Abstract
The demand for microbial pectinase has increased due to its vast applications in different industries. The current study dealt with the synthesis of pectinase by a novel native strain Aspergillus cervinus ARS2 using agro-industrial waste. Comparative studies conducted on pectinase production by submerged fermentation (SmF) and solid-state fermentation (SSF) showed that pectinase activity was more increased in SSF (44.51 ± 1.33 IU/mL) than in SmF (40.60 ± 1.15 IU/mL) when using orange peel as a substrate. Using SSF, one-factor-at-a-time (OFAT) studies were conducted, considering different process variables such as inoculum size, initial pH, incubation time, moisture content, incubation temperature, and substrate particle size, all of which affected the pectinase activity. OFAT results showed the highest pectinase activity at an inoculum size of 106 spores/mL (43.11 ± 1.06 U/mL), an incubation time of 6 days (43.81 ± 1.21 U/mL), a moisture content of 100% (44.30 ± 1.69 U/mL), a substrate particle size of 1.7 mm (42.06 ± 1.20 U/mL), an incubation temperature of 37 °C (45.90 ± 1.33 U/mL), and an initial pH of 4 (43.31 ± 0.89 U/mL). The identified significant process variables were then optimized by response surface methodology (RSM)-central composite design (CCD). The results showed optimum pectinase activity of 107.14 ± 0.71 IU/mL for a substrate particle size of 2 mm, an incubation temperature of 31.5 °C, an initial pH of 4.9, and a moisture content of 107%, which was obtained from the Minitab optimizer. By using statistical optimization, the pectinase production from the isolated novel fungal strain A. cervinus ARS2 was increased 2.38-fold. Therefore, the A. cervinus ARS2 strain can be further explored for large-scale pectinase production which could meet the growing industrial demands.
1. Introduction
Enzymes are substances which speed up chemical reactions without undergoing any change themselves during the reaction. Enzymes have many advantages over chemical catalysts, such as adjustable activity, high specificity, and high catalytic efficiency, which encourages their use in pharmaceutical, chemical, and food industries [1]. Due to their vast applications, the demand for industrial enzymes has increased, which has led to continuous research and development in optimizing the production of enzymes and in reducing this production cost [2]. Amongst industrially vital enzymes, pectinases have gained importance owing to their various applications in different sectors [1]. Pectinase is the group of enzymes which catalyzes the hydrolysis of substances containing pectin. Pectinases are grouped into polygalacturonase (EC 3.2.15), pectate lyase (EC 4.2.2.2), pectinesterase (EC 3.1.1.11), and pectin lyase (EC 4.2.2.10), depending on the mode of action on pectic substances [3]. The share of pectinase in the global enzyme production is around 10%, and around 25% in the worldwide enzyme market [4,5]. Pectinases have an important role in retting flax and vegetable fibers, coffee, and tea fermentation, biosouring cotton fibers, fruit juice clarification, degumming plant blast fibers, extraction of vegetable oil, haze removal from wines, and wastewater treatment [6,7,8,9,10,11].
Different sources such as microbes, plants, insects, and nematodes are used to produce pectinase. The microbial source is significant because of rapid growth, extensively widespread, less fermentation time, and easy genetic modifications. Microbial pectinases play a very important role in plant pathogenesis, symbiosis, and decomposition of plant deposits [12]. Among microbial sources, fungi, bacteria, and yeasts are used to produce pectinase using submerged fermentation (SmF) and solid-state fermentation (SSF). Fungi are a common source of acidic pectinases; Aspergillus sp. is considered an important fungal strain for the production of pectinase and is generally regarded as a safe (GRAS) microbe [13,14]. Many researchers have worked on the production of pectinase using different fungal strains such as Aspergillus fumigatus [15], Aspergillus oryzae [16], Aspergillus niger [17], Aspergillus tubingensis [18], Penicillium oxalicum [19], Penicillium chrysogenum [20], Fusarium proliferatum [21], Schizophyllum commune [22], Trichoderma harzianum [23], Geotricumcandidum [24], Rhizopus sp. [25], and Candida sp. [26]. The production of pectinase using Aspergillus cervinus species has not been reported by any researcher till now [1], which was confirmed by the detailed literature review. The ability of Aspergillus cervinus species to produce pectinase should be studied using SSF; moreover, the enzyme activity should be increased and production cost should be decreased by optimizing the fermentation conditions and by using low-cost agro-industrial waste, respectively.
Numerous studies have been conducted on different process variables such as moisture content, initial pH, incubation temperature, inoculum size, substrate particle size, incubation time, and agitation for enhanced pectinase production [27]. The incubation time, a vital process variable used for pectinase production, varied depending on the microorganism and the nature of the substrate used. Wong et al. [28] found the highest pectinase activity for the incubation period of 129 h. Demir and Tari [29] reported the maximum polygalacturonase activity (136.9 U/gds) by Aspergillus sojae on the fourth day of incubation. Incubation temperature also plays an important role in pectinase production. Wong et al. [28] found the highest activity of polygalacturonase by Aspergillus strains at an incubation temperature of 33 °C. Jahan et al. [30] found that pectinase activity decreased by of 29% and 32% when fermentation was carried out at 40 °C and 45 °C, respectively.
Moisture content is another vital aspect in SSF for pectinase production using different fungal strains. Mahmoodi et al. [31] obtained the highest polygalacturonase activity at a moisture content of 70% (w/w). Ortiz et al. [32] found that a moisture content of 130% (w/w) gave the maximum pectinase activity. The particle size is an essential parameter for SSF because of its effect on the specific area, aeration, and porosity of the substrate. A substrate particle size of between 150 and 250 µm was observed for high levels of pectinase activity by Demir and Tari [29]. Therefore, the cultural variables play a vital role in microbial pectinase production. Thus, the optimization of these variables helps in increasing the production yield and decreasing the production expenditure [33].
Owing to the vast industrial demand for pectinase enzyme, emphasis is on continuous research for low-cost high production of the enzyme. The use of potent novel strains, less expensive agro-industrial waste as substrates, optimum fermentation conditions, and effective downstream processing technologies can decrease the expenditure of enzyme production [34]. The stability of the enzymes for longer duration of time against wide range of pH and temperature has to be improved along with cost-effective production [35]. The immobilization of the enzyme onto the cheap materials has vast potential in specific applications and also makes the process more cost-effective [36]. It is necessary to find the strain which produces pectinase along with the other enzymes and the use of its blend for specific application which significantly reduces the enzyme production cost [1].
Therefore, keeping all these challenges in view, the present paper mainly focuses on the use of a novel native isolated strain, Aspergillus cervinus ARS2, for the production of pectinase using economical agro-industrial waste, a comparison of different fermentation methods for the synthesis of pectinase, and the statistical optimization of the process variables for enhanced enzyme activity using RSM-CCD.
2. Materials and Methods
2.1. Media and Chemicals Used
All of the chemicals used, such as buffer components, mineral salts, and organic solvents, were procured from Sigma-Aldrich Chemicals Private Limited, India. All media components were procured from HiMedia laboratories, India. The optimization studies of cultural conditions for pectinase production were carried out using Minitab 17 statistical software (Minitab, LLC, State College, PA, USA).
2.2. Pectinase Productionusing Submerged Fermentation and Solid-State Fermentation
Decayed fruit wastes (lemon peel, banana peel, and orange peel) and soil containing decomposed vegetables (City market, Durgadbail, Hubballi, India) (the latitude and longitude coordinates are: 15.340854, 75.148397) were used as sources for the isolation of pectinolytic fungi. The serial dilution and spread plate method was used for the isolation of fungi. Zone of clearance (pectin agar medium) and pectinase activity was used for the screening of the potent isolate. This isolate was molecularly characterized by 18S rRNA sequencing at the National Collection of Industrial Microorganisms (NCIM), NCL, Pune [37].
Comparison studies of pectinase production using SmF and SSF was carried out using different fruit peels such as banana, carrot, orange, lemon, sweet lime, and apple. Many researchers have reported on the nutritional value (approximate) of these different carbon sources expressed in g/100 g. Orange peel: protein, 9.73; lipids, 8.70; fibers, 14.19; ash, 5.17; carbohydrates, 53.27 [38]. Apple peel: protein, 2.80; lipids, 9.96; fibers, 13.95; ash, 1.39; carbohydrates, 59.96 [38]. Banana peel: protein, 10.44; ash, 12.45; lipid, 8.40; carbohydrates, 43.40 [38]. Sweet lime peel: protein, 12.26; fibers, 4.34; fat, 0.16; carbohydrates, 12.07 [39]. Carrot peel: protein, 1.65; ash, 0.6; carbohydrates, 9.68; fat, 0.3 [40]. Lemon peel: protein, 3.59; fibers, 4.78; fat, 0.3; ash, 1.79; carbohydrates, 30.74 [41]. The fruit peels (substrates) were dried in hot air oven for 24 h at 50 °C. The dried peels were powdered using a laboratory grinder and sieved for 1.5 mm particle size [42]. Submerged fermentation was conducted by taking the media of composition: (g/100 mL) different fruit peel powder (in different conical flask), 3.0; yeast extract (YE), 0.3; peptone, 0.5; FeSO4.7H2O, 0.01; NaH2PO4, 0.5; CaCl2.2H2O, 0.01; KCl, 0.5; KH2PO4, 0.1; ZnSO4.7H2O, 0.002; MgSO4.7H2O, 0.5. Media were sterilized at 121 °C at 15 psi. After sterilization, inoculum of 3% was added to the cultural media and incubated at room temperature in rotary shaker for 6 days at 150 rpm. The fungal biomass was separated using muslin cloth. The filtrate obtained was then centrifuged at 10,000 rpm for 10 min at 4 °C and the supernatant was assayed for pectinase activity.
For the production of pectinase by SSF, 5% (w/v) of previously prepared inoculum was inoculated to 10 g of 1.5 mm sized dried fruit peels (substrate) taken in different conical flask to which 15 mL moistening agent at pH 5.0 of composition (g/100 mL): yeast extract (YE), 0.3; peptone, 0.5; FeSO4.7H2O, 0.01; NaH2PO4, 0.5; CaCl2.2H2O, 0.01; KCl, 0.5; KH2PO4, 0.1; ZnSO4.7H2O, 0.002; MgSO4.7H2O, 0.5, was added. It was incubated at 37 °C for 5 days and further extracted the enzyme by adding 100 mL of 0.1 M citrate buffer (pH 4.5) and keeping it in incubator shaker at 37 °C and 130 rpm for 3 h. The filtrate obtained from muslin cloth was centrifuged at 10,000 rpm for 15 min at 4 °C. The supernatant obtained was considered for enzyme activity. Pectinase activity was estimated by measuring the release of reducing groups (galacturonic acid) in the reaction mixture containing the pectinase enzyme and 1% citrus pectin as a substrate. The pectin was hydrolyzed by pectinase into galacturonic acid units, and the quantification of hydrolysis of the pectin substrate was carried out by 3, 5-Dinitrosalicylic acid (DNS) reagent assay method described by Miller [43]. The reaction mixture consisted of 1% pectin in 0.1 M citrate buffer (pH 4.5) and pectinase enzyme, which was incubated for 1 h. DNS reagent was added to this reaction mixture and kept in a water bath for 10 min. The reaction mixture was then cooled, measured the absorbance using a UV-visible spectrophotometer (Shimadzu 1900, Kyoto, Japan) at 540 nm. The reducing sugars convert 3, 5-Dinitrosalicylic acid under the alkaline condition to 3-amino-5-nitrosalicylic acid, an orange-yellowish compound that has an absorption maximum at 540 nm. A standard graph of D-galacturonic acid was prepared to check the enzyme activity. One international unit of enzymatic activity (IU) was defined as the quantity of enzyme which produced one µmol of galacturonic acid per minute under assay conditions. Enzyme activity in (IU/mL) was determined using Equation (1) [44].
where ‘v’ is the volume of the enzyme, ‘194.1’ is molecular weight of galacturonic acid, and ‘t’ is the reaction time.
2.3. One-Factor-at-a-Time Studies of Process Variables
To know the effects of different process variables and their concentration levels on pectinase production, OFAT studies were conducted. The effect of different process variables on the production of pectinase was studied taking orange peel as substrate due to its high pectin content and has showed maximum pectinase activity. Substrate particle size (0.6 to 2 mm) was obtained using a sieve apparatus (Universal Instruments, India). Moisture content (60% to 120%) was fixed by the moistening solution of composition (g/100 mL): yeast extract (YE), 0.3; peptone, 0.5; MgSO4.7H2O, 0.5; NaH2PO4, 0.5; KCl, 0.5; FeSO4.7H2O, 0.01; CaCl2.2H2O, 0.01; KH2PO4, 0.1; ZnSO4.7H2O, 0.002. The initial pH (3 to 10) was maintained using the moistening solution with NaOH and HCl. The incubation temperature (25 °C to 60 °C) was maintained by using the incubator. Both inoculum size (102 to 109 spores/mL) and incubation time (1 to 8 days) were used in the study. All of the experiments were conducted in triplicates.
2.4. Response Surface Methodology-Central Composite Design Optimization of Pectinase Production
Statistical optimization of the process variables which affects the production of pectinase was carried out using CCD to increase the enzyme activity. The CCD is an experimental design, which is helpful in RSM for framing a second-order (quadratic) model for the response variable without using an entire three-level factorial experiment. From the results of the OFAT studies and an extensive literature review, the significant process parameters (substrate particle size, initial pH, incubation temperature, and moisture content) affecting the production of pectinase were optimized using RSM-CCD. RSM-CCD comprised 31 runs at five levels (−α, −1, 0, 1, +α) involving four significant process variables based on the OFAT results. The pectinase activity (response) was fitted by a second order model to correlate the response to the independent parameters. A general quadratic equation for RSM-CCD using four factors is given by Equation (2).
where Y represents the response of the design, β0 represents the model intercept, β1 to β4 represents the coefficients for linear terms, β11 to β44 represents quadratic coefficients, β12 to β34 represents cross-product coefficient indicating the interaction between variables, A, B, C and D represents the process variables considered for optimization. The values of different levels of independent parameters are shown in Table 1. The analysis of variance (ANOVA), pareto chart, 3D response surface diagram, and 2D contour plot were obtained using Minitab 17 software.
Y = β0 + β1A + β2B + β3C + β4D + β11A2 + β22B2 + β33C2 + β44D2 + β12AB + β13AC + β14AD + β23BC + β24BD + β34CD

Table 1.
Process variables and its levels used in RSM-CCD.
2.5. Model Validation
The validation of the model equation obtained by optimization of process variables was carried out by performing the experiments in triplicates. The prediction error was calculated by comparing the experimental and predicted enzyme activity values.
3. Results and Discussion
3.1. Production of Pectinase Using Submerged Fermentation and Solid-State Fermentation
Seven fungal isolates were screened for pectinolytic activity out of twenty-seven isolated from soil and rotten fruit peels, based on the zone of clearance and enzyme activity. The isolate ARS2 obtained from decomposed orange peel was found to be potent fungal isolate, which was identified as Aspergillus cervinus by 18S rRNA sequencing [45,46] at NCIM, NCL, Pune. The sequence was submitted to NCBI Gen bank for which accession number MN238704 was allotted as reported in our previous work [37].
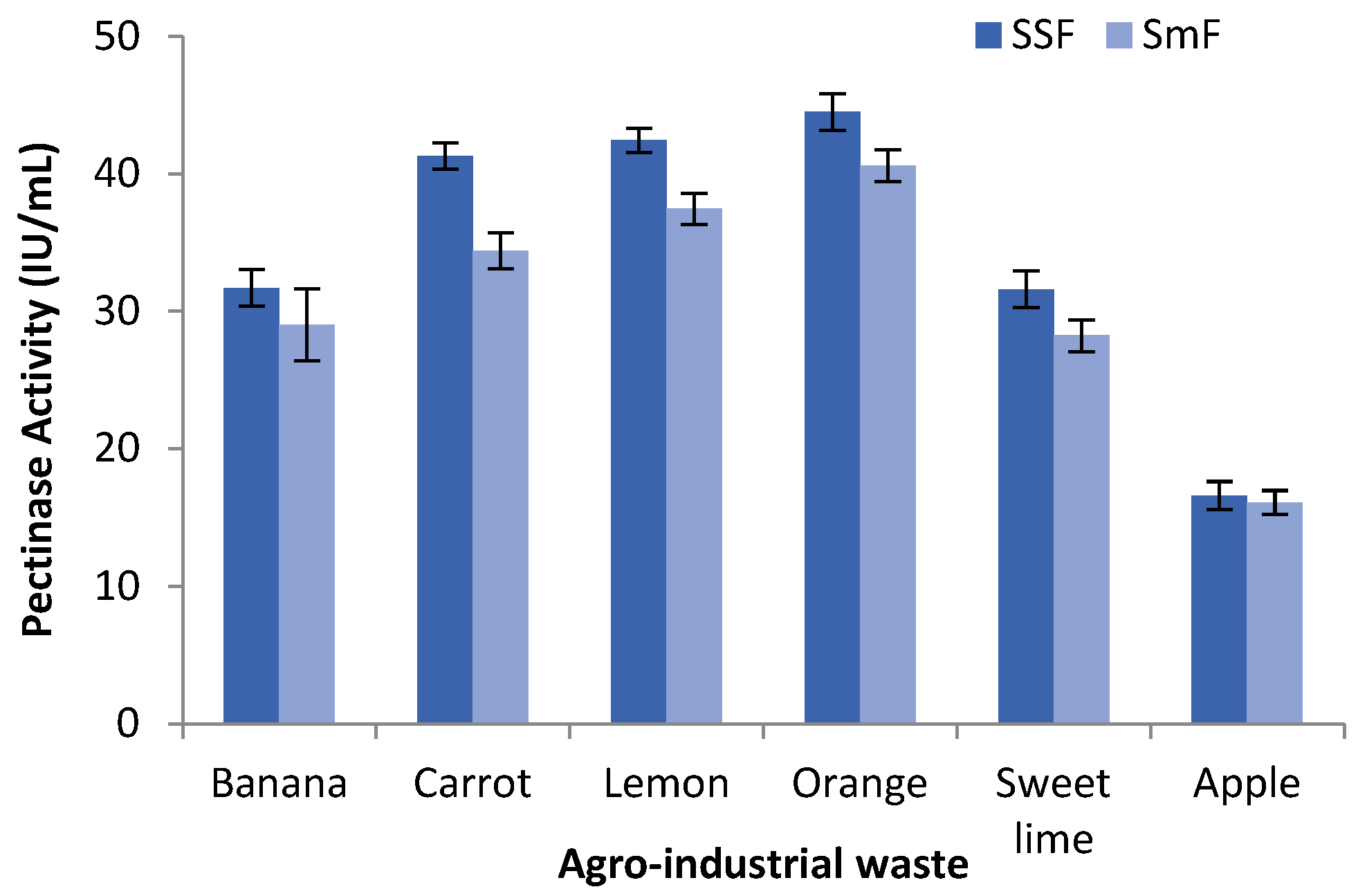
In order to assess the ability of Aspergillus cervinus ARS2 for pectinase production, different agro-industrial residues such as peels of orange, banana, carrot, lemon, sweet lime, and apple as a carbon source were examined by both SSF and SmF. All the substrates used, supported the fungal growth and also induced the production of pectinase. During the fermentation process, the maximum pectinase activity of 44.51 ± 1.33 IU/mL and 40.60 ± 1.15 IU/mL was obtained for SSF and SmF, respectively, taking orange peel as a substrate. The orange peel, lemon peel, and carrot peel substrates showed higher pectinase activity, whereas banana peel, sweet lime peel, and apple peel showed lower pectinase activity in both SmF and SSF. When pectinase activity was compared between SSF and SmF, it was found to be higher in SSF (16.61 ± 1.01 IU/mL to 44.51 ± 1.33 IU/mL) than in SmF (16.10 ± 0.87 IU/mL to 40.60 ± 1.15 IU/mL), as shown in Figure 1. From these results, SSF was found to be more suitable for fungi than SmF for the production of pectinase. This may be due to the ability of fungi hypha to grow on substrate particle surfaces and enter into the space between the particles, thereby colonizing the solid substrates. With respect to the agro-industrial waste, orange peel was found to be the potential substrate giving the maximum pectinase activity in both SSF and SmF. This might be due to the presence of proteins, minerals, carbohydrates, fats, and higher pectin content in orange peels [47].
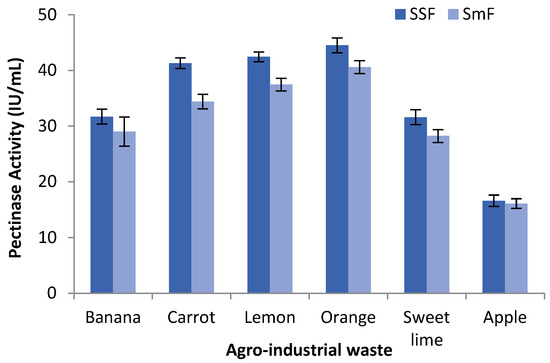
Figure 1.
Comparison studies of pectinase activity for different agro-industrial waste using SmF and SSF.
Al Mousa et al. [48] reported a maximum pectinase activity of 38.02 U/mL and 39.76 U/mL by Mucor circinelloides and Mucor hiemalis, respectively under SmF. Heidarizadeh et al. [49] found the highest pectinase yield of 10.47 U/mL by Piriformospora indica under SmF. Junior et al. [21] observed an optimal pectinase activity of 137.36 ± 2.21 U/mL by Fusarium proliferatum using SSF. Patil and Dayanand [50] compared SSF and SmF for the production of pectinase by Aspergillus niger and found that SSF (34.2 U/g) was better than SmF (12.6 U/mL) under optimum conditions. Maldonado and Strasser de Saad [51] compared the polygalacturonase activity of Aspergillus niger using submerged and solid-state fermentation and found that SSF produced six times more enzymes and took less time for enzyme production than SmF. Govindaraji and Vuppu [52] reported the highest pectinase activity of 45.93 U/mL using a novel strain, Streptomyces fumigates cleroticus VIT-SP4, and orange peel as a substrate, which is in concurrence with the present findings. Ahmed and Awad [20], in their research work, found the maximum pectinase activity by Pencillium chrysogenum MF318506 using orange peels as the substrate. The results of the present study were supported by the literature review; hence, SSF (fermentation method) and orange peels (substrate) were taken for further studies.
3.2. One-Factor-at-a-Time Studies of Process Variables
One-factor-at-a-time studies of process variables were performed to know the concentration level of each variable affecting the pectinase activity, which was further used in optimization studies by RSM-CCD. The use of purified pectin as the only carbon source is uneconomical; thus, the agro-industrial residues were considered economical and used for the production of pectinase. At the same time, the optimization of media components and process variables are required for low-cost, high production of pectinase. Therefore, the current study aimed to optimize the process parameters by using optimized media components for high enzyme activity from our previous study [37].
3.2.1. Effect of Substrate Particle Size
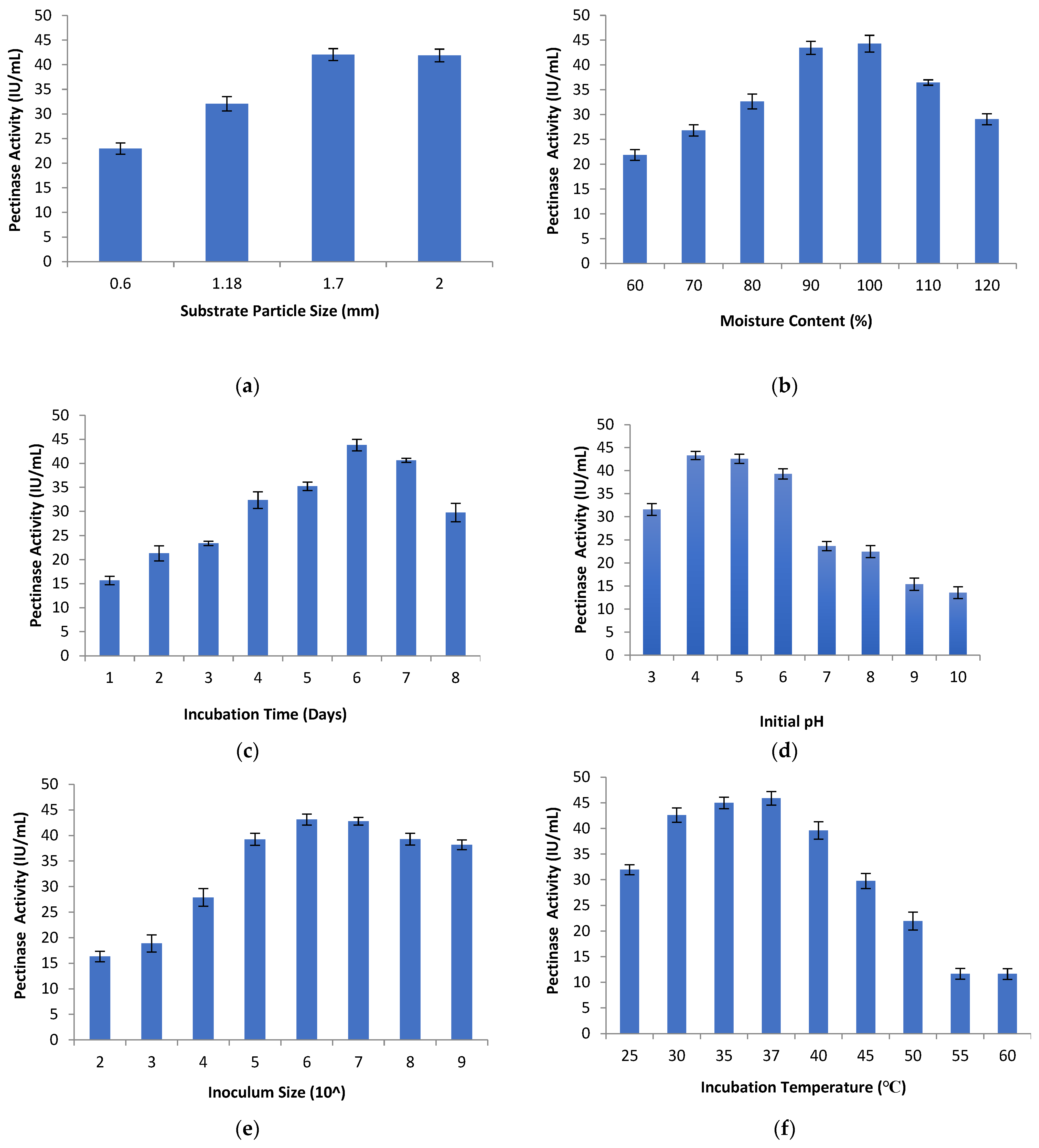
The effect of different substrate particle size on pectinase activity was studied using orange peel as the substrate. Different-sized substrates ranging between 0.6 mm and 2 mm were used in the study. Substrate particle size plays a crucial role in the production of enzymes because of the microbial growth on the damp surface of the particles and between the particles. The gap between the particles is filled by a gas phase which depends on the shape, size, and tortuosity of the interconnected pores [53]. In the current study, the pectinase activity increased with the increase in substrate particle size ranging from 0.6 mm to 1.7 mm. A substrate particle size of 1.7 mm gave the maximum enzyme activity of (42.06 ± 1.20 IU/mL), as shown in Figure 2a. Pectinase activity at a substrate particle size of 2 mm was (41.88 ± 1.28 IU/mL). This shows that the pectinase activity decreased with the increase in particle size above 1.7 mm and also decreased with the decrease in particle size below 1.7 mm. This might be due to the larger particle size resulting in a loose texture of substrate which allows oxygen to pass between the particles. However, smaller particles form a packed and dense substrate bed which restricts oxygen passage and interferes with microbial respiration. This results in poor microbial growth and low enzyme activity [54,55]. Membrillo et al., 2011 [56], reported that the substrate size and shape, along with the geometrical ratio, strongly influenced the packing density for the SSF substrate with an impact on the production of extracellular enzymes and microbial growth rate. Darah et al. [57] found that a substrate particle size (pomela peel) of 0.75 mm achieved the maximum level of pectinase activity using Aspergillus niger LFP-1 under SSF.
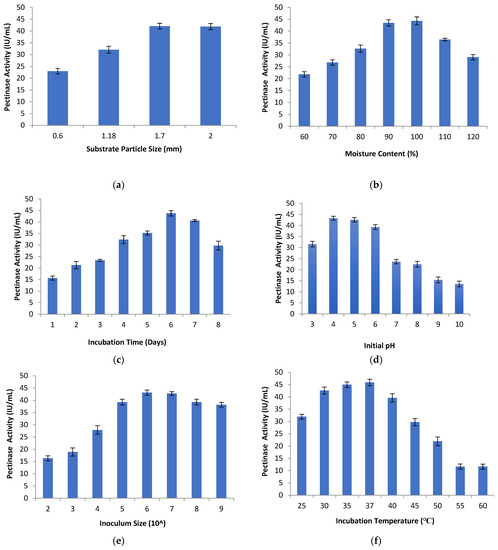
Figure 2.
Pectinase activity for different (a) substrate particle size, (b) moisture content, (c) incubation time, (d) initial pH, (e) inoculum size, (f) incubation temperature.
3.2.2. Effect of Moisture Content
To study the moisture content effect on pectinase activity, the moistening solution was used to obtain different amounts of moisture content (60% to 120%). Enzyme activity was determined after 6 days of incubation. The levels of pectinase activity increased with the increase in moisture content from 60% to 100%. At lower levels of moisture content, the pectinase activity was minimum, which may be due to lower swelling of the substrate, reduced nutrient solubility and higher water tension [57,58]. Further increase in the moisture content from 100% to 120% decreased the pectinase activity, as shown in Figure 2b. This might be due to the increased substrate diffusion barrier, decreased substrate porosity, reductions in the oxygen transfer rate, the development of stickiness, and loss of particle structure, which may lead to decrease in microbial growth and cell metabolic activity [28,59]. The optimum moisture content depends on the microorganism and the substrate nature used in the fermentation process. In the present work, maximum pectinase activity of (44.30 ± 1.69 IU/mL) was observed for 100% moisture content considering orange peel as substrate. The optimal moisture content favors the nutrients’ entry into the cell membrane which leads to the highest levels of enzyme activity [42].
Zehra et al. [60] found the highest pectinase activity for moisture contents of 65% using banana peels as a substrate for Aspergillus fumigates MS 16 under SSF. Darah et al. [57] reported a 1:1 ratio (w/v), i.e., 100% moisture content giving maximum pectinase activity for Aspergillus niger LFP-1, which is in line with our results. Wong et al. [28] observed the maximum pectinase activity for 40% moisture content using Aspergillus fumigates R6 under SSF. Botella et al. [61] found the highest pectinase activity for a moisture content of 65% and grape pomace as the substrate using Aspergillus awamori. Amin et al. [58] showed 40% and 60% as the favorable moisture contents for maximum pectinase activity using Penicillium notatum and Coriolus versicolor, respectively.
3.2.3. Effect of Incubation Time
Solid-state fermentation was carried out at different incubation times to study its effect on pectinase activity. The incubation times considered were between 1 and 8 days. It was observed that the maximum pectinase activity of (43.81 ± 1.21 IU/mL) was observed on the sixth day of incubation, as shown in Figure 2c. Increase in the incubation period up to six days increased the pectinase activity. Gradual decrease in enzyme activity were observed after the sixth day of incubation, which might be due to the non-availability of nutrients and metabolite accumulation in the medium. A shorter incubation period may lead to less enzyme synthesis and thus lower levels of enzyme activity [28,59,62]. The incubation time depends on the physiological conditions of the process, the concentration of the nutrients, microorganisms, and the nature of the medium [63].
Different researchers studied the effect of incubation time on pectinase activity. Abdullah et al. [64] found the maximum pectinase activity of 8 IU/mL on the third day using Aspergillus niger ABT-5. Zehra et al. [60] reported the maximum pectinase activity on the fifth day using banana peel as the substrate for Aspergillus fumigates MS 16 under SSF. Darah et al. [57], in their study, showed the maximum pectinase activity of 1.27 IU/mL using Aspergillus niger HFD5A-1 on the sixth day of incubation, which is in concurrence with our results. Sethi et al. [65] found the highest pectinase activity using Aspergillus terreus NCFT 4269.10 on the fourth day of incubation. Wong et al. [28] found the incubation time of 4 days as the optimal for maximum pectinase activity using Aspergillus fumigates R6. The study conducted by Amin et al. [58] showed an incubation time of 3 and 4 days for maximum pectinase activity using Penicillium notatum and Coriolus versicolor, respectively. The results obtained by Satapathy et al. [63] indicated the maximum enzyme activity by Aspergillus parvisclerotigenus KX928754 for an incubation time of 7 days.
3.2.4. Effect of Initial pH
Solid-state fermentation was performed to check the effect of initial pH on pectinase activity. At different initial pH levels (3 to 10), fermentation production of pectinase enzymes was conducted using orange peel as a substrate. A medium pH level affects the enzyme activity by maintaining the cell membrane permeability, nutrient solubility, and the ionization of amino acids and proteins [60,64]. Therefore, a medium pH level regulates the culture growth and affects the enzyme activity [65]. During SSF, no significant changes in the pH levels of the media were observed. Maximum pectinase activity of (43.31 ± 0.89 IU/mL) was observed at an initial pH of 4, as shown in Figure 2d. The increase in pH from 4 to 8 decreased the pectinase activity. A further increase in pH from 9 to 10 led to a further decrease in enzyme activity. The reason might be that the fungi can grow in acidic conditions, and any change in the pH levels of the media had significant effects on enzyme activity. pH levels regulate and promote the synthesis of extracellular enzymes by using microorganisms [63,66]. The enzyme activity is significantly influenced by the increase in hydrogen ion concentration, as it alters the ionic charges at the active sites on amino acids [59]. By studying the effect of pH on pectinase production, it is clear that the isolated strain produces acidic pectinase.
Wong et al. [28] found a pH of 4.0 to be the optimum level for maximum pectinase activity using Aspergillus fumigates R6 under SSF, which is in concurrence with our results. Abdullah et al. [64] found the maximum enzyme activity at a pH of 6.0 for Aspergillus niger ABT-5. Darah et al. [57] observed an optimal pH of 4.0 for the maximum pectinase activity of 2.32 IU/mL using Aspergillus niger HFD5A-1. Sethi et al. [65] showed an optimal pH of 5.0, resulting in maximal pectinase activity using Aspergillus terreus NCFT 4692.10 under SSF. The research work carried out by Amin et al. [58] indicated that initial pH levels of 3.0 and 5.0 favored utmost pectinase activity by Penicillium notatum and Coriolus versicolor, respectively. Satapathy et al. [63] observed an initial pH of 7.0 leading to the maximum pectinase activity using Aspergillus parvisclerotigenus KX928754.
3.2.5. Effect of Inoculum Size
An inoculum size between 102 and 109 spores/mL of the moistening solution was considered to study its effects on enzyme activity. The pectinase activity increased with the increase in inoculum size from 102 to 106 spores/mL. At a low inoculum size of 102 spores/mL, the pectinase activity was minimal because a smaller inoculum size may not be sufficient to initiate the microbial growth and enzyme production. Therefore, the inoculum size influenced the mycelia growth and enzyme activity [53,64]. An inoculum size of 106 spores/mL gave the highest pectinase activity of (43.11 ± 1.06 IU/mL). Further increase in inoculum size led to a decrease in pectinase activity, as indicated in Figure 2e. A large inoculum size may cause over-accumulation of spores, which may result in the increase in moisture content and competition for nutrients among the cells, resulting in less microbial growth and less enzyme activity [65]. Darah et al. [53] found that an inoculum size of 1 × 106 spores/mL gave the maximum pectinase activity using Aspergillus niger HFD5A-1, which is in consensus with our results. Abdullah et al. [64] reported that an inoculum size of 0.5% (v/v) led to the maximum pectinase activity using Aspergillus niger ABT-5. Darah et al. [57] observed that an inoculum size of 1 × 107 spores/mL resulted in the maximum pectinase activity using Aspergillus niger LFP-1 under SSF.
3.2.6. Effect of Incubation Temperature
The effect of incubation temperature on pectinase activity was studied using SSF. An incubation temperature of between 25 °C and 60 °C was considered for the study. The increase in incubation temperature between 25 °C and 37 °C increased the pectinase activity. Furthermore, the increase in the incubation temperature between 37 °C and 60 °C decreased the enzyme activity as indicated in Figure 2f. The decline in enzyme activity at high temperature may be due to the reduction in the organism metabolic rate, as well as the denaturation of the enzyme [28,64,67]. Alternatively, it might be the case that with the increase in temperature, more energy is received by the enzyme molecule to overcome the weak forces of globular protein structure and deactivate the enzyme. The low temperature results in the freezing of the protoplasmic membrane, which may lead to stress conditions in the solute transport systems in the cells [53,64]. In the present work, the maximum pectinase activity of (45.90 ± 1.33 IU/mL) was found at an incubation temperature of 37 °C.
Demir and Tari [29] observed the maximum pectinase activity at an incubation temperature of 37 °C using Aspergillus sojae, which is in line with the results obtained in the present study. Abdullah et al. [64], in their research, found that 30 °C was the optimum temperature for maximum pectinase activity using Aspergillus niger ABT-5. Zehra et al. [60] determined 25 °C to be the optimum incubation temperature for maximum pectinase activity using Aspergillus fumigates MS16 under SSF. Darah et al. [53] observed maximal pectinase activity of 1.40 IU/mL using Aspergillus niger HFD5A-1 for an incubation temperature of 30 °C. Sethi et al. [65] determined 30 °C to be the optimal temperature for maximum enzyme activity using Aspergillus terreus NCFT 4692.10. Wong et al. [28], in their research, determined 30 °C to be the optimal temperature for maximum pectinase activity using Aspergillus fumigates R6. In a study reported on by Darah et al. [57], the highest pectinase activity was observed at a temperature of 30 °C using Aspergillus niger LFP-1 under SSF. Amin et al. [58] reported 35 °C and 30 °C to be favorable temperatures for the maximum pectinase activity using Penicillium notatum and Coriolus versicolor, respectively. Satapathy et al. [63] found 30 °C to be the optimum incubation temperature for the highest levels of pectinase activity by Aspergillus parvisclerotigenus KX928754.
3.3. Optimization of Pectinase Production
Optimization studies of the process variables for highest pectinase activity by Aspergillus cervinus ARS2 were performed by considering the following significant process variables: substrate particle size, incubation temperature, initial pH, and moisture content. CCD matrix and experimental results are shown in Table 2. The model (uncoded form) specified by Equation (3) indicated pectinase activity as a function of substrate particle size (A), initial pH (B), moisture content (C), and incubation temperature (D).
Pectinase Activity (IU/mL) = 0.5300 + 0.0130 A+ 0.06020 B+ 0.008089 C + 0.00873 D − 0.00739 A × A − 0.005860 B × B − 0.000037 C × C − 0.000118 D × D − 0.00365 A × B + 0.000214 A × C + 0.000391 A × D + 0.000028 B × C + 0.000064 B × D − 0.000023 C × D

Table 2.
CCD matrix for process variables.
The goodness of the model was verified by the coefficient of determination (R2). A R2 value nearer to 1 signifies the model accuracy. The regression coefficient (R2) value indicates the amount of variability in the observed response values. The variability is explained by experimental variables and their interactions. The R2 value in terms of a percentage of 85.47% indicates variation in enzyme activity due to the independent parameters; the remaining 14.53% of the total variation cannot be explained by the model. The nearer the R2 value to 1, the better the correlation between the experimental and predicted values, and the prediction of the response by the model is also better. The predicted R2 value of 0.7826 indicated a better correlation between the experimental and predicted enzyme activity. The predicted R2 value of 0.7826 is in agreement with the adjusted R2 value of 0.8287. The adjusted R2 value fixes the R2 value based on the sample size and the number of parameters in the model. The adjusted R2 value (0.8287) is smaller than R2 (0.8547) because of the small sample size in the model.
A probability value (p) < 0.05 indicates significant model variables, whereas the p-value > 0.05 indicates non-significant model variables. The p-value indicates the significance of each coefficient and the interaction strength of each parameter. The p-value < 0.05 indicates a significant correlation of the coefficients. The smaller the p-value, the greater is the significance of the coefficient. From the ANOVA Table 3, it was observed that the linear effect except particle size (0.483) and squared effect of all the process variables and interaction effects of substrate particle size and pH, moisture content and temperature were observed to be highly significant for pectinase production. The square effects of the process variables were found to be more significant than the individual process parameters. The interaction effects of substrate particle size and moisture content (0.111), substrate particle size and temperature (0.145), pH and moisture content (0.617), pH and temperature (0.563) were found to have less significance.

Table 3.
ANOVA table for RSM-CCD.
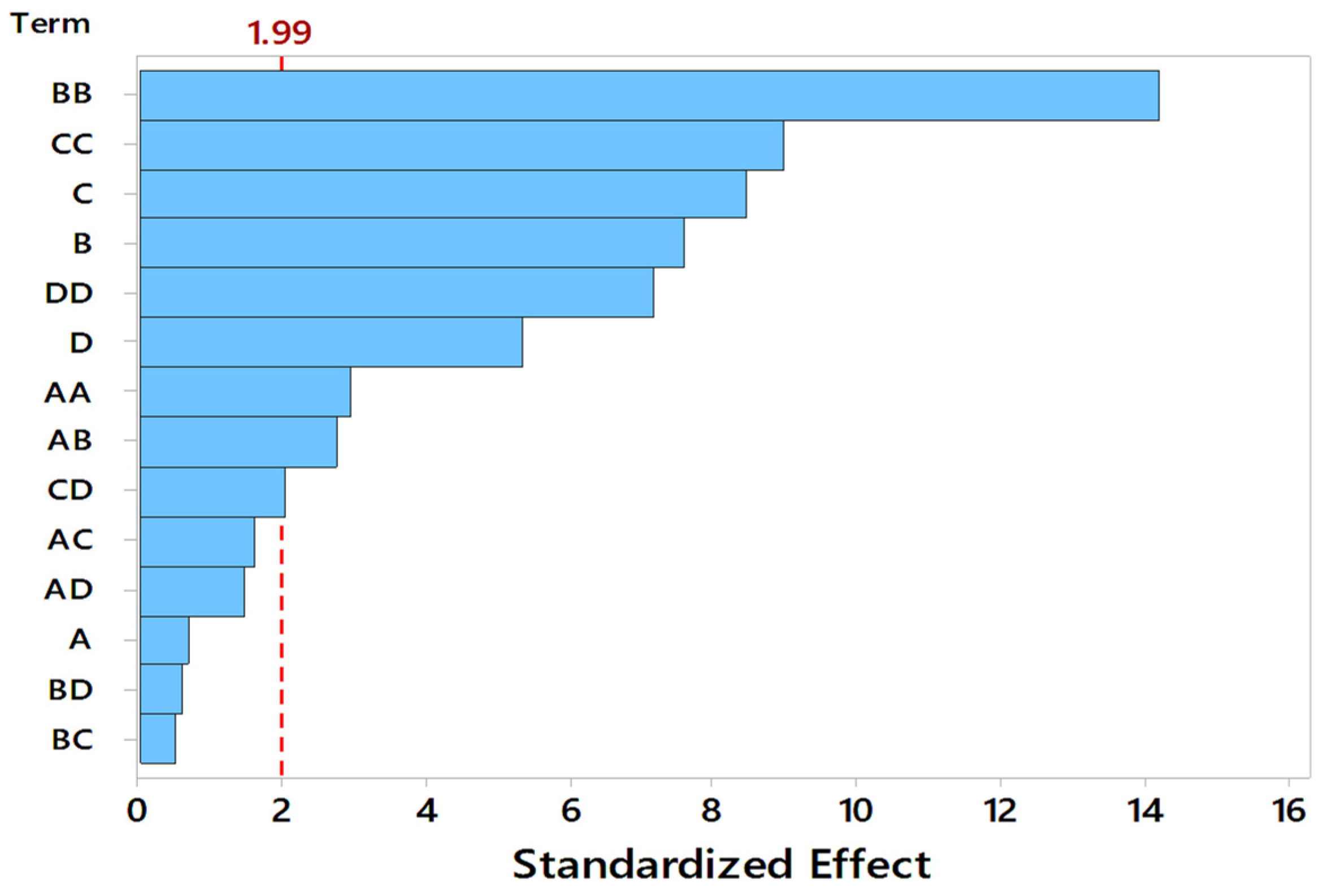
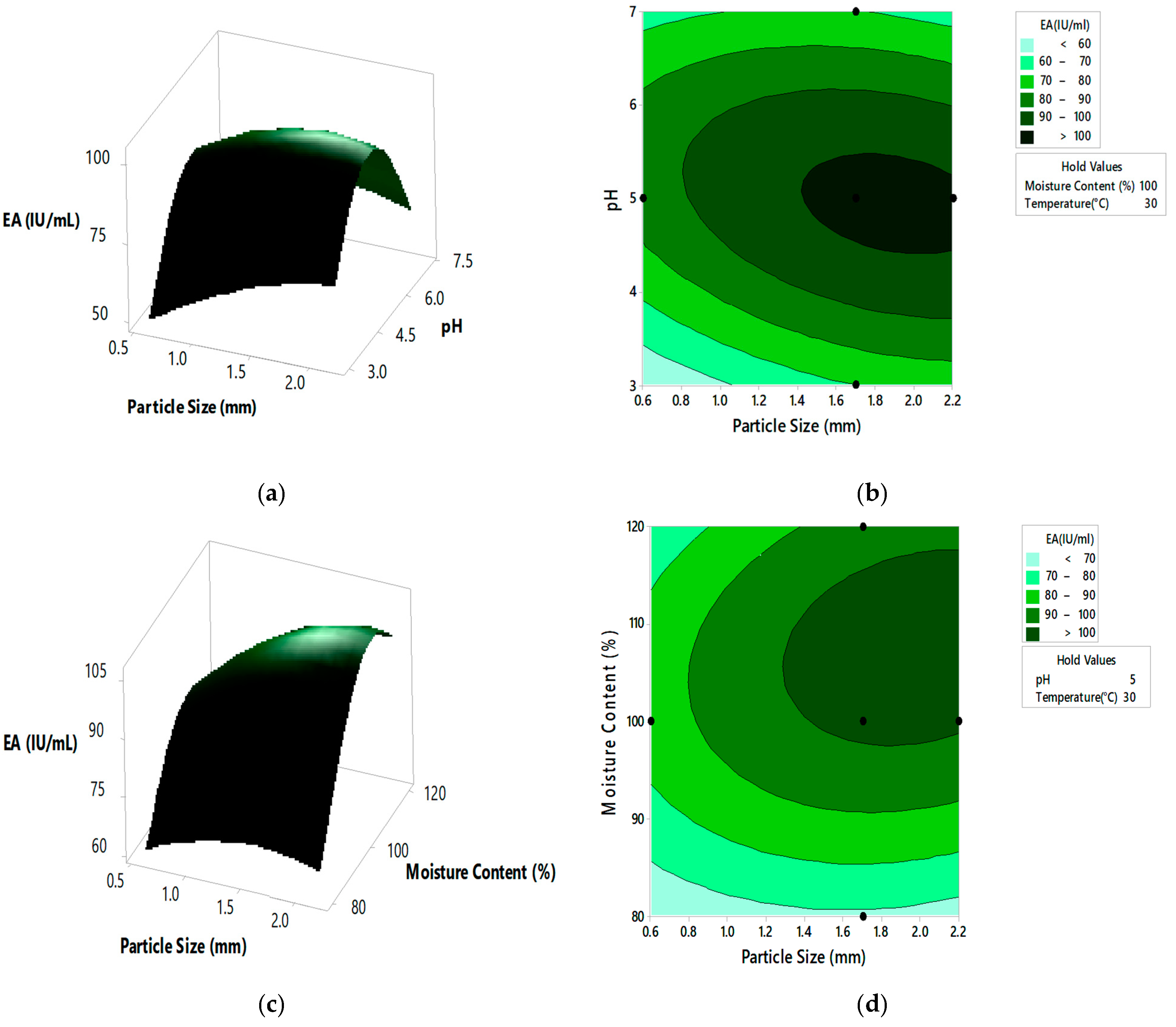
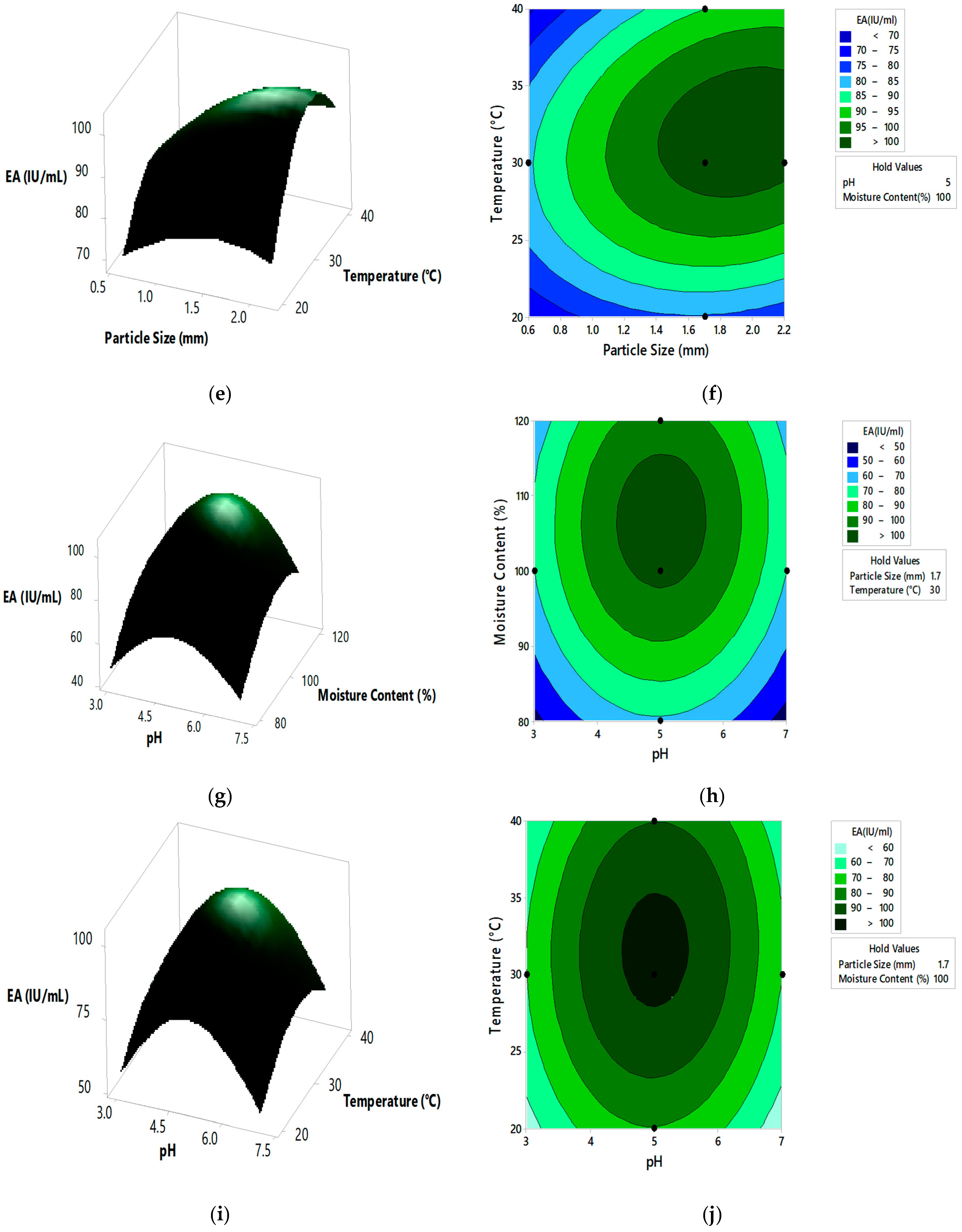
Response surface curves are used to determine the optimized values of the factors for highest enzyme activity and to know the interaction between the variables. The effect of two parameters on the enzyme activity is indicated by pareto chart, 3D surface plots, and 2D contour plots, as shown in Figure 3 and Figure 4, respectively. A number of 3D surface plots were plotted in order to study the relative effects of process variables on pectinase activity. In the 3D surface plots, the vertical axis represents the response (enzyme activity), and the two horizontal axes represent the coded levels of two independent process variables by keeping other variables at their control level. From the pareto chart, as shown in Figure 3, the interaction effect between substrate particle size and initial pH, moisture content and incubation temperature were significant, and the interaction effect between the substrate particle size and moisture content, substrate particle size and incubation temperature, initial pH and moisture content, and initial pH and incubation temperature, was insignificant.
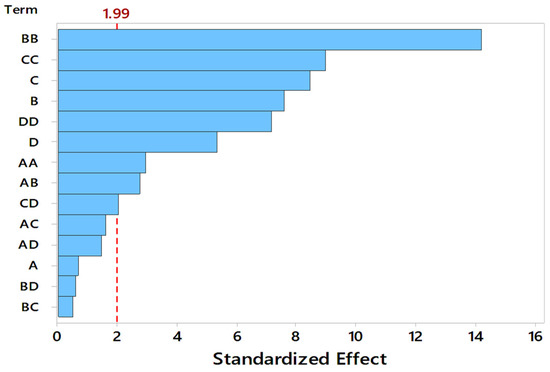
Figure 3.
Pareto chart indicating the significance of process variables, A: Substrate Particle Size (mm), B: Initial pH, C: Moisture Content (%), D: Incubation Temperature (°C).
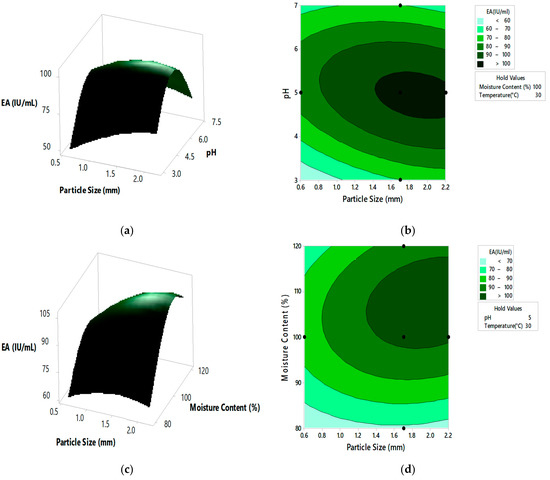
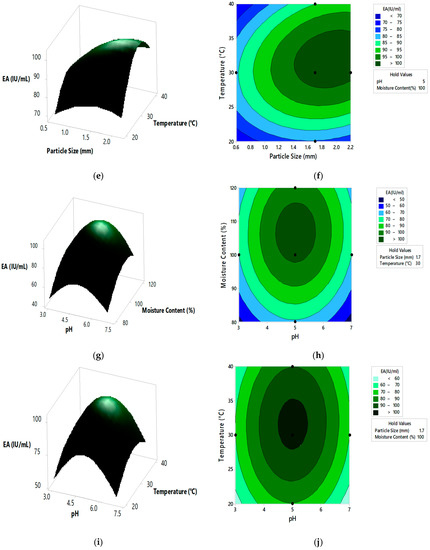
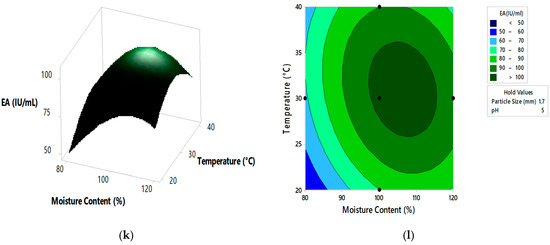
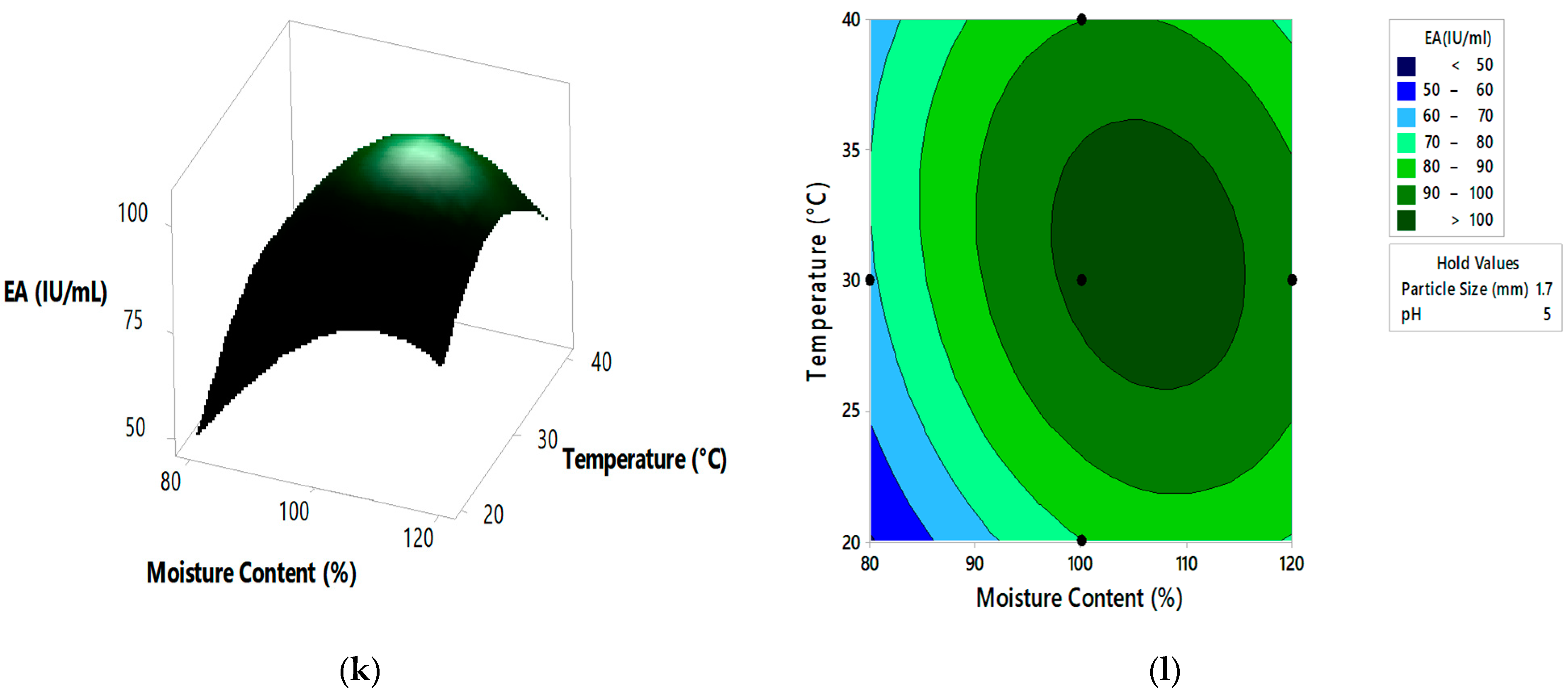
Figure 4.
The 3D surface plots and 2D contour plots indicating relative effect for pectinase activity (a,b) substrate particle size and initial pH, (c,d) substrate particle size and moisture content, (e,f) substrate particle size and incubation temperature, (g,h) initial pH and moisture content, (i,j) initial pH and incubation temperature, and (k,l) moisture content and incubation temperature.
From the 3D surface plots and 2D contour plots, as indicated in Figure 4a,b, the relative effects of substrate particle size (0.6 to 2.20 mm) and pH (3 to 7) on pectinase activity was observed, keeping temperature and moisture content constant at 30 °C and 100%, respectively. Pectinase activity increased with the increase in substrate particle size, reaching maximal levels at the highest value of the particle size. With the increase in pH, there was an increase in enzyme activity which reached maximal levels at mid pH values and decreased at high pH values. Figure 4c,d represents the relative effects of substrate particle size (0.6 to 2.20 mm) and moisture content (80 to 120%) on pectinase activity, keeping pH and temperature constant at 5 and 30 °C, respectively. Enzyme activity increased with the increase in moisture content, reaching maximal levels at the highest levels of moisture content. With the increase in substrate particle size, there was an increase in enzyme activity which reached the maximum and then decreased at the highest values of particle size. Figure 4e,f shows the relative effects of substrate particle size (0.6 to 2.20 mm) and incubation temperature (20 to 40 °C) on pectinase activity, keeping pH and moisture content at 5 and 100%, respectively. Enzyme activity increased with the increase in incubation temperature, reaching its maximum at high temperatures. With the increase in substrate particle size, there was an increase in enzyme activity, reaching its maximum and then decreasing at high values of particle size. Figure 4g,h shows the relative effects of pH and moisture content on enzyme activity, keeping substrate particle size and incubation temperature constant at 1.7 mm and 30 °C, respectively. Enzyme activity increased with the increase in pH and moisture content reached maximum at mid values of pH and moisture content and then decreased at high values of both the parameters. Similar effect on pectinase activity is shown by relative effects of pH and incubation temperature as shown in Figure 4i,j. The relative effects of moisture content and incubation temperature on pectinase activity is shown in Figure 4k,l. The enzyme activity increases with the increase in moisture content and temperature, reached maximum at mid values of both the parameters. The optimum pectinase activity was obtained from Minitab optimizer as 107.14 ± 0.71 IU/mL for substrate particle size of 2 mm, initial pH of 4.9, moisture content of 107% and incubation temperature of 31.5 °C. In the current study the pectinase activity by A. cervinus ARS2 increased by nearly 2.38-fold after statistical optimization.
Esawy et al. [68] reported 40 U/mL pectinase activity by Aspergillus niger NRC1ami on the optimization of the incubation temperature and incubation period. Junior et al. [21] obtained optimum pectinase activity of 137.36 ± 2.21 U/mL using Fusarium proliferatum at the optimal values of pH 3.6 and 43.4 °C. Pagarra et al. [69] used CCD for optimization and found optimum exo-polygalacturonase activity of 54.64 U/g by Aspergillus niger for 120 h incubation time, 34 °C incubation temperature, and 75.26% moisture content. Wong et al. [28] showed highest polygalacturonase activity of 565 U/g by Aspergillus fumigates R6 resulting in 2.65-fold increase at 49.6% moisture content, 33 °C temperature and 129 h incubation time. In their research work on pectinase production by Rhizopus sp. using RSM and CCD, Handa et al. [25] observed the highest levels of enzyme activity of 11.63 IU/mL for 1:3.5 moisture ratios, 7 days of incubation, and 30 °C incubation temperature. Barman et al. [62] reported maximum pectinase activity of 6.6 U/mL by Aspergillus niger using CCD at an incubation time of 65.82 h and an incubation temperature of 32.37 °C.
3.4. Validation of the Model
To validate the model adequacy, experiments were performed in triplicates to verify the predicted optima. The experimental valves were in consensus with the predicted values, which showed that the model is adequate. The optimal values of different process variables were obtained from the Minitab 17 software optimizer, which is shown in Table 4. The experimental pectinase activity of (107.14 ± 0.71 IU/mL) was in line with the predicted pectinase activity of (106.31 IU/mL) obtained using optimal values. The enzyme activity increased 2.38-fold after optimization of process variables using RSM-CCD, and the model was found to be valid for the optimization process.

Table 4.
Process variables optimal values for maximum pectinase activity.
4. Conclusions
According to an exhaustive literature survey, the current investigation on pectinase production employing A. cervinus has been reported for the first time. Comparative studies showed that SSF is better than SmF for pectinase production. OFAT studies were followed by optimization studies using RSM-CCD for improved pectinase activity. The optimum pectinase activity of 107.14 ± 0.71 IU/mL was obtained for the optimal values of a substrate particle size of 2 mm, an initial pH of 4.9, a moisture content of 107%, and an incubation temperature of 31.5 °C. Optimization studies have shown an increase in pectinase activity by 2.38-fold. Compared with the earlier reports on pectinase production by other strains, the isolated fungal strain A. cervinus ARS2 has shown better enzyme activity. The use of economical agro-industrial waste and optimization techniques has shown an increase in the enzyme activity. Future work should be focused on the application of protein and genetic engineering, along with immobilization techniques for the development of robust, versatile, and stable enzymes for different industrial applications and economical processes.
Author Contributions
Conceptualization, A.R.S., S.V.D. and U.M.M.; Data curation, A.R.S. and S.A.; Formal analysis, A.B.M., M.H.M., U.M.M. and A.Y.A.; Funding acquisition, A.B.M., A.Y.A., M.H.M. and A.A.K.; Investigation, A.R.S. and S.V.D.; Methodology, A.R.S., S.A. and S.V.D.; Project administration, A.B.M., A.Y.A., I.A.S. and A.A.K.; Resources, M.H.M. and I.A.S.; Software, A.R.S. and S.A.; Supervision, S.V.D. and U.M.M.; Visualization, A.Y.A. and A.A.K.; Writing—original draft, A.R.S., S.A. and U.M.M.; Writing—review and editing, A.Y.A., S.V.D., M.H.M. and I.A.S. All authors have read and agreed to the published version of the manuscript.
Funding
The authors extend their appreciation to the Deanship of Scientific Research at King Khalid University for funding this work through the Large Groups Project under grant number (RGP. 2/159/43/2022).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data generated and analyzed during this study are included in the article.
Acknowledgments
The authors thank King Khalid University, Abha, Saudi Arabia; Department of Biotechnology, KLE Technological University, Hubballi, India and ISNC, Jeddah, Saudi Arabia for providing support for this research work.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Haile, S.; Ayele, A. Pectinase from microorganisms and its industrial applications. Sci. World J. 2022, 1881305. [Google Scholar] [CrossRef]
- Arnau, J.; Yaver, D.; Hjort, C.M. Strategies and challenges for the development of industrial enzymes using fungal cell factories. In Grand Challenges in Fungal Biotechnology; Springer: Cham, Switzerland, 2019; pp. 179–210. [Google Scholar] [CrossRef]
- Combo, A.M.M.; Aguedo, M.; Goffin, D.; Wathelet, B.; Paquot, M. Enzymatic production of pectic oligosaccharides from polygalacturonic acid with commercial pectinase preparations. Food Bioprod. Process. 2012, 90, 588–596. [Google Scholar] [CrossRef]
- Rebello, S.; Anju, M.; Aneesh, E.M.; Sindhu, R.; Binod, P.; Pandey, A. Recent advancements in the production and applications of microbial pectinases-an overview. Rev. Environ. Sci. Biotechnol. 2017, 16, 381–394. [Google Scholar] [CrossRef]
- Oumer, O.J.; Abate, D. Screening and molecular identification of pectinase producing microbes from coffee pulp. BioMed Res. Int. 2018, 2018, 2961767. [Google Scholar] [CrossRef]
- Kashyap, D.R.; Vohra, P.K.; Chopra, S.; Tewari, R. Applications of pectinases in the commercial sector: A review. Bioresour. Technol. 2001, 77, 215–227. [Google Scholar] [CrossRef]
- Singh, S.; Gupta, R. Apple juice clarification using fungal pectinolytic enzyme and gelatin. Indian J. Biotechnol. 2004, 3, 573–576. [Google Scholar]
- Jayani, R.S.; Saxena, S.; Gupta, R. Microbial Pectinolytic enzymes: A review. Process Biochem. 2005, 40, 2931–2944. [Google Scholar] [CrossRef]
- Rashmi, R.; Murthy, K.R.S.; Sneha, G.; Shabana, S.; Syama, A.; Radhika, V. Partial purification and biochemical characterization of extracellular pectinase from Aspergillus niger isolated from groundnut seeds. J. Appl. Biosci. 2008, 9, 378–384. [Google Scholar]
- Singh, S.; Mandal, S.K. Optimization of processing parameters for production of pectinolytic enzymes from fermented pineapple residue of mixed Aspergillus species. Jordan J. Biol. Sci. 2012, 5, 307–314. [Google Scholar]
- Demir, H.; Gogus, N.; Tari, C.; Heerd, D.; Lahore, M.F. Optimization of the process parameters for the utilization of orange peel to produce polygalacturonase by solid-state fermentation from an Aspergillus sojae mutant strain. Turk. J. Biol. 2012, 36, 394–404. Available online: https://dergipark.org.tr/en/pub/tbtkbiology/issue/11697/139664 (accessed on 10 October 2022). [CrossRef]
- Amin, S.A.; Gopinarayanan, V.E.; Nair, N.U.; Hassoun, S. Establishing synthesis pathway-hostcompatibility via enzyme solubility. Biotechnol. Bioeng. 2019, 116, 1405–1416. [Google Scholar] [CrossRef] [PubMed]
- Bhardwaj, V.; Degrassi, G.; Bhardwaj, R.K. Microbial pectinases and their applications in industries: A review. Int. J. Eng. Res. 2017, 4, 829–836. [Google Scholar] [CrossRef]
- John, J.; Kaimal, K.K.S.; Smith, M.L.; Rahman, P.K.S.M.; Chellam, P.V. Advances in upstream and downstream strategies of pectinase bioprocessing: A review. Int. J. Biol. Macromol. 2020, 162, 1086–1099. [Google Scholar] [CrossRef] [PubMed]
- Sandri, I.G.; Fontana, R.C.; Da silveira, M.M. Influence of pH and temperature on the production of polygalacturonases by Aspergillus fumigates. LWT-Food Sci. Technol. 2015, 61, 430–436. [Google Scholar] [CrossRef]
- Biz, A.; Finkler, A.T.J.; Pitol, L.O.; Medina, B.S.; Krieger, N.; Mitchell, D.A. Production of pectinases by solid state fermentation of a mixture of citrus waste and sugarcane bagasse in a pilot-scale packed bed bioreactor. Biochem. Eng. J. 2016, 111, 54–62. [Google Scholar] [CrossRef]
- Anand, G.; Yadav, S.; Yadav, D. Production, purification and biochemical characterization of an exo-polygalacturonase from Aspergillus niger MTCC478 suitable for clarification of orange juice. 3 Biotech 2017, 7, 122. [Google Scholar] [CrossRef]
- Huang, D.; Song, Y.; Lin, Y.; Qin, Y. A new strain of Aspergillus tubingensis for high activity pectinase production. Braz. J. Microbiol. 2019, 50, 53–65. [Google Scholar] [CrossRef]
- Li, Y.; Wang, Y.; Tu, T.; Zhang, D.; Ma, R.; You, S.; Wang, X.; Yao, B.; Luo, H.; Xu, B. Two acidic, thermophilic GH28 polygalacturonase from Talaromycesleycettanus JCM 12802 with application potentials for grape juice clarification. Food Chem. 2017, 237, 997–1003. [Google Scholar] [CrossRef]
- Ahmed, N.E.; Awad, H.M. Optimizing the production of pectinase of orange peel waste by penicillium chrysogenum MF318506 using response surface methodology in submerged fermentation. J. Microbiol. Biotechnol. Food Sci. 2021, 11, e3931. [Google Scholar] [CrossRef]
- Junior, A.N.; Mansoldo, F.R.P.; Godoy, M.G.; Firpo, R.M.; Cedrola, S.M.L.; Vermelho, A.B. Production of an endo-polygalacturonase from Fusarium proliferatum isolated from agro-industrial waste. Biocatal. Agric. Biotechnol. 2021, 38, 102199. [Google Scholar] [CrossRef]
- Mehmood, T.; Saman, T.; Irfan, M.; Anwar, F.; Ikram, M.S.; Tabassam, Q. Pectinase production from Schizophyllum commune through central composite design using citrus waste and its immobilization for industrial exploitation. Waste Biomass Valorization 2019, 10, 2527–2536. [Google Scholar] [CrossRef]
- Nabi, N.G.; Nabi, M.; Asgher, A.; Shah, H.; Sheikh, M.A.; Asad, M.J. Production of pectinase by Trichoderma harzianum in solid state fermentation of citrus peels. Pak. J. Agric. Sci. 2003, 40, 193–201. [Google Scholar]
- Ejaz, U.; Ahmed, A.; Sohail, M. Statistical optimization of immobilization of yeast cells on corncob for pectinase production. Biocatal. Agric. Biotechnol. 2018, 14, 450–456. [Google Scholar] [CrossRef]
- Handa, S.; Sharma, N.; Pathania, S. Multiple parameter optimization for maximization of pectinase production by Rhizopus sp. C4 under solid state fermentation. Fermentation 2016, 2, 10. [Google Scholar] [CrossRef]
- Aggarwal, R.; Dutta, T.; Sheikh, J. Extraction of pectinase from Candida isolated from textile mill effluent and its application in bio-scouring of cotton. Sustain. Chem. Pharm. 2020, 17, 100291. [Google Scholar] [CrossRef]
- Patidar, M.K.; Nighojkar, S.; Kumar, A.; Nighojkar, A. Pectinolytic enzymes- solid state fermentation, assay methods and applications in fruit juice industries: A review. 3 Biotech. 2018, 8, 199. [Google Scholar] [CrossRef]
- Wong, L.Y.; Saad, W.Z.; Mohamad, R.; Tahir, P.M. Optimization of cultural conditions for polygalacturonase production by a newly isolated Aspergillus fumigates R6 capable of retting kenaf. Ind. Crops Prod. 2017, 97, 175–183. [Google Scholar] [CrossRef]
- Demir, H.; Tari, C. Valorization of wheat bran for the production of polygalacturonase in SSF of Aspergillus sojae. Ind. Crops Prod. 2014, 54, 302–309. [Google Scholar] [CrossRef]
- Jahan, N.; Shahid, F.; Aman, A.; Mujahid, T.Y.; Qader, S.A.U. Utilization of agro waste pectin for the production of industrially important polygalacturonase. Heliyon 2017, 3, e00330. [Google Scholar] [CrossRef] [PubMed]
- Mahmoodi, M.; Najafpour, G.D.; Mohammadi, M. Productionof pectinases for quality apple juice through fermentation of orange pomace. J. Food Sci. Technol. 2017, 54, 4123–4128. [Google Scholar] [CrossRef] [PubMed]
- Ortiz, G.E.; Maria, C.; Mora, P.; Noseda, D.G.; Cazabat, G.; Saravalli, C.; Lopez, M.C.; Gil, G.P.; Blasco, M.; Alberto, E.O. Pectinase production by Aspergillus giganteus in solid state fermentation: Optimization, scale up, biochemical characterization and its application in olive-oil extraction. J. Ind. Microbiol. Biotechnol. 2017, 44, 197–211. [Google Scholar] [CrossRef] [PubMed]
- Bhavikatti, J.S.; Bodducharl, S.M.; Kamagond, R.S.; Desai, S.V.; Shet, A.R. Statistical optimisation of protease production using a freshwater bacterium Chryseobacterium cucumeris SARJS-2 for multiple industrial applications. 3 Biotech. 2020, 10, 279. [Google Scholar] [CrossRef] [PubMed]
- Satapathy, S.; Rout, J.R.; Kerry, R.G.; Thatoi, H.; Sahoo, S.L. Biochemical prospects of various microbial pectinase and pectin: An approachable concept in pharmaceutical bioprocessing. Front. Nutr. 2020, 7, 117. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, S.; Rahman, M.S.; Qin, W. New insights in pectinase production development and industrial applications. Appl. Microbiol. Biotechnol. 2021, 105, 9069–9087. [Google Scholar] [CrossRef] [PubMed]
- Patel, V.B.; Chatterjee, S.; Dhoble, A.S. A review on pectinase properties, application in juiceclarification, and membranes as immobilization support. J. Food Sci. 2022, 87, 3338–3354. [Google Scholar] [CrossRef] [PubMed]
- Shet, A.R.; Muhsinah, A.B.; Alsayari, A.; Achappa, S.; Desai, S.V.; Mahnashi, M.H.; Muddapur, U.M.; Shaikh, I.A.; Mannasaheb, B.A.; Khan, A.A. Media optimization by response surface methodology for the enhanced production of acidic extracellular pectinase by the indigenously isolated novel strain Aspergillus cervinus ARS2 using solid-state fermentation. Fermentation 2022, 8, 485. [Google Scholar] [CrossRef]
- Romelle, F.D.; Ashwini Rani, P.; Manohar, R.S. Chemical composition of some selected fruit peels. Eur. Food Res. Technol. 2016, 4, 12–21. [Google Scholar]
- Priyadarshini, S.; John, S. Analysis of nutrient content and physicochemical properties of newly developed sweet lime peel vinegar and sweet lime fruit-peel combo vinegar. Indian J. Appl. Res. 2014, 4, 260–262. [Google Scholar]
- Muhammad, A.; Muhammad, S.H.; Abdullah, I. Effect of pre-treatments and drying methods on dehydration and rehydration characteristics of carrot. Univers. J. Food Nutr. Sci. 2015, 3, 23–28. [Google Scholar] [CrossRef]
- Pham, T.; Nguyen, N.T.P.; Dinh, D.V.; Kieu, N.T.; Bach, L.G.; Phong, H.X.; Muoi, N.V.; Truc, T.T. Evaluate the chemical composition of peels and juice of seedless lemon (Citrus latifolia) grown in haugiang province, Vietnam. IOP Conf. Ser. Mater. Sci. Eng. 2020, 991, 012127. [Google Scholar] [CrossRef]
- Almowallad, S.A.; Aljobair, M.O.; Alkuraieef, A.N.; Aljahani, A.H. Utilization of agro-industrial orange peel and sugar beet pulp wastes for fungal endo-ploygalacturonase production. Saudi J. Biol. Sci. 2022, 29, 963–969. [Google Scholar] [CrossRef] [PubMed]
- Miller, G.L. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Li, Q.; Coffman, A.M.; Ju, L.K. Development of reproducible assays for polygalacturonase and pectinase. Enzyme Microb. Technol. 2015, 72, 42–48. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Karlin, S.; Altschul, S.F. Methods for assessing the statistical significance of molecular sequence features by using general scoring schemes. Proc. Natl. Acad. Sci. USA 1990, 87, 2264–2268. [Google Scholar] [CrossRef]
- Kute, A.B.; Mohapatra, D.; Kotwaliwale, N.; Giri, S.K.; Sawant, B.P. Characterization of pectin extracted from orange peel powder using microwave-assisted and acid extraction methods. Agric. Res. 2020, 9, 241–248. [Google Scholar] [CrossRef]
- Al Mousa, A.A.; Hassane, A.M.A.; Gomaa, A.E.-R.F.; Aljuriss, J.A.; Dahmash, N.D.; Abo-Dahab, N.F. Response-surface statistical optimization of submerged fermentation for pectinase and cellulase production by Mucor circinelloides and M. hiemalis. Fermentation 2022, 8, 205. [Google Scholar] [CrossRef]
- Heidarizadeh, M.; Fathi Rezaei, P.; Shahabivand, S. Novel pectinase from Piriformospora indica optimization of growth parameters and enzyme production in submerged culture condition. Turkish J. Biochem. 2018, 43, 289–295. [Google Scholar] [CrossRef]
- Patil, S.R.; Dayanand, A. Optimization of process for the production of fungal pectinases from deseeded sunflower head in submerged and solid-state conditions. Bioresour. Technol. 2006, 97, 2340–2344. [Google Scholar] [CrossRef]
- Maldonado, M.C.; Strasser de Saad, A.M. Production of pectinesterase and polygalacturonase by Aspergillus niger in submerged and solid-state systems. J. Ind. Microbiol. Biotechnol. 1998, 20, 34–38. [Google Scholar] [CrossRef]
- Govindaraji, P.K.; Vuppu, S. Characterisation of pectin and optimization of pectinase enzyme from novel Streptomyces fumigatiscleroticus VIT-SP4 for drug delivery and concrete crack-healing applications: An eco-friendly approach. Saudi J. Biol. Sci. 2020, 27, 3529–3540. [Google Scholar] [CrossRef] [PubMed]
- Darah, I.; Haritharan, W.; Lim, S.H. Involvement of physicochemical parameters on pectinase production by Aspergillus niger HFD5A-1. J. Pure Appl. Microbiol. 2013, 7, 2541–2549. [Google Scholar]
- Pandey, A.; Benjamin, S.; Soccol, C.R.; Nigam, P.; Krieger, N.; Soccol, V.T. The realm of microbial lipases in biotechnology. Biotechnol. Appl. Biochem. 1999, 29, 119–131. [Google Scholar]
- Darah, I.; Salikin, N.H.; Hong, L.S.; Ahmad, R.; Weloosamy, H. Pomelo peels as alternative substrate for extracellular pectinase production by Aspergillus niger HFM-8. Malays. J. Microbiol. 2013, 9, 308–316. [Google Scholar]
- Membrillo, I.; Sanchez, C.; Meneses, M.; Favela, E.; Loera, O. Particle geometry affects differentially substrate composition and enzyme profiles by Pleurotusostreatus growing on sugar cane bagasse. Bioresour. Technol. 2011, 102, 1581–1586. [Google Scholar] [CrossRef]
- Darah, L.; Taufiq, M.M.J.; Lim, S.H. Pomelo Citrus grandis (L) Osbeck Peel as an Economical Alternative Substrate for Fungal Pectinase Production. Food Sci. Biotechnol. 2013, 22, 1683–1690. [Google Scholar] [CrossRef]
- Amin, F.; Bhatti, H.N.; Bilal, M.; Asgher, M. Multiple parameter optimizations for enhanced biosynthesis of exo-polygalacturonase enzyme and its application in fruit juice clarification. Int. J. Food Eng. 2016, 13, 20160256. [Google Scholar] [CrossRef]
- Rehman, S.; Bhatti, H.N.; Bilal, M.; Asgher, M. Optimization of process variables for enhanced production of extracellular lipase by Pleurotusostreatus IBL-02 in solid-state fermentation. Pak. J. Pharm. Sci. 2019, 32, 617–624. [Google Scholar]
- Zehra, M.; Syed, M.N.; Sohail, M. Banana Peels: A promising substrate for the coproduction of Pectinase and Xylanase from Aspergillus fumigates MS16. Pol. J. Microbiol. 2020, 69, 19–26. [Google Scholar] [CrossRef]
- Botella, C.; Diaz, A.B.; Ory, I.D.; Webb, C. Xylanase and pectinase production by Aspergillus awamori on grape pomace in solid state fermentation. Process Biochem. 2007, 42, 98–101. [Google Scholar] [CrossRef]
- Barman, S.; Sit, N.; Badwaik, L.S.; Deka, S.C. Pectinase production by Aspergillus niger using banana (Musa balbisiana) peel as substrate and its effect on clarification of banana juice. J. Food Sci. Technol. 2015, 52, 3579–3589. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Satapathy, S.; Soren, J.P.; Mondal, K.C.; Srivastava, S.; Pradhan, C.; Sahoo, S.L.; Thatoi, H.; Rout, J.R. Industrially relevant pectinase production from Aspergillus parvisclerotigenus KX928754 using apple pomace as the promising substrate. J. Taibah Univ. Sci. 2021, 15, 347–356. [Google Scholar] [CrossRef]
- Abullah, R.; Jafer, A.; Nisar, K.; Kaleem, A.; Iqtedar, M.; Iftikhar, T.; Saleem, F.; Naz, S. Process optimization for pectinase production by locally isolated fungal strain using submerged fermentation. J. Biosci. 2018, 34, 1025–1032. [Google Scholar] [CrossRef]
- Sethi, B.K.; Nanda, P.K.; Sahoo, S. Enhanced production of pectinase by Aspergillus terreus NCFT 4269.10 using banana peels as substrate. 3 Biotech 2016, 6, 36. [Google Scholar] [CrossRef]
- Azzaz, H.H.; Murad, H.A.; Kholif, A.M.; Morsy, T.A.; Mansour, A.M.; El-Sayed, H.M. Pectinase production optimization and its application in banana fiber degradation. Egypt. J. Nutr. Health 2013, 16, 117–125. [Google Scholar]
- Mulluye, K.; Kebede, A.; Bussa, N. Production and optimization of pectinase from pectinolytic fungi cultivated on mango peels and pectin subjected to submerged fermentation. Biol. Med. Nat. Prod. Chem. 2021, 10, 15–21. [Google Scholar] [CrossRef]
- Esawy, M.A.; Gamal, A.A.; Kamel, Z. Optimization of Aspergillus niger NRC1ami pectinase using citrus peel pectin, purification, and thermodynamic characterization of the free and modified enzyme. Waste Biomass Valorization 2022, 13, 4823–4837. [Google Scholar] [CrossRef]
- Pagarra, H.; Rahman, R.A.; Azelee, N.I.W.; Illias, R.M. Optimization and characterization of exo-polygalacturonase by Aspergillus niger cultured via solid state fermentation. J. Teknol. 2018, 81, 59–67. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).