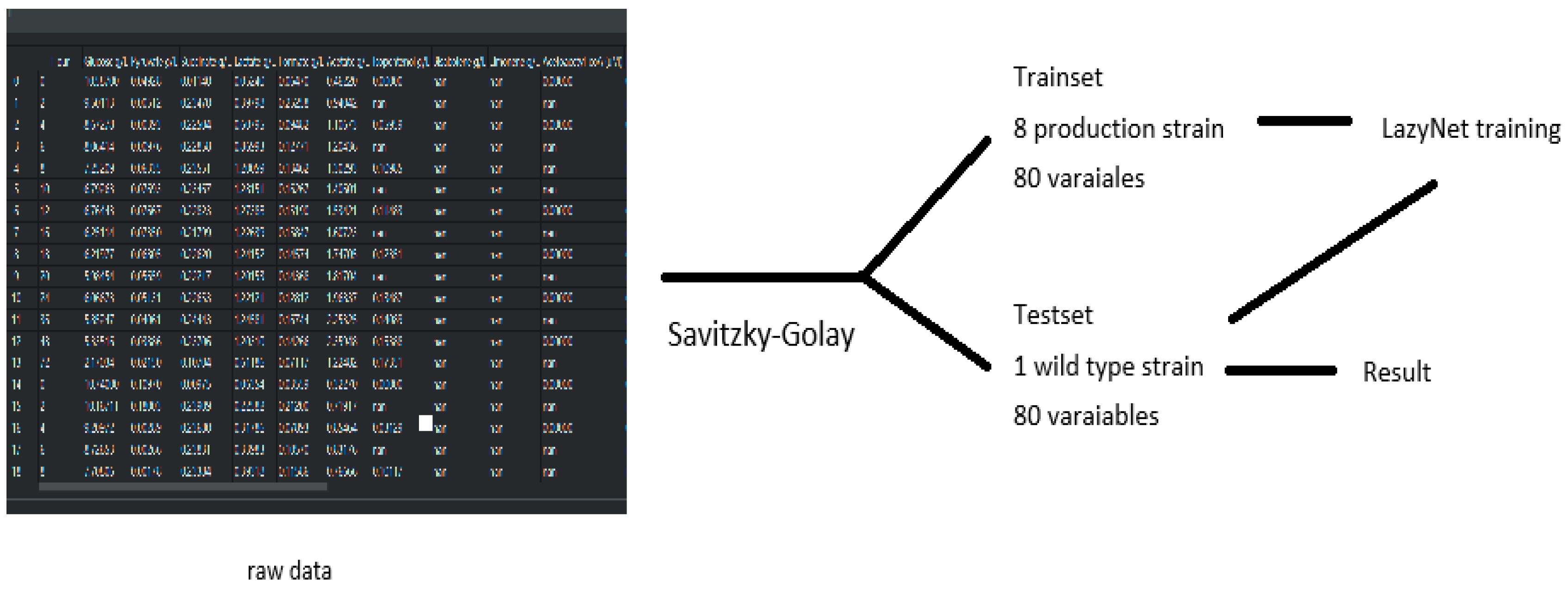

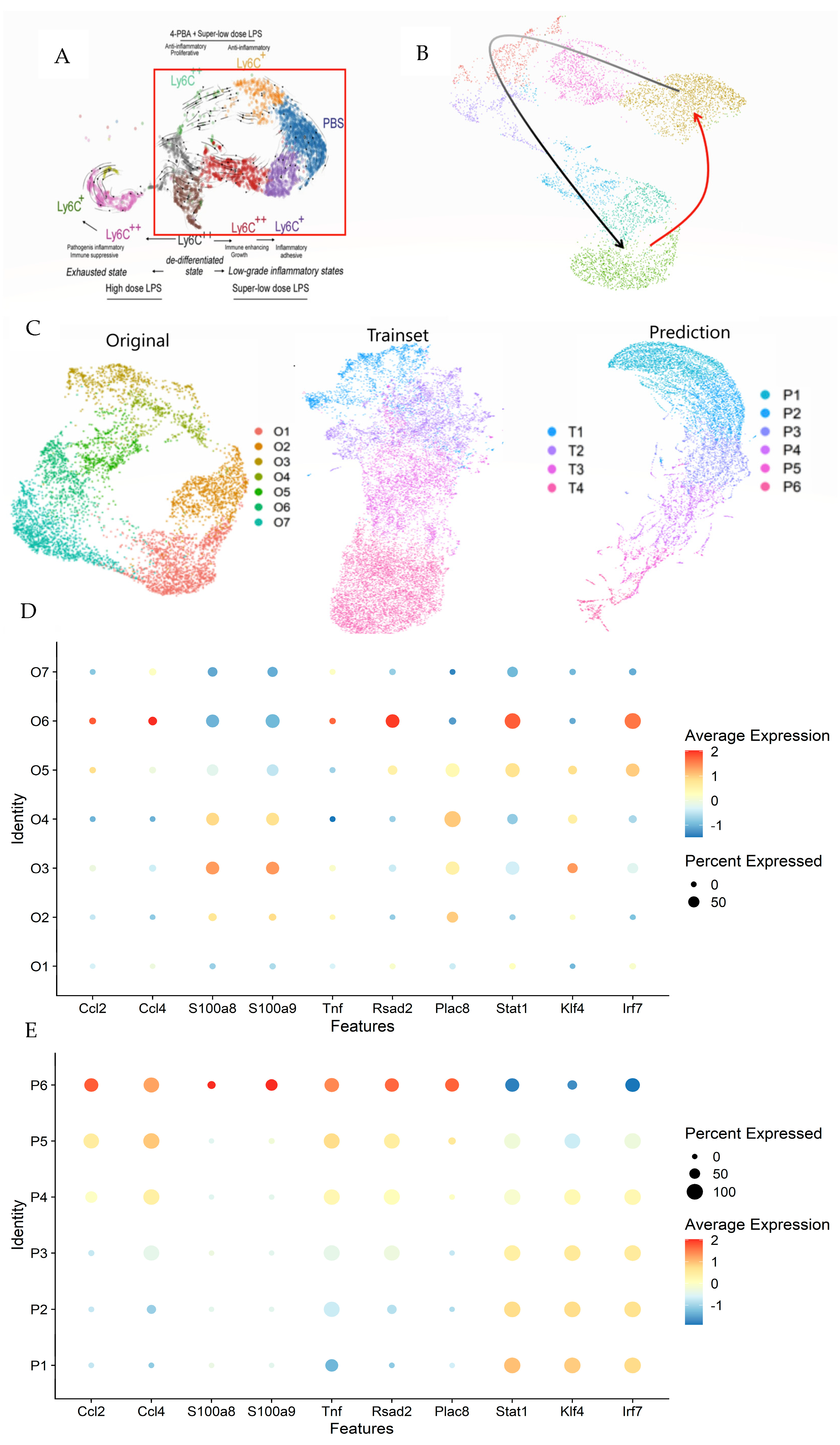
| Case | Trainset | Features 1 | RMSD 2 | AUC | Pearson Correlation | Training Time (hr) | Params | Purpose |
|---|---|---|---|---|---|---|---|---|
| HIV | 894 | 20 | 0.0079 | 0.75 | 0.998 | ~8 | 2280 | Dynamics |
| GRN | 26,450 | 8 | 0.02 | 0.93 | 0.997 | ~16 | 624 | Dynamics |
| Metabolomics | 8 | 80 | 0.019 | 0.75 | 0.96 | 0.75 | 25,920 | Dynamics |
| scRNA | 1 | 718 | NA | NA | NA | ~4 | 3,088,836 | Extension |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yi, Z. Integrating Equation Coding with Residual Networks for Efficient ODE Approximation in Biological Research. Math. Comput. Appl. 2025, 30, 47. https://doi.org/10.3390/mca30030047
Yi Z. Integrating Equation Coding with Residual Networks for Efficient ODE Approximation in Biological Research. Mathematical and Computational Applications. 2025; 30(3):47. https://doi.org/10.3390/mca30030047
Chicago/Turabian StyleYi, Ziyue. 2025. "Integrating Equation Coding with Residual Networks for Efficient ODE Approximation in Biological Research" Mathematical and Computational Applications 30, no. 3: 47. https://doi.org/10.3390/mca30030047
APA StyleYi, Z. (2025). Integrating Equation Coding with Residual Networks for Efficient ODE Approximation in Biological Research. Mathematical and Computational Applications, 30(3), 47. https://doi.org/10.3390/mca30030047






