Adverse Effects of Arsenic Uptake in Rice Metabolome and Lipidome Revealed by Untargeted Liquid Chromatography Coupled to Mass Spectrometry (LC-MS) and Regions of Interest Multivariate Curve Resolution
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Plant Growth, Arsenic Treatments, and Extraction Protocols
2.2.1. General Growing Conditions and Harvesting
2.2.2. Watering and Soil Treatments
2.2.3. Lipid Extraction
2.2.4. Metabolite Extraction
2.3. LC-MS Analysis
2.3.1. Lipidomic Analysis
2.3.2. Metabolomic Analysis
2.4. Data Analysis
2.4.1. Data Compression, Filtering, and Normalization
2.4.2. Statistical Assessment, Exploratory Analysis, and Discovery of Markers of the Exposure
2.4.3. Compound Identification
3. Results
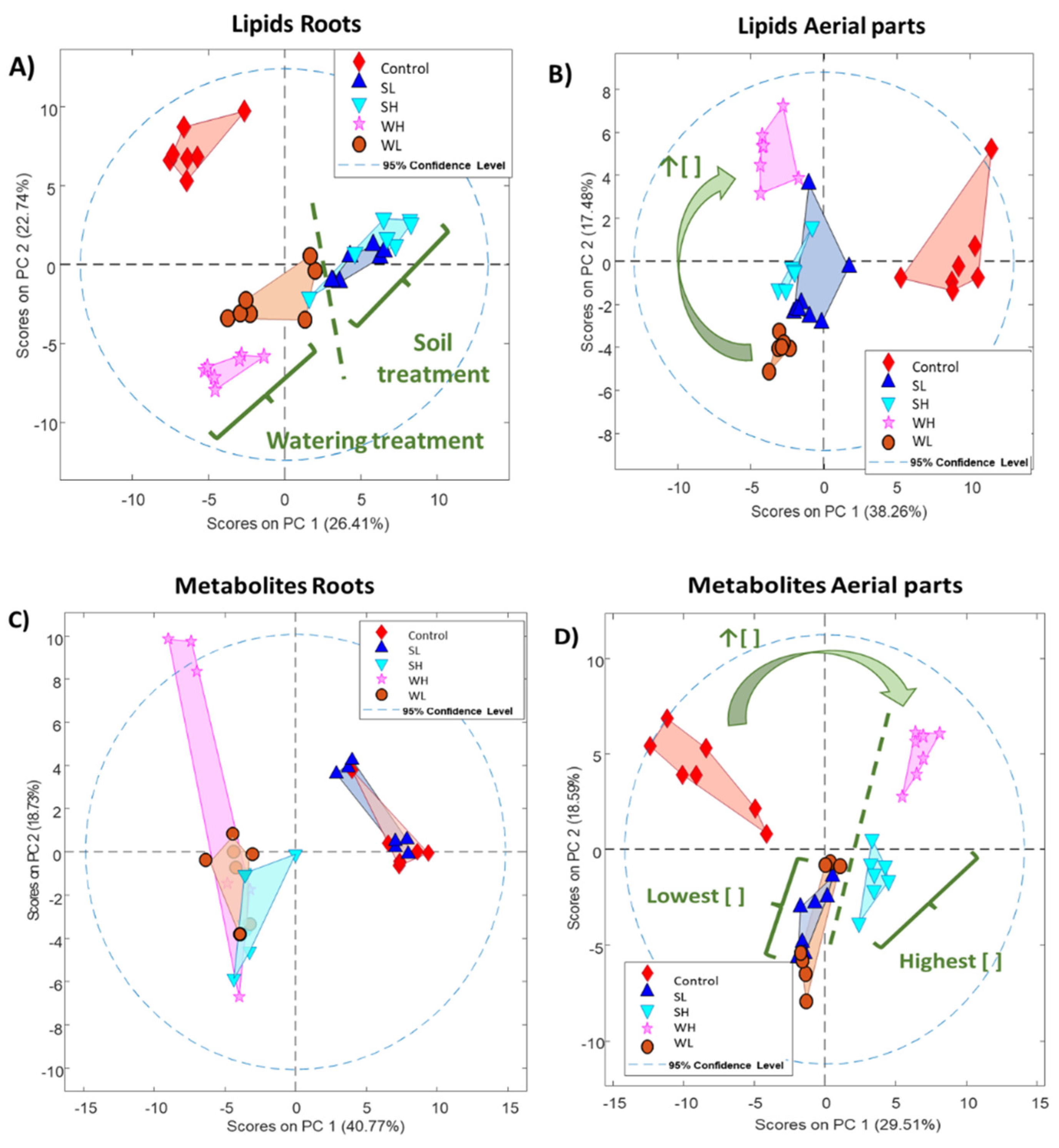
3.1. Statistical Assessment and Exploratory Analysis of Arsenic Exposure
3.2. MCR-ALS Component Selection and Annotation
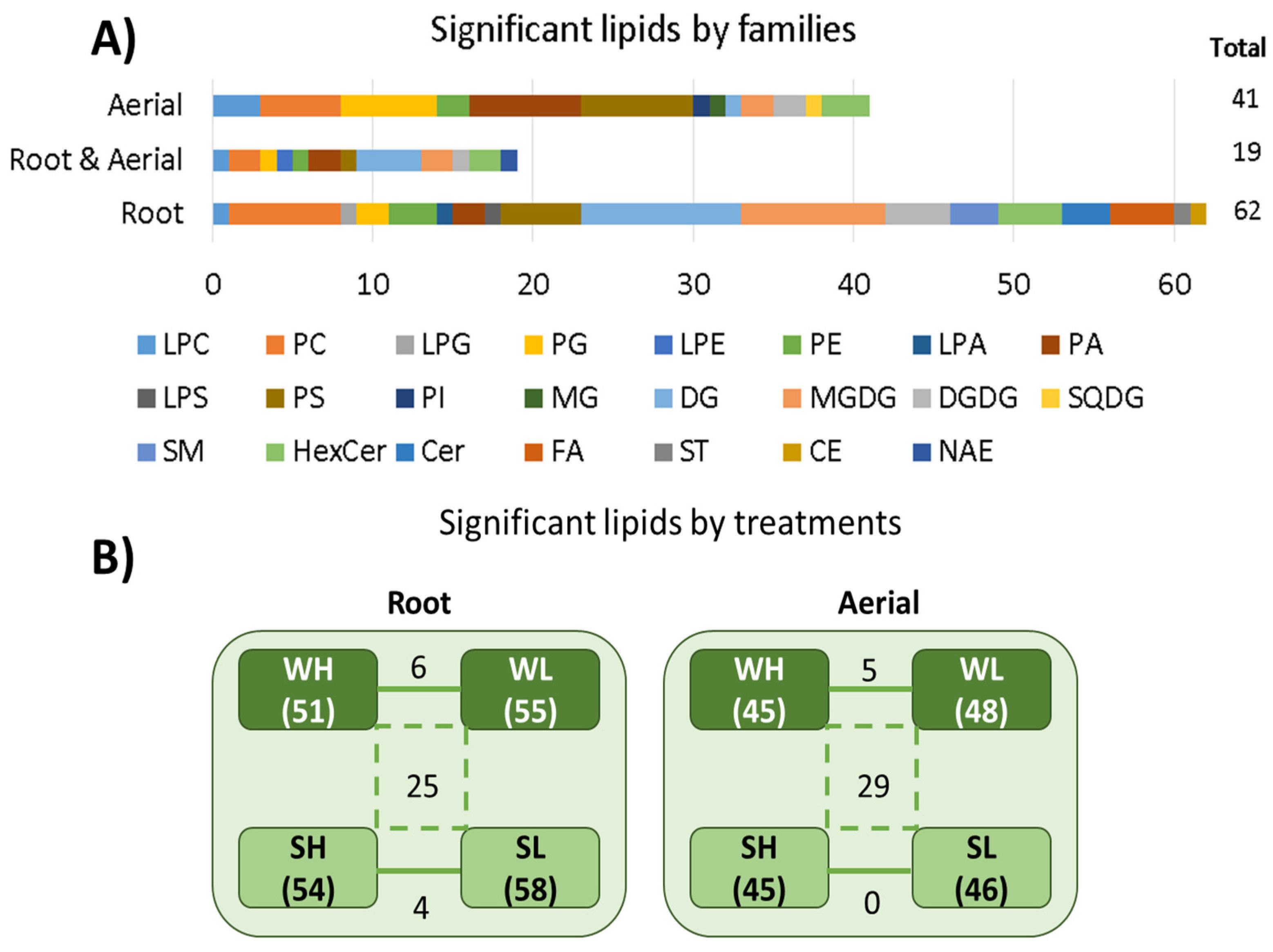
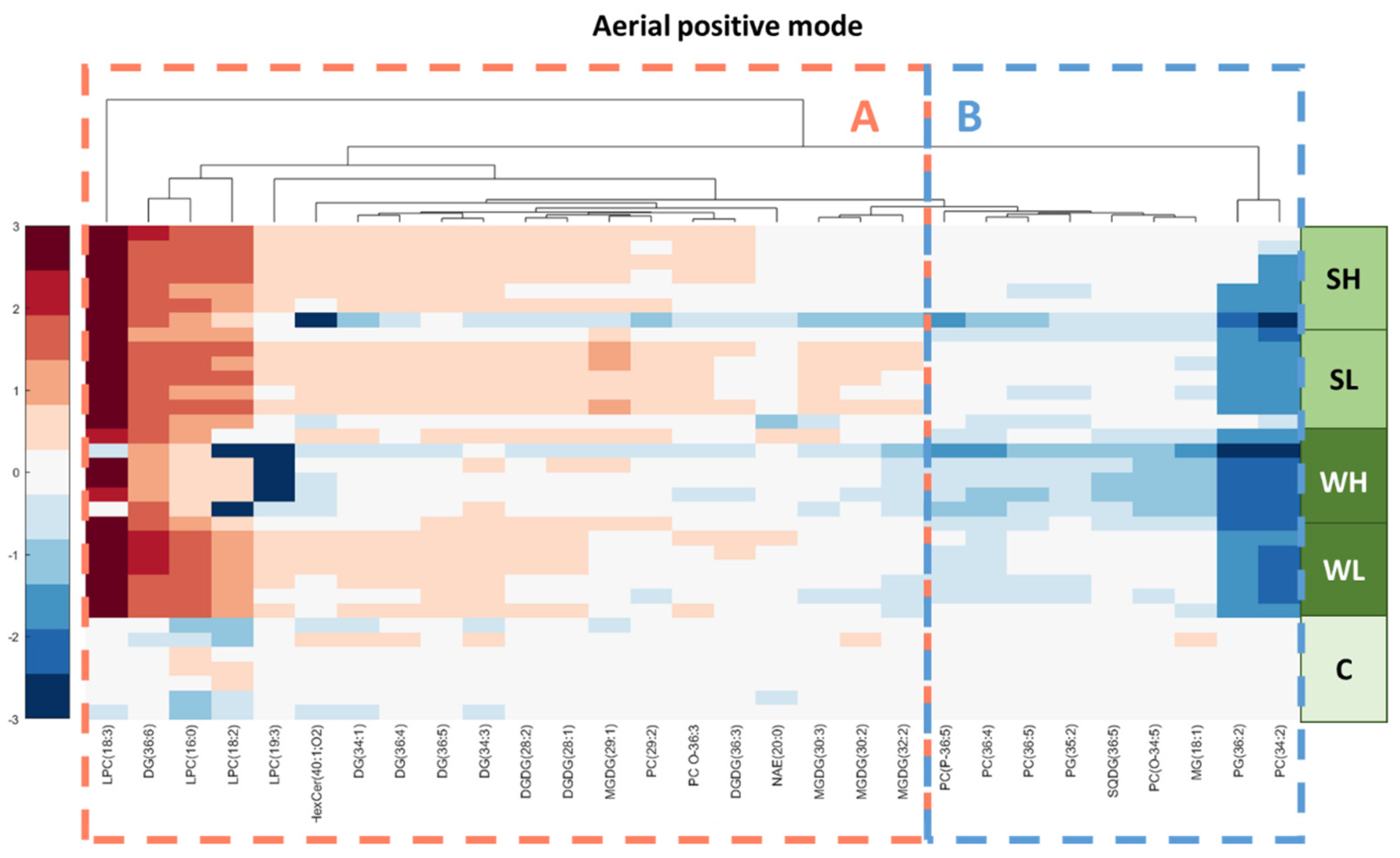
3.3. Lipidomic Results
3.4. Metabolomic Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Awika, J.M. Major Cereal Grains Production and Use around the World. ACS Symp. Ser. 2011, 1089, 1–13. [Google Scholar] [CrossRef]
- Kaur, B.; Sandhu, K.S.; Kamal, R.; Kaur, K.; Singh, J.; Röder, M.S.; Muqaddasi, Q.H. Omics for the Improvement of Abiotic, Biotic, and Agronomic Traits in Major Cereal Crops: Applications, Challenges, and Prospects. Plants 2021, 10, 1989. [Google Scholar] [CrossRef]
- Tarpley, L.; Duran, A.L.; Kebrom, T.H.; Sumner, L.W. Biomarker Metabolites Capturing the Metabolite Variance Present in a Rice Plant Developmental Period. BMC Plant Biol. 2005, 5, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xue, J.; Lu, D.; Wang, S.; Lu, Z.; Liu, W.; Wang, X.; Fang, Z.; He, X. Integrated Transcriptomic and Metabolomic Analysis Provides Insight into the Regulation of Leaf Senescence in Rice. Sci. Rep. 2021, 11, 14083. [Google Scholar] [CrossRef] [PubMed]
- Hall, R.D.; Brouwer, I.D.; Fitzgerald, M.A. Plant Metabolomics and Its Potential Application for Human Nutrition. Physiol. Plant. 2008, 132, 162–175. [Google Scholar] [CrossRef]
- Yang, Y.; Saand, M.A.; Huang, L.; Abdelaal, W.B.; Zhang, J.; Wu, Y.; Li, J.; Sirohi, M.H.; Wang, F. Applications of Multi-Omics Technologies for Crop Improvement. Front. Plant Sci. 2021, 12, 1846. [Google Scholar] [CrossRef]
- Thi, K.; Vo, X.; Rahman, M.; Rahman, M.; Trinh, T.; Kim, S.T.; Jeon, J.-S. Proteomics and Metabolomics Studies on the Biotic Stress Responses of Rice: An Update. Rice 2021, 14, 30. [Google Scholar] [CrossRef]
- Glaubitz, U.; Li, X.; Schaedel, S.; Erban, A.; Sulpice, R.; Kopka, J.; Hincha, D.K.; Zuther, E. Integrated Analysis of Rice Transcriptomic and Metabolomic Responses to Elevated Night Temperatures Identifies Sensitivity- and Tolerance-Related Profiles. Plant Cell Environ. 2017, 40, 121–137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, P.; De, B. Metabolomics Analysis of Rice Responses to Salinity Stress Revealed Elevation of Serotonin, and Gentisic Acid Levels in Leaves of Tolerant Varieties. Plant Signal. Behav. 2017, 12, e1335845. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Hou, H.; Liu, Y.; Yin, S.; Bian, S.; Liang, S.; Wan, C.; Yuan, S.; Xiao, K.; Liu, B.; et al. Microplastics Affect Rice (Oryza sativa L.) Quality by Interfering Metabolite Accumulation and Energy Expenditure Pathways: A Field Study. J. Hazard. Mater. 2022, 422, 126834. [Google Scholar] [CrossRef] [PubMed]
- Saeed, M.; Quraishi, U.M.; Malik, R.N. Arsenic Uptake and Toxicity in Wheat (Triticum aestivum L.): A Review of Multi-Omics Approaches to Identify Tolerance Mechanisms. Food Chem. 2021, 355, 129607. [Google Scholar] [CrossRef] [PubMed]
- Jamla, M.; Khare, T.; Joshi, S.; Patil, S.; Penna, S.; Kumar, V. Omics Approaches for Understanding Heavy Metal Responses and Tolerance in Plants. Curr. Plant Biol. 2021, 27, 100213. [Google Scholar] [CrossRef]
- Oikawa, A.; Matsuda, F.; Kusano, M.; Okazaki, Y.; Saito, K. Rice Metabolomics. Rice 2008, 1, 63–71. [Google Scholar] [CrossRef] [Green Version]
- Kim, T.J.; Kim, S.Y.; Park, Y.J.; Lim, S.H.; Ha, S.H.; Park, S.U.; Lee, B.; Kim, J.K. Metabolite Profiling Reveals Distinct Modulation of Complex Metabolic Networks in Non-Pigmented, Black, and Red Rice (Oryza sativa L.) Cultivars. Metabolites 2021, 11, 367. [Google Scholar] [CrossRef] [PubMed]
- Kusano, M.; Yang, Z.; Okazaki, Y.; Nakabayashi, R.; Fukushima, A.; Saito, K. Using Metabolomic Approaches to Explore Chemical Diversity in Rice. Mol. Plant 2015, 8, 58–67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Z.; Nakabayashi, R.; Okazaki, Y.; Mori, T.; Takamatsu, S.; Kitanaka, S.; Kikuchi, J.; Saito, K. Toward Better Annotation in Plant Metabolomics: Isolation and Structure Elucidation of 36 Specialized Metabolites from Oryza sativa (Rice) by Using MS/MS and NMR Analyses. Metabolomics 2014, 10, 543–555. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.; Lan, M.M.; He, E.K.; Yao, A.J.; Wang, G.B.; Tang, Y.T.; Qiu, R.L. Phenomic and Metabolomic Responses of Roots to Cadmium Reveal Contrasting Resistance Strategies in Two Rice Cultivars (Oryza sativa L.). Soil Ecol. Lett. 2021, 3, 220–229. [Google Scholar] [CrossRef]
- Booth, S.C.; Workentine, M.L.; Weljie, A.M.; Turner, R.J. Metabolomics and Its Application to Studying Metal Toxicity. Metallomics 2011, 3, 1142–1152. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.; Ji, S.; Ping, J.; Cui, D. Recent Advances in Metabolomics for Studying Heavy Metal Stress in Plants. TrAC—Trends Anal. Chem. 2021, 143, 116402. [Google Scholar] [CrossRef]
- World Health Organization Web Page. Available online: https://Www.Who.Int/News-Room/Fact-Sheets/Detail/Arsenic (accessed on 14 February 2022).
- Shankar, S.; Shanker, U. Shikha Arsenic Contamination of Groundwater: A Review of Sources, Prevalence, Health Risks, and Strategies for Mitigation. Sci. World J. 2014, 2014, 304524. [Google Scholar] [CrossRef] [PubMed]
- Bundschuh, J.; Schneider, J.; Alam, M.A.; Niazi, N.K.; Herath, I.; Parvez, F.; Tomaszewska, B.; Guilherme, L.R.G.; Maity, J.P.; López, D.L.; et al. Seven Potential Sources of Arsenic Pollution in Latin America and Their Environmental and Health Impacts. Sci. Total Environ. 2021, 780, 146274. [Google Scholar] [CrossRef] [PubMed]
- Ren, S.; Song, C.; Ye, S.; Cheng, C.; Gao, P. The Spatiotemporal Variation in Heavy Metals in China’s Farmland Soil over the Past 20 years: A Meta-Analysis. Sci. Total Environ. 2022, 806, 150322. [Google Scholar] [CrossRef] [PubMed]
- Nilkarnjanakul, W.; Watchalayann, P.; Chotpantarat, S. Spatial Distribution and Health Risk Assessment of As and Pb Contamination in the Groundwater of Rayong Province, Thailand. Environ. Res. 2022, 204, 111838. [Google Scholar] [CrossRef]
- Varol, M.; Gündüz, K.; Sünbül, M.R. Pollution Status, Potential Sources and Health Risk Assessment of Arsenic and Trace Metals in Agricultural Soils: A Case Study in Malatya Province, Turkey. Environ. Res. 2021, 202, 111806. [Google Scholar] [CrossRef]
- Lima, J.Z.; Ferreira da Silva, E.; Patinha, C.; Durães, N.; Vieira, E.M.; Rodrigues, V.G.S. Sorption of Arsenic by Composts and Biochars Derived from the Organic Fraction of Municipal Solid Wastes: Kinetic, Isotherm and Oral Bioaccessibility Study. Environ. Res. 2022, 204, 111988. [Google Scholar] [CrossRef]
- Li, Y.; Bi, Y.; Mi, W.; Xie, S.; Ji, L. Land-Use Change Caused by Anthropogenic Activities Increase Fluoride and Arsenic Pollution in Groundwater and Human Health Risk. J. Hazard. Mater. 2021, 406, 124337. [Google Scholar] [CrossRef] [PubMed]
- US Food and Drug Administration. Arsenic in Rice and Rice Products Risk Assessment Report; Center for Food Safety and Applied Nutrition of the Food and Drug Administration, 2016; Volume 1, pp. 1–284. Available online: http://www.fda.gov/Food/FoodScienceResearch/RiskSafetyAssessment/default.htm (accessed on 15 March 2022).
- Roel, A.; Campos, F.; Verger, M.; Huertas, R.; Carracelas, G. Regional Variability of Arsenic Content in Uruguayan Polished Rice. Chemosphere 2022, 288, 132426. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.J.; McGrath, S.P.; Meharg, A.A. Arsenic as a Food Chain Contaminant: Mechanisms of Plant Uptake and Metabolism and Mitigation Strategies. Annu. Rev. Plant Biol. 2010, 61, 535–559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mishra, S.; Mattusch, J.; Wennrich, R. Accumulation and Transformation of Inorganic and Organic Arsenic in Rice and Role of Thiol-Complexation to Restrict Their Translocation to Shoot. Sci. Rep. 2017, 7, 40522. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Dubey, R.S.; Tripathi, R.D.; Chakrabarty, D.; Trivedi, P.K. Omics and Biotechnology of Arsenic Stress and Detoxification in Plants: Current Updates and Prospective. Environ. Int. 2015, 74, 221–230. [Google Scholar] [CrossRef] [PubMed]
- Tuli, R.; Chakrabarty, D.; Trivedi, P.K.; Tripathi, R.D. Recent Advances in Arsenic Accumulation and Metabolism in Rice. Mol. Breed. 2010, 26, 307–323. [Google Scholar] [CrossRef]
- Perez de Souza, L.; Alseekh, S.; Naake, T.; Fernie, A. Mass Spectrometry-Based Untargeted Plant Metabolomics. Curr. Protoc. Plant Biol. 2019, 4, e20100. [Google Scholar] [CrossRef]
- Akram, M.I.; Vincent, I.M.; Siddiqui, A.J.; Musharraf, S.G. Polymeric Hydrophilic Interaction Liquid Chromatography Coupled with Orbitrap Mass Spectrometry and Chemometric Analysis for Untargeted Metabolite Profiling of Natural Rice Variants. J. Cereal Sci. 2017, 73, 165–173. [Google Scholar] [CrossRef]
- Xiao, R.; Ma, Y.; Zhang, D.; Qian, L. Discrimination of Conventional and Organic Rice Using Untargeted LC-MS-Based Metabolomics. J. Cereal Sci. 2018, 82, 73–81. [Google Scholar] [CrossRef]
- Concepcion, J.C.T.; Calingacion, M.; Garson, M.J.; Fitzgerald, M.A. Lipidomics Reveals Associations between Rice Quality Traits. Metabolomics 2020, 16, 54. [Google Scholar] [CrossRef]
- Navas-Iglesias, N.; Carrasco-Pancorbo, A.; Cuadros-Rodríguez, L. From Lipids Analysis towards Lipidomics, a New Challenge for the Analytical Chemistry of the 21st Century. Part II: Analytical Lipidomics. TrAC—Trends Anal. Chem. 2009, 28, 393–403. [Google Scholar] [CrossRef]
- Navarro-Reig, M.; Jaumot, J.; Tauler, R. An Untargeted Lipidomic Strategy Combining Comprehensive Two-Dimensional Liquid Chromatography and Chemometric Analysis. J. Chromatogr. A 2018, 1568, 80–90. [Google Scholar] [CrossRef] [PubMed]
- Matyash, V.; Liebish, G.; Kurzchalia, T.V.; Shevchenko, A.; Schwudke, D. Lipid Extraction by Methyl-Tert-Butyl Ether for High-Throughput Lipidomics. J Lipid Res. 2008, 49, 1137–1146. [Google Scholar] [CrossRef] [Green Version]
- Navarro-Reig, M.; Jaumot, J.; Piña, B.; Moyano, E.; Galceran, M.T.; Tauler, R. Metabolomic Analysis of the Effects of Cadmium and Copper Treatment in: Oryza sativa L. Using Untargeted Liquid Chromatography Coupled to High Resolution Mass Spectrometry and All-Ion Fragmentation. Metallomics 2017, 9, 660–675. [Google Scholar] [CrossRef]
- 2006/118/EC. Directive 2006/118/EC of the European Parliment and of the Council of 12 December 2006 on the Protection of Groundwater against Pollution and Deterioration. Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/?uri=celex%3A32006L0118 (accessed on 15 March 2022).
- Akhter, H.; Cartledge, F.K.; Miller, J.; McLearn, M. Treatment of Arsenic-Contaminated Soils. I: Soil Characterization. J. Environ. Eng. 2000, 126, 999–1003. [Google Scholar] [CrossRef]
- Menéndez-Pedriza, A.; Jaumot, J.; Bedia, C. Lipidomic analysis of single and combined effects of polyethylene microplastics and polychlorinated biphenyls on human hepatoma cells. J. Hazard. Mater. 2022, 421, 126777. [Google Scholar] [CrossRef]
- Ortiz-Villanueva, E.; Jaumot, J.; Martínez, R.; Navarro-Martín, L.; Piña, B.; Tauler, R. Assessment of Endocrine Disruptors Effects on Zebrafish (Danio Rerio) Embryos by Untargeted LC-HRMS Metabolomic Analysis. Sci. Total Environ. 2018, 635, 156–166. [Google Scholar] [CrossRef] [PubMed]
- Navarro-Reig, M.; Jaumot, J.; García-Reiriz, A.; Tauler, R. Evaluation of Changes Induced in Rice Metabolome by Cd and Cu Exposure Using LC-MS with XCMS and MCR-ALS Data Analysis Strategies. Anal. Bioanal. Chem. 2015, 407, 8835–8847. [Google Scholar] [CrossRef] [PubMed]
- Puig-Castellví, F.; Bedia, C.; Alfonso, I.; Piña, B.; Tauler, R. Deciphering the Underlying Metabolomic and Lipidomic Patterns Linked to Thermal Acclimation in Saccharomyces Cerevisiae. J. Proteome Res. 2018, 17, 2034–2044. [Google Scholar] [CrossRef] [PubMed]
- Chambers, M.C.; Maclean, B.; Burke, R.; Amode, D.; Ruderman, D.L.; Neumann, S.; Gatto, L.; Fischer, B.; Pratt, B.; Egertson, J.; et al. A Cross-Platform Toolkit for Mass Spectrometry and Proteomics. Nat. Biotechnol. 2012, 30, 918–920. [Google Scholar] [CrossRef]
- Gorrochategui, E.; Jaumot, J.; Tauler, R. ROIMCR: A Powerful Analysis Strategy for LC-MS Metabolomic Datasets. BMC Bioinform. 2019, 20, 256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pérez-Cova, M.; Bedia, C.; Stoll, D.R.; Tauler, R.; Jaumot, J. MSroi: A Pre-Processing Tool for Mass Spectrometry-Based Studies. Chemom. Intell. Lab. Syst. 2021, 215, 104333. [Google Scholar] [CrossRef]
- Smilde, A.K.; Jansen, J.J.; Hoefsloot, H.C.J.; Lamers, R.J.A.N.; van der Greef, J.; Timmerman, M.E. ANOVA-Simultaneous Component Analysis (ASCA): A New Tool for Analyzing Designed Metabolomics Data. Bioinformatics 2005, 21, 3043–3048. [Google Scholar] [CrossRef]
- Joliffe, I.T.; Morgan, B. Principal Component Analysis and Exploratory Factor Analysis. Stat. Methods Med. Res. 1992, 1, 69–95. [Google Scholar] [CrossRef] [PubMed]
- Mangiameli, P.; Chen, S.K.; West, D. A Comparison of SOM Neural Network and Hierarchical Clustering Methods. Eur. J. Oper. Res. 1996, 93, 402–417. [Google Scholar] [CrossRef]
- Gromski, P.S.; Muhamadali, H.; Ellis, D.I.; Xu, Y.; Correa, E.; Turner, M.L.; Goodacre, R. A Tutorial Review: Metabolomics and Partial Least Squares-Discriminant Analysis—A Marriage of Convenience or a Shotgun Wedding. Anal. Chim. Acta 2015, 879, 10–23. [Google Scholar] [CrossRef]
- Lee, L.C.; Liong, C.Y.; Jemain, A.A. Partial Least Squares-Discriminant Analysis (PLS-DA) for Classification of High-Dimensional (HD) Data: A Review of Contemporary Practice Strategies and Knowledge Gaps. Analyst 2018, 143, 3526–3539. [Google Scholar] [CrossRef] [PubMed]
- Chicco, D.; Jurman, G. The Advantages of the Matthews Correlation Coefficient (MCC) over F1 Score and Accuracy in Binary Classification Evaluation. BMC Genom. 2020, 21, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fahy, E.; Sud, M.; Cotter, D.; Subramaniam, S. LIPID MAPS Online Tools for Lipid Research. Nucleic Acids Res. 2007, 35, W606–W612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wishart, D.S.; Guo, A.C.; Oler, E.; Wang, F.; Anjum, A.; Peters, H.; Dizon, R.; Sayeeda, Z.; Tian, S.; Lee, B.L.; et al. HMDB 5.0: The Human Metabolome Database for 2022. Nucleic Acids Res. 2022, 50, D622–D631. [Google Scholar] [CrossRef]
- MS-DIAL Web Page. Available online: http://Prime.Psc.Riken.Jp/Compms/MS-DIAL/Main.Html (accessed on 14 February 2022).
- Schulze, T.; Meier, R.; Alygizakis, N.; Schymanski, E.; Bach, E.; LI, D.H.; lauperbe; raalizadeh; Tanaka, S.; Witting, M. MassBank/MassBank-Data: Release Version 2021.12. 2021. Available online: https://doi.org/10.5281/ZENODO.5775684 (accessed on 17 March 2022).
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and Community Curation of Mass Spectrometry Data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hawkins, C.; Ginzburg, D.; Zhao, K.; Dwyer, W.; Xue, B.; Xu, A.; Rice, S.; Cole, B.; Paley, S.; Karp, P.; et al. Plant Metabolic Network 15: A Resource of Genome-Wide Metabolism Databases for 126 Plants and Algae. J. Integr. Plant Biol. 2021, 63, 1888–1905. [Google Scholar] [CrossRef] [PubMed]
- Tsugawa, H.; Ikeda, K.; Takahashi, M.; Satoh, A.; Mori, Y.; Uchino, H.; Okahashi, N.; Yamada, Y.; Tada, I.; Bonini, P.; et al. A Lipidome Atlas in MS-DIAL 4. Nat. Biotechnol. 2020, 38, 1159–1163. [Google Scholar] [CrossRef] [PubMed]
- Blaženović, I.; Kind, T.; Ji, J.; Fiehn, O. Software Tools and Approaches for Compound Identification of LC-MS/MS Data in Metabolomics. Metabolites 2018, 8, 31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pang, Z.; Chong, J.; Zhou, G.; de Lima Morais, D.A.; Chang, L.; Barrette, M.; Gauthier, C.; Jacques, P.É.; Li, S.; Xia, J. MetaboAnalyst 5.0: Narrowing the Gap between Raw Spectra and Functional Insights. Nucleic Acids Res. 2021, 49, W388–W396. [Google Scholar] [CrossRef] [PubMed]
- Batista, B.L.; Nigar, M.; Mestrot, A.; Rocha, B.A.; Júnior, F.B.; Price, A.H.; Raab, A.; Feldmann, J. Identification and Quantification of Phytochelatins in Roots of Rice to Long-Term Exposure: Evidence of Individual Role on Arsenic Accumulation and Translocation. J. Exp. Bot. 2014, 65, 1467–1479. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, D.; Zhao, L.; Wang, W.; Wang, Q.; Liu, J.; Wang, Y.; Liu, H.; Shang, B.; Duan, X.; Sun, H. Lipidomics Reveals the Changes in Non-Starch and Starch Lipids of Rice (Oryza sativa L.) during Storage. J. Food Compos. Anal. 2022, 105, 104205. [Google Scholar] [CrossRef]
- Liu, L.; Waters, D.L.E.; Rose, T.J.; Bao, J.; King, G.J. Phospholipids in Rice: Significance in Grain Quality and Health Benefits: A Review. Food Chem. 2013, 139, 1133–1145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rizov, I.; Doulis, A. Separation of Plant Membrane Lipids by Multiple Solid-Phase Extraction. J. Chromatogr. A 2001, 922, 347–354. [Google Scholar] [CrossRef]
- Kobayashi, K. Role of Membrane Glycerolipids in Photosynthesis, Thylakoid Biogenesis and Chloroplast Development. J. Plant Res. 2016, 129, 565–580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Basnet, R.; Zhang, J.; Hussain, N.; Shu, Q. Characterization and Mutational Analysis of a Monogalactosyldiacylglycerol Synthase Gene OsMGD2 in Rice. Front. Plant Sci. 2019, 10, 992. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kadotani, N.; Akagi, A.; Takatsuji, H.; Miwa, T.; Igarashi, D. Exogenous Proteinogenic Amino Acids Induce Systemic Resistance in Rice. BMC Plant Biol. 2016, 16, 60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, N.; Zhang, S.; Gu, M.; Xu, G. Function, Transport, and Regulation of Amino Acids: What Is Missing in Rice? Crop J. 2021, 9, 530–542. [Google Scholar] [CrossRef]
- Tripathi, P.; Tripathi, R.D.; Singh, R.P.; Dwivedi, S.; Chakrabarty, D.; Trivedi, P.K.; Adhikari, B. Arsenite Tolerance in Rice (Oryza sativa L.) Involves Coordinated Role of Metabolic Pathways of Thiols and Amino Acids. Environ. Sci. Pollut. Res. 2013, 20, 884–896. [Google Scholar] [CrossRef]
- de Juan, A.; Jaumot, J.; Tauler, R. Multivariate Curve Resolution (MCR). Solving the Mixture Analysis Problem. Anal. Methods 2014, 6, 4964–4976. [Google Scholar] [CrossRef]
- Windig, W.; Stephenson, D.A. Self-Modeling Mixture Analysis of Second-Derivative Near-Infrared Spectral Data using the Simplisma Approach. Anal. Chem. 1992, 64, 2735–2742. [Google Scholar]
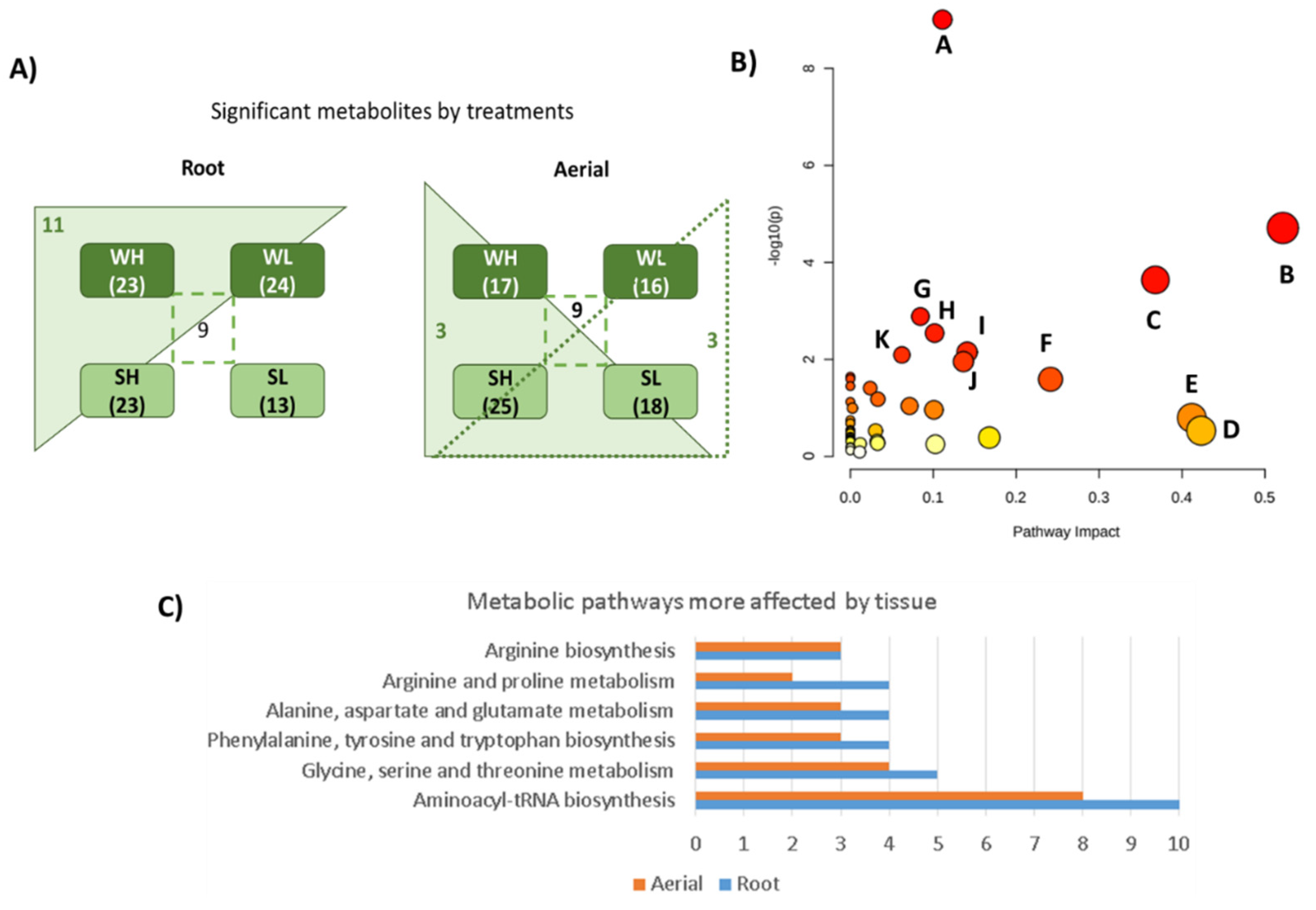
| Result from Pathway Analysis | Total | Expected | Hits | Raw p | −Log10(p) | Holm Adjust | FDR | Impact |
|---|---|---|---|---|---|---|---|---|
| Aminoacyl-tRNA biosynthesis | 46 | 1.31 | 12 | 9.86 × 10−10 | 9.01 | 9.37 × 10−8 | 9.37 × 10−8 | 0.11 |
| Alanine, aspartate, and glutamate metabolism | 22 | 0.63 | 6 | 1.951 × 0−5 | 4.71 | 1.83 × 10-3 | 9.26 × 10-4 | 0.52 |
| Glycine, serine, and threonine metabolism | 33 | 0.94 | 6 | 2.29 × 10−4 | 3.64 | 2.13 × 10-2 | 7.26 × 10−3 | 0.37 |
| Arginine biosynthesis | 18 | 0.51 | 4 | 1.30 × 10−3 | 2.89 | 1.20 × 10-1 | 3.09 × 10−2 | 0.08 |
| Phenylalanine, tyrosine, and tryptophan biosynthesis | 22 | 0.63 | 4 | 2.86 × 10−3 | 2.54 | 2.61 × 10-1 | 5.44 × 10−2 | 0.10 |
| Arginine and proline metabolism | 28 | 0.80 | 4 | 7.08 × 10−3 | 2.15 | 6.37 × 10−1 | 1.09 × 10−1 | 0.14 |
| Glyoxylate and dicarboxylate metabolism | 29 | 0.83 | 4 | 8.04 × 10−3 | 2.09 | 7.16 × 10−1 | 1.09 × 10−1 | 0.06 |
| Butanoate metabolism | 17 | 0.48 | 3 | 1.11 × 10−2 | 1.96 | 9.74 × 10−1 | 1.31 × 10−1 | 0.14 |
| Valine, leucine, and isoleucine biosynthesis | 22 | 0.63 | 3 | 2.27 × 10−2 | 1.64 | 1.00 | 2.21 × 10−1 | 0.00 |
| Lysine biosynthesis | 9 | 0.26 | 2 | 2.51 × 10−2 | 1.60 | 1.00 | 2.21 × 10−1 | 0.00 |
| Tryptophan metabolism | 23 | 0.66 | 3 | 2.56 × 10−2 | 1.59 | 1.00 | 2.21 × 10−1 | 0.24 |
| Cyanoamino acid metabolism | 26 | 0.74 | 3 | 3.54 × 10−2 | 1.45 | 1.00 | 2.81 × 10−1 | 0.00 |
| Cysteine and methionine metabolism | 46 | 1.31 | 4 | 3.90 × 10−2 | 1.41 | 1.00 | 2.85 × 10−1 | 0.02 |
| Sulfur metabolism | 15 | 0.43 | 2 | 6.59 × 10−2 | 1.18 | 1.00 | 4.47 × 10−1 | 0.03 |
| Phenylpropanoid biosynthesis | 35 | 1.00 | 3 | 7.48 × 10−2 | 1.13 | 1.00 | 4.74 × 10−1 | 0.00 |
| beta-Alanine metabolism | 18 | 0.51 | 2 | 9.10 × 10−2 | 1.04 | 1.00 | 5.40 × 10−1 | 0.07 |
| Purine metabolism | 63 | 1.80 | 4 | 1.01 × 10−1 | 9.9710−1 | 1.00 | 5.63 × 10−1 | 0.00 |
| Citrate cycle (TCA cycle) | 20 | 0.57 | 2 | 1.09 × 10−1 | 9.6210−1 | 1.00 | 5.76 × 10−1 | 0.10 |
| Isoquinoline alkaloid biosynthesis | 6 | 0.17 | 1 | 1.60 × 10−1 | 7.9710−1 | 1.00 | 7.98 × 10−1 | 0.41 |
| Galactose metabolism | 27 | 0.77 | 2 | 1.78 × 10−1 | 7.4910−1 | 1.00 | 8.47 × 10−1 | 0.00 |
| Monobactam biosynthesis | 8 | 0.23 | 1 | 2.07 × 10−1 | 6.8410−1 | 1.00 | 8.94 × 10−1 | 0.00 |
| Tropane, piperidine, and pyridine alkaloid biosynthesis | 8 | 0.23 | 1 | 2.07 × 10−1 | 6.8410−1 | 1.00 | 8.94 × 10−1 | 0.00 |
| Valine, leucine, and isoleucine degradation | 37 | 1.05 | 2 | 2.85 × 10−1 | 5.4510−1 | 1.00 | 1.00 | 0.00 |
| Nitrogen metabolism | 12 | 0.34 | 1 | 2.94 × 10−1 | 5.3110−1 | 1.00 | 1.00 | 0.00 |
| Phenylalanine metabolism | 12 | 0.34 | 1 | 2.94 × 10−1 | 5.3110−1 | 1.00 | 1.00 | 0.42 |
| Pyrimidine metabolism | 38 | 1.08 | 2 | 2.96 × 10−1 | 5.2910−1 | 1.00 | 1.00 | 0.03 |
| Nicotinate and nicotinamide metabolism | 13 | 0.37 | 1 | 3.15 × 10−1 | 5.0210−1 | 1.00 | 1.00 | 0.00 |
| Cutin, suberine, and wax biosynthesis | 14 | 0.40 | 1 | 3.34 × 10−1 | 4.76 × 10−1 | 1.00 | 1.00 | 0.00 |
| Sphingolipid metabolism | 17 | 0.48 | 1 | 3.90 × 10−1 | 4.09 × 10−1 | 1.00 | 1.00 | 0.00 |
| Ascorbate and aldarate metabolism | 18 | 0.51 | 1 | 4.08 × 10−1 | 3.90 × 10−1 | 1.00 | 1.00 | 0.00 |
| Tyrosine metabolism | 18 | 0.51 | 1 | 4.08 × 10−1 | 3.90 × 10−1 | 1.00 | 1.00 | 0.17 |
| Fructose and mannose metabolism | 20 | 0.57 | 1 | 4.42 × 10−1 | 3.55 × 10−1 | 1.00 | 1.00 | 0.00 |
| Propanoate metabolism | 20 | 0.57 | 1 | 4.42 × 10−1 | 3.55 × 10−1 | 1.00 | 1.00 | 0.00 |
| Carbon fixation in photosynthetic organisms | 21 | 0.60 | 1 | 4.58 × 10−1 | 3.39 × 10−1 | 1.00 | 1.00 | 0.00 |
| Zeatin biosynthesis | 21 | 0.60 | 1 | 4.58 × 10−1 | 3.39 × 10−1 | 1.00 | 1.00 | 0.00 |
| Biosynthesis of unsaturated fatty acids | 22 | 0.63 | 1 | 4.73 × 10−1 | 3.25 × 10−1 | 1.00 | 1.00 | 0.00 |
| Fatty acid elongation | 23 | 0.66 | 1 | 4.89 × 10−1 | 3.11 × 10−1 | 1.00 | 1.00 | 0.00 |
| Pantothenate and CoA biosynthesis | 23 | 0.66 | 1 | 4.89 × 10−1 | 3.11 × 10−1 | 1.00 | 1.00 | 0.03 |
| Phosphatidylinositol signaling system | 26 | 0.74 | 1 | 5.32 × 10−1 | 2.74 × 10−1 | 1.00 | 1.00 | 0.03 |
| Glutathione metabolism | 27 | 0.77 | 1 | 5.45 × 10−1 | 2.63 × 10−1 | 1.00 | 1.00 | 0.01 |
| Inositol phosphate metabolism | 28 | 0.80 | 1 | 5.59 × 10−1 | 2.53 × 10−1 | 1.00 | 1.00 | 0.10 |
| Ubiquinone and other terpenoid-quinone biosynthesis | 35 | 1.00 | 1 | 6.41 × 10−1 | 1.93 × 10−1 | 1.00 | 1.00 | 0.00 |
| Fatty acid degradation | 37 | 1.05 | 1 | 6.62 × 10−1 | 1.79 × 10−1 | 1.00 | 1.00 | 0.00 |
| Flavonoid biosynthesis | 47 | 1.34 | 1 | 7.49 × 10−1 | 1.25 × 10−1 | 1.00 | 1.00 | 0.00 |
| Fatty acid biosynthesis | 56 | 1.60 | 1 | 8.08 × 10−1 | 9.23 × 10−2 | 1.00 | 1.00 | 0.01 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pérez-Cova, M.; Tauler, R.; Jaumot, J. Adverse Effects of Arsenic Uptake in Rice Metabolome and Lipidome Revealed by Untargeted Liquid Chromatography Coupled to Mass Spectrometry (LC-MS) and Regions of Interest Multivariate Curve Resolution. Separations 2022, 9, 79. https://doi.org/10.3390/separations9030079
Pérez-Cova M, Tauler R, Jaumot J. Adverse Effects of Arsenic Uptake in Rice Metabolome and Lipidome Revealed by Untargeted Liquid Chromatography Coupled to Mass Spectrometry (LC-MS) and Regions of Interest Multivariate Curve Resolution. Separations. 2022; 9(3):79. https://doi.org/10.3390/separations9030079
Chicago/Turabian StylePérez-Cova, Miriam, Romà Tauler, and Joaquim Jaumot. 2022. "Adverse Effects of Arsenic Uptake in Rice Metabolome and Lipidome Revealed by Untargeted Liquid Chromatography Coupled to Mass Spectrometry (LC-MS) and Regions of Interest Multivariate Curve Resolution" Separations 9, no. 3: 79. https://doi.org/10.3390/separations9030079
APA StylePérez-Cova, M., Tauler, R., & Jaumot, J. (2022). Adverse Effects of Arsenic Uptake in Rice Metabolome and Lipidome Revealed by Untargeted Liquid Chromatography Coupled to Mass Spectrometry (LC-MS) and Regions of Interest Multivariate Curve Resolution. Separations, 9(3), 79. https://doi.org/10.3390/separations9030079








