Simultaneous Determination of C18 Fatty Acids in Milk by GC-MS
Abstract
:1. Introduction
2. Materials and Methods
2.1. Milk Samples
2.2. Chemicals and Reagents
2.3. Sample Pretreatment
2.4. GC-MS Analysis
2.5. Method Validation
2.6. Statistical Analysis
3. Results and Discussion
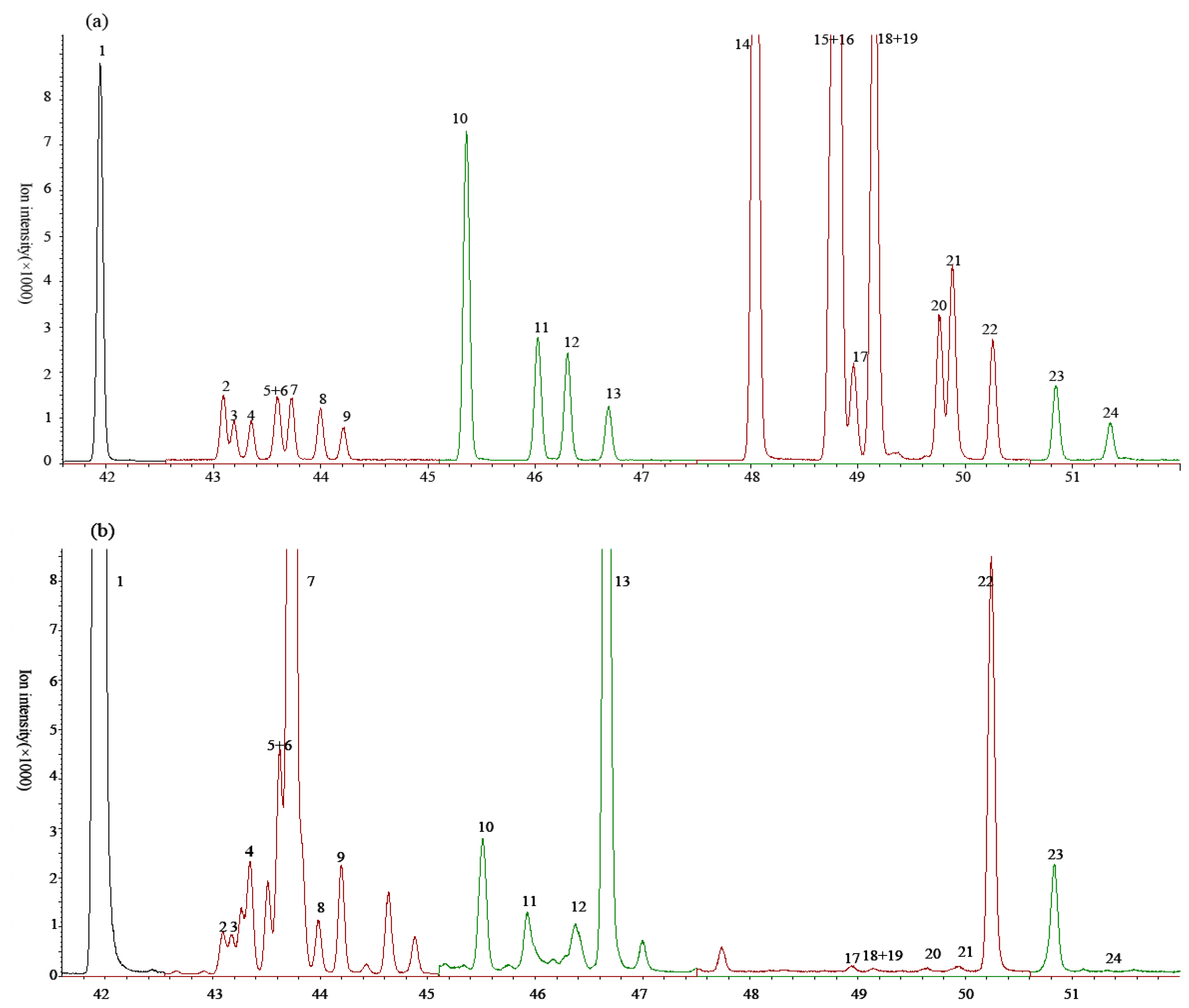
3.1. The Species of Detected C18 FAs
3.2. Methylation
3.3. Method Validation
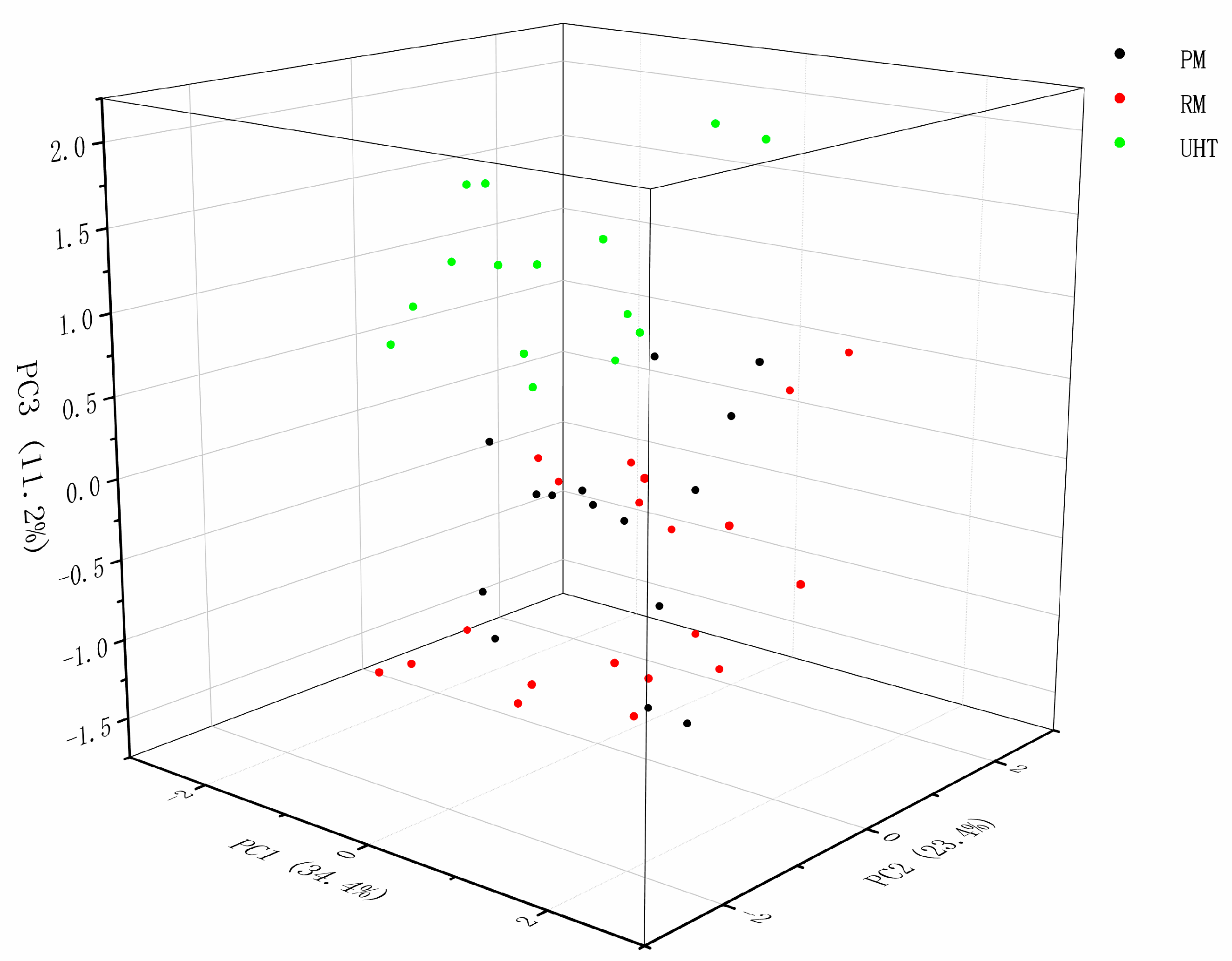
3.4. The Analysis of C18 FAs in Milk Samples
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gómez-Cortés, P.; Juárez, M.; de la Fuente, M.A. Milk fatty acids and potential health benefits: An updated vision. Trends Food Sci. Technol. 2018, 81, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Wei, W.; Jin, Q.; Wang, X. Human milk fat substitutes: Past achievements and current trends. Prog. Lipid Res. 2019, 74, 69–86. [Google Scholar] [CrossRef] [PubMed]
- Amores, G.; Virto, M. Total and Free Fatty Acids Analysis in Milk and Dairy Fat. Separations 2019, 6, 14. [Google Scholar] [CrossRef] [Green Version]
- Yoshinaga, K.; Asanuma, M.; Mizobe, H.; Kojima, K.; Nagai, T.; Beppu, F.; Gotoh, N. Characterization of cis- and trans-octadecenoic acid positional isomers in edible fat and oil using gas chromatography–flame ionisation detector equipped with highly polar ionic liquid capillary column. Food Chem. 2014, 160, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Yoshinaga, K.; Asanuma, M.; Xu, C.; Mizobe, H.; Kojima, K.; Nagai, T.; Beppu, F.; Gotoh, N. Resolution Behavior of cis- and trans-Octadecenoic Acid Isomers by AOCS Official Method Using SP-2560 Column. J. Oleo Sci. 2013, 62, 781–788. [Google Scholar] [CrossRef] [Green Version]
- Buccioni, A.; Rapaccini, S.; Antongiovanni, M.; Minieri, S.; Conte, G.; Mele, M. Conjugated linoleic acid and C18:1 isomers content in milk fat of sheep and their transfer to Pecorino Toscano cheese. Int. Dairy J. 2010, 20, 190–194. [Google Scholar] [CrossRef]
- Kramer, J.K.G.; Cruz-Hernandez, C.; Deng, Z.; Zhou, J.; Jahreis, G.; Dugan, M.E.R. Analysis of conjugated linoleic acid and trans 18:1 isomers in synthetic and animal products. Am. J. Clin. Nutr. 2004, 79, 1137S–1145S. [Google Scholar] [CrossRef] [Green Version]
- Shingfield, K.J.; Bernard, L.; Leroux, C.; Chilliard, Y. Role of trans fatty acids in the nutritional regulation of mammary lipogenesis in ruminants. Animal 2010, 4, 1140–1166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lanier, J.S.; Corl, B.A. Challenges in enriching milk fat with polyunsaturated fatty acids. J. Anim. Sci. Biotechnol. 2015, 6, 26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanuš, O.; Samková, E.; Křížová, L.; Hasoňová, L.; Kala, R. Role of Fatty Acids in Milk Fat and the Influence of Selected Factors on Their Variability—A Review. Molecule 2018, 23, 1636. [Google Scholar] [CrossRef] [Green Version]
- Dewanckele, L.; Toral, P.; Vlaeminck, B.; Fievez, V. Invited review: Role of rumen biohydrogenation intermediates and rumen microbes in diet-induced milk fat depression: An update. J. Dairy Sci. 2020, 103, 7655–7681. [Google Scholar] [CrossRef] [PubMed]
- Waktola, H.D.; Zeng, A.X.; Chin, S.-T.; Marriott, P.J. Advanced gas chromatography and mass spectrometry technologies for fatty acids and triacylglycerols analysis. TrAC Trends Anal. Chem. 2020, 129, 115957. [Google Scholar] [CrossRef]
- Hewavitharana, G.G.; Perera, D.; Navaratne, S.; Wickramasinghe, I. Extraction methods of fat from food samples and preparation of fatty acid methyl esters for gas chromatography: A review. Arab. J. Chem. 2020, 13, 6865–6875. [Google Scholar] [CrossRef]
- Wei, G.-L.; Zeng, E.Y. Gas chromatography-mass spectrometry and high-performance liquid chromatography-tandem mass spectrometry in quantifying fatty acids. TrAC Trends Anal. Chem. 2011, 30, 1429–1436. [Google Scholar] [CrossRef]
- Perna, A.M.; Intaglietta, I.; Simonetti, A.; Gambacorta, E. The influence of casein haplotype on morphometric characteristics of fat globules and fatty acid composition of milk in Italian Holstein cows. J. Dairy Sci. 2016, 99, 2512–2519. [Google Scholar] [CrossRef] [Green Version]
- Trigueros, L.; Sendra, E. Fatty acid and conjugated linoleic acid (CLA) content in fermented milks as assessed by direct methylation. LWT Food Sci. Technol. 2015, 60, 315–319. [Google Scholar] [CrossRef]
- Cattani, M.; Mantovani, R.; Schiavon, S.; Bittante, G.; Bailoni, L. Recovery of n-3 polyunsaturated fatty acids and conjugated linoleic acids in ripened cheese obtained from milk of cows fed different levels of extruded flaxseed. J. Dairy Sci. 2014, 97, 123–135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tzamaloukas, O.; Orford, M.; Miltiadou, D.; Papachristoforou, C. Partial suckling of lambs reduced the linoleic and conjugated linoleic acid contents of marketable milk in Chios ewes. J. Dairy Sci. 2015, 98, 1739–1749. [Google Scholar] [CrossRef]
- Firl, N.; Kienberger, H.; Rychlik, M. Validation of the sensitive and accurate quantitation of the fatty acid distribution in bovine milk. Int. Dairy J. 2014, 35, 139–144. [Google Scholar] [CrossRef]
- Ariko, T.; Kass, M.; Henno, M.; Fievez, V.; Kärt, O.; Kaart, T.; Ots, M. The effect of replacing barley with glycerol in the diet of dairy cows on rumen parameters and milk fatty acid profile. Anim. Feed Sci. Technol. 2015, 209, 69–78. [Google Scholar] [CrossRef]
- Kramer, J.K.G.; Hernandez, M.; Cruz-Hernandez, C.; Kraft, J.; Dugan, M.E.R. Combining Results of Two GC Separations Partly Achieves Determination of All cis and trans 16:1, 18:1, 18:2 and 18:3 Except CLA Isomers of Milk Fat as Demonstrated Using Ag-Ion SPE Fractionation. Lipids 2008, 43, 259–273. [Google Scholar] [CrossRef] [PubMed]
- De la Fuente, M.A.; Rodríguez-Pino, V.; Juárez, M. Use of an extremely polar 100-m column in combination with a cyanoalkyl polysiloxane column to complement the study of milk fats with different fatty acid profiles. Int. Dairy J. 2015, 47, 52–63. [Google Scholar] [CrossRef] [Green Version]
- Dreiucker, J.; Vetter, W. Fatty acids patterns in camel, moose, cow and human milk as determined with GC/MS after silver ion solid phase extraction. Food Chem. 2011, 126, 762–771. [Google Scholar] [CrossRef]
- Ecker, J.; Scherer, M.; Schmitz, G.; Liebisch, G. A rapid GC–MS method for quantification of positional and geometric isomers of fatty acid methyl esters. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2012, 897, 98–104. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Li, P.; Sun, X.; Hu, W.; Wang, X.; Zhang, Q.; Ding, X. Untargeted fatty acid profiles based on the selected ion monitoring mode. Anal. Chim. Acta 2014, 839, 44–50. [Google Scholar] [CrossRef]
- Liu, Z.; Rochfort, S.; Cocks, B. Milk lipidomics: What we know and what we don’t. Prog. Lipid Res. 2018, 71, 70–85. [Google Scholar] [CrossRef]
- Liu, Z.; Ezernieks, V.; Rochfort, S.; Cocks, B. Comparison of methylation methods for fatty acid analysis of milk fat. Food Chem. 2018, 261, 210–215. [Google Scholar] [CrossRef]
- Association of Official Analytical Chemists. AOAC Fat (Total, Saturated, and Unsaturated) in Foods. Hydrolytic Extraction Gas Chromatographic Method; AOAC Official Method 996.06; AOAC International: Arlington VA, USA, 2002. [Google Scholar]
- Serafim, V.; Tiugan, D.A.; Andreescu, N.; Mihailescu, A.; Paul, C.; Velea, I.; Puiu, M.; Niculescu, M.D. Development and Validation of a LC-MS/MS-Based Assay for Quantification of Free and Total Omega 3 and 6 Fatty Acids from Human Plasma. Molecules 2019, 24, 360. [Google Scholar] [CrossRef] [Green Version]
- ICH Harmonized Tripartite Guideline. Validation of Analytical Procedures: Text and Methodology Q2(R1); ICH: Geneva, Switzerland, 2005. [Google Scholar]
- Li, T.; Guo, Q.; Qu, Y.; Li, Y.; Wang, X.; Sun, Z.; Wang, Q. An improved gas chromatography-based approach for characterisation of fatty acids in fresh basil seed oil. Int. J. Food Sci. Technol. 2021, 56, 2492–2503. [Google Scholar] [CrossRef]
- Teng, F.; Wang, P.; Yang, L.; Ma, Y.; Day, L. Quantification of Fatty Acids in Human, Cow, Buffalo, Goat, Yak, and Camel Milk Using an Improved One-Step GC-FID Method. Food Anal. Methods 2017, 10, 2881–2891. [Google Scholar] [CrossRef]
- Ratnayake, W.M.N.; Hansen, S.L.; Kennedy, M.P. Evaluation of the CP-Sil 88 and SP-2560 GC columns used in the recently approved AOCS official method Ce 1h-05: Determination of cis-, trans-, saturated, monounsaturated, and polyunsaturated fatty acids in vegetable or non-ruminant animal oils and fats by capillary GLC method. J. Am. Oil Chem. Soc. 2006, 83, 475–488. [Google Scholar]
- Hauff, S.; Vetter, W. Quantitation of cis- and trans-Monounsaturated Fatty Acids in Dairy Products and Cod Liver Oil by Mass Spectrometry in the Selected Ion Monitoring Mode. J. Agric. Food Chem. 2009, 57, 3423–3430. [Google Scholar] [CrossRef] [PubMed]
- Delmonte, P.; Kia, A.-R.F.; Kramer, J.K.; Mossoba, M.M.; Sidisky, L.; Rader, J.I. Separation characteristics of fatty acid methyl esters using SLB-IL111, a new ionic liquid coated capillary gas chromatographic column. J. Chromatogr. A 2011, 1218, 545–554. [Google Scholar] [CrossRef]
- Delmonte, P.; Fardin-Kia, A.R.; Kramer, J.K.; Mossoba, M.M.; Sidisky, L.; Tyburczy, C.; Rader, J.I. Evaluation of highly polar ionic liquid gas chromatographic column for the determination of the fatty acids in milk fat. J. Chromatogr. A 2012, 1233, 137–146. [Google Scholar] [CrossRef]
- Liu, Z.; Wang, J.; Li, C.; Rochfort, S. Development of one-step sample preparation methods for fatty acid profiling of milk fat. Food Chem. 2020, 315, 126281. [Google Scholar] [CrossRef] [PubMed]
- Haug, A.; Høstmark, A.T.; Harstad, O.M. Bovine milk in human nutrition—A review. Lipids Health Dis. 2007, 6, 25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fuke, G.; Nörnberg, J.L. Systematic evaluation on the effectiveness of conjugated linoleic acid in human health. Crit. Rev. Food Sci. Nutr. 2015, 57, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Shokryazdan, P.; Rajion, M.A.; Meng, G.Y.; Boo, L.J.; Ebrahimi, M.; Royan, M.; Sahebi, M.; Azizi, P.; Abiri, R.; Jahromi, M.F. Conjugated linoleic acid: A potent fatty acid linked to animal and human health. Crit. Rev. Food Sci. Nutr. 2015, 57, 2737–2748. [Google Scholar] [CrossRef]
- Ajmal, M.; Nadeem, M.; Imran, M.; Junaid, M. Lipid compositional changes and oxidation status of ultra-high temperature treated Milk. Lipids Health Dis. 2018, 17, 227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodríguez-Alcalá, L.M.; Alonso, L.; Fontecha, J. Stability of fatty acid composition after thermal, high pressure, and microwave processing of cow milk as affected by polyunsaturated fatty acid concentration. J. Dairy Sci. 2014, 97, 7307–7315. [Google Scholar] [CrossRef]

| No. | FAMEs | Retention Time (min) | Windows | Quantification Ion (m/z) | Qualification Ion (m/z) | Dwell Time (ms) | ||
|---|---|---|---|---|---|---|---|---|
| 1 | C18:0 | 42.032 | 1 | 74 | 87 | 143 | 298 | 20 |
| 2 | C18:1 t6 | 43.203 | 2 | 264 | 97 | 222 | 180 | 20 |
| 3 | C18:1 t9 | 43.305 | ||||||
| 4 | C18:1 t11 | 43.466 | ||||||
| 5 | C18:1 c6 | 43.710 | ||||||
| 6 | C18:1 c8 | 43.712 | ||||||
| 7 | C18:1 c9 | 43.848 | ||||||
| 8 | C18:1 c11 | 44.118 | ||||||
| 9 | C18:1 c12 | 44.337 | ||||||
| 10 | C18:2 t9,t12 | 45.502 | 3 | 81 | 95 | 294 | 263 | 20 |
| 11 | C18:2 c9,t12 | 46.17 | ||||||
| 12 | C18:2 t9,c12 | 46.446 | ||||||
| 13 | C18:2 c9,c12 | 46.839 | ||||||
| 14 | C18:3 t9,t12,t15 | 48.215 | 4 | 79 | 93 | 121 | 292 | 20 |
| 15 | C18:3 t9,t12,c15 | 48.988 | ||||||
| 16 | C18:3 t9,c12,t15 | |||||||
| 17 | C18:3 c6,c9,c12 | 49.139 | ||||||
| 18 | C18:3 c9,t12,t15 | 49.325 | ||||||
| 19 | C18:3 c9,c12,t15 | |||||||
| 20 | C18:3 c9,t12,c15 | 49.937 | ||||||
| 21 | C18:3 t9,c12,c15 | 50.057 | ||||||
| 22 | C18:3 c9,c12,c15 | 50.438 | ||||||
| 23 | C18:2 c9,t11 | 51.015 | 5 | 81 | 95 | 294 | 263 | 20 |
| 24 | C18:2 t10,c12 | 51.525 | ||||||
| FAMEs | Regression Equation | Linear Range (μg/mL) | R2 | LOD (μg/L) | LOQ (μg/L) | Intra-Day RSD (%) | Inter-Day RSD (%) |
|---|---|---|---|---|---|---|---|
| C18:0 | y = 117,066x − 53,301 | 0.60~400 | 1.0000 | 9.0 | 30.1 | 2.5 | 3.8 |
| C18:1 t6 | y = 12,851x + 592.45 | 0.60~60 | 1.0000 | 156.7 | 522.3 | 3.7 | 2.7 |
| C18:1 t9 | y = 12,895x + 1385.3 | 0.60~60 | 0.9999 | 157.3 | 524.4 | 4.8 | 4.6 |
| C18:1 t11 | y = 12,430x − 955.05 | 0.60~60 | 0.9993 | 149.4 | 498.0 | 3.6 | 2.8 |
| C18:1 c6/C18:1 c8 | y = 12,696x + 505.85 | 0.60~60 | 0.9999 | 158.9 | 529.6 | 1.9 | 2.1 |
| C18:1 c9 | y = 12,446x − 858.17 | 0.60~400 | 1.0000 | 161.4 | 538.0 | 2.1 | 3.3 |
| C18:1 c11 | y = 12,254x + 1154 | 0.60~60 | 0.9999 | 162.3 | 541.2 | 2.1 | 2.1 |
| C18:1 c12 | y = 12072x − 296.45 | 0.60~60 | 0.9997 | 168.8 | 562.7 | 2.7 | 2.9 |
| C18:2 t9,t12 | y = 49,383x + 8749 | 0.35~60 | 0.9999 | 84.9 | 283.0 | 2.5 | 3.4 |
| C18:2 c9,t12 | y = 43,275x − 5604.1 | 0.35~60 | 0.9989 | 95.8 | 319.3 | 2.1 | 2.5 |
| C18:2 t9,c12 | y = 41,591x − 6525.4 | 0.35~60 | 0.9999 | 98.1 | 326.9 | 4.9 | 3.9 |
| C18:2 c9,c12 | y = 36,584x − 1898.1 | 0.35~60 | 1.0000 | 103.0 | 343.4 | 2.0 | 2.4 |
| C18:2 c9,t11 | y = 43,267x − 6093.4 | 0.35~60 | 0.9989 | 96.5 | 321.6 | 1.9 | 2.8 |
| C18:2 t10,c12 | y = 41,715x − 7973 | 0.35~60 | 0.9986 | 100.6 | 335.2 | 2.2 | 3.2 |
| C18:3 t9,t12,t15 | y = 84,286x − 9165.8 | 0.15~20 | 0.9999 | 37.8 | 125.8 | NA | NA |
| C18:3 t9,t12,c15/C18:3 t9,c12,t15 | y = 86254x − 7942.8 | 0.15~20 | 1.0000 | 37.1 | 123.6 | NA | NA |
| C18:3 c6,c9,c12 | y = 88193x − 13659 | 0.15~20 | 0.9998 | 43.7 | 145.5 | 1.8 | 2.6 |
| C18:3 c9,t12,t15/C18:3 c9,c12,t15 | y = 72766x − 5435.2 | 0.15~20 | 1.0000 | 44.7 | 148.9 | 2.6 | 3.9 |
| C18:3 c9,t12,c15 | y = 50915x + 688.36 | 0.15~20 | 0.9999 | 42.8 | 142.5 | 4.6 | 3.9 |
| C18:3 t9,c12,c15 | y = 72954x − 1137.6 | 0.15~20 | 0.9997 | 41.6 | 138.8 | 4.2 | 3.9 |
| C18:3 c9,c12,c15 | y = 85866x + 16879 | 0.15~20 | 0.9986 | 30.4 | 101.2 | 1.9 | 2.6 |
| Spiked Level (μg/mL) | ||||||
|---|---|---|---|---|---|---|
| Tags | 20 | 100 | 400 | |||
| Recovery (%) | CV (%) | Recovery (%) | CV (%) | Recovery (%) | RSD (%) | |
| C18:1 c9 | 103.3 | 2.0 | 104.1 | 3.3 | 95.5 | 5.1 |
| C18:2 c9,c12 | 102.7 | 3.3 | 105.1 | 2.5 | 105.8 | 1.8 |
| C18:3 c6,c9,c12 | 98.9 | 3.1 | 99.5 | 4.5 | 103.2 | 4.2 |
| C18:3 c9,c12,c15 | 104.4 | 3.7 | 99.3 | 3.7 | 100.2 | 4.3 |
| Raw Milk | UHT Milk | PAS Milk | |
|---|---|---|---|
| C18:0 | 3026.3 ± 374.1 | 2707.5 ± 230.6 | 2717.34 ± 240.4 |
| C18:1 t6 | 78.5 ± 10.6 | 80.7 ± 11.7 | 78.2 ± 8.9 |
| C18:1 t9 | 79.2 ± 15.0 | 70.9 ± 10.9 | 77.9 ± 12.4 |
| C18:1 t11 | 234.9 ± 36.5 | 227.0 ± 38.6 | 250.8 ± 54.6 |
| C18:1 c6/C18:1 c8 | 241.6 ± 35.6 | 219.3 ± 27.2 | 232.8 ± 32.2 |
| C18:1 c9 | 6086.4 ± 631.2 | 5805.3 ± 543.7 | 5722.6 ± 466.7 |
| C18:1 c11 | 180.6 ± 34.0 | 199.2 ± 8.1 | 196.8 ± 20.2 |
| C18:1 c12 | 135.4 ± 20.7 | 123.2 ± 15.9 | 130.5 ± 18.1 |
| C18:2 t9,t12 | 39.7 ± 8.1 | 37.4 ± 4.1 | 35.4 ± 6.8 |
| C18:2 c9,t12 | 24.3 ± 5.6 | 23.9 ± 3.0 | 20.3 ± 4.0 |
| C18:2 t9,c12 | 9.2 ± 1.9 | 13.6 ± 2.8 | 10.2 ± 1.7 |
| C18:2 c9,c12 | 898.1 ± 112.7 | 773.9 ± 76.9 | 805.1 ± 53.5 |
| C18:2 c9,t11 (CLA) | 114.4 ± 18.7 | 106.2 ± 14.4 | 112.0 ± 17.7 |
| C18:2 t10,c12 (CLA) | 21.3 ± 4.4 | 19.7 ± 2.3 | 25.4 ± 0.7 |
| C18:3 t9,t12,t15 | NA | NA | NA |
| C18:3 t9,t12,c15/C18:3 t9,c12,t15 | NA | NA | NA |
| C18:3 c6,c9,c12 | 22.7 ± 2.1 | 19.6 ± 1.4 | 23.2 ± 1.0 |
| C18:3 c9,t12,t15/C18:3 c9,c12,t15 | 4.7 ± 0.9 | 4.5 ± 0.5 | 5.6 ± 0.17 |
| C18:3 c9,t12,c15 | 3.3 ± 0.8 | 3.9 ± 0.4 | 3.6 ± 0.3 |
| C18:3 t9,c12,c15 | 3.5 ± 0.5 | 3.3 ± 0.3 | 3.3 ± 0.2 |
| C18:3 c9,c12,c15 | 82.3 ± 15.1 | 70.8 ± 8.0 | 73.3 ± 10.7 |
| Cis/Trans C18:1 | 17.3 ± 1.4 | 17.0 ± 1.7 | 15.1 ± 3.8 |
| ∑ C18:1 | 7036.5 ± 689.4 | 6725.4 ± 601.7 | 6689.5 ± 497.4 |
| ∑ C18:2 | 971.2 ± 123.4 | 848.7 ± 84.2 | 871.1 ± 58.1 |
| ∑ C18:3 | 118.9 ± 12.7 | 102.5 ± 8.1 | 102.5 ± 25.1 |
| ∑ CLA | 135.7 ± 17.7 | 125.9 ± 14.8 | 137.4 ± 17.7 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, M.; Zhang, Y.; Wang, F.; Zheng, N.; Wang, J. Simultaneous Determination of C18 Fatty Acids in Milk by GC-MS. Separations 2021, 8, 118. https://doi.org/10.3390/separations8080118
Chen M, Zhang Y, Wang F, Zheng N, Wang J. Simultaneous Determination of C18 Fatty Acids in Milk by GC-MS. Separations. 2021; 8(8):118. https://doi.org/10.3390/separations8080118
Chicago/Turabian StyleChen, Meiqing, Yangdong Zhang, Fengen Wang, Nan Zheng, and Jiaqi Wang. 2021. "Simultaneous Determination of C18 Fatty Acids in Milk by GC-MS" Separations 8, no. 8: 118. https://doi.org/10.3390/separations8080118
APA StyleChen, M., Zhang, Y., Wang, F., Zheng, N., & Wang, J. (2021). Simultaneous Determination of C18 Fatty Acids in Milk by GC-MS. Separations, 8(8), 118. https://doi.org/10.3390/separations8080118