Identification of Block-Structured Covariance Matrix on an Example of Metabolomic Data
Abstract
:1. Introduction
2. Materials and Methods
2.1. Hierarchical Clustering
2.1.1. Distance Functions
- Euclidean—,
- Maximum—,
- Manhattan—,
- Canberra—,
- Binary—Jaccard index —ratio of number of common elements in both sets to number of all elements,
- Minkowski—.
2.1.2. Linkage Criteria
- Ward.D and Ward.D2—procedures applied the variance analysis to compute the cluster distances. The difference of the two Ward linkage algorithms is that the additional Ward’s clustering criterion is not implemented in “Ward.D” (1963), whereas the option “ward.D2” implements that criterion, with the latter, the dissimilarities are squared before cluster updating; cf. [17],
- Single—procedure computing the distance between two clusters as the minimum distance between each observation from one cluster and each observation from the other cluster,
- Complete—procedure computing the distance between two clusters as the maximum distance between each observation from one cluster and each observation from the other cluster,
- Average (UPGMA—Unweighted Pair-Group Method using Arithmetic Averages)—procedure computing the distance between two clusters as the average distance between each observation from one cluster and each observation from the other cluster,
- Mcquitty (WPGMA—Weighted Pair Group Method with Arithmetic Mean)—procedure based on UPGMA using the size of clusters (number of elements) as weights,
- Centroid (UPGMC—Unweighted Pair-Group Method using the Centroid Average)—the distance between two clusters is the distance between the cluster centroids,
- Median (WPGMC—Weighted Pair-Group Method using the Centroid Average)—procedure based on UPGMC using the size of clusters as weights.
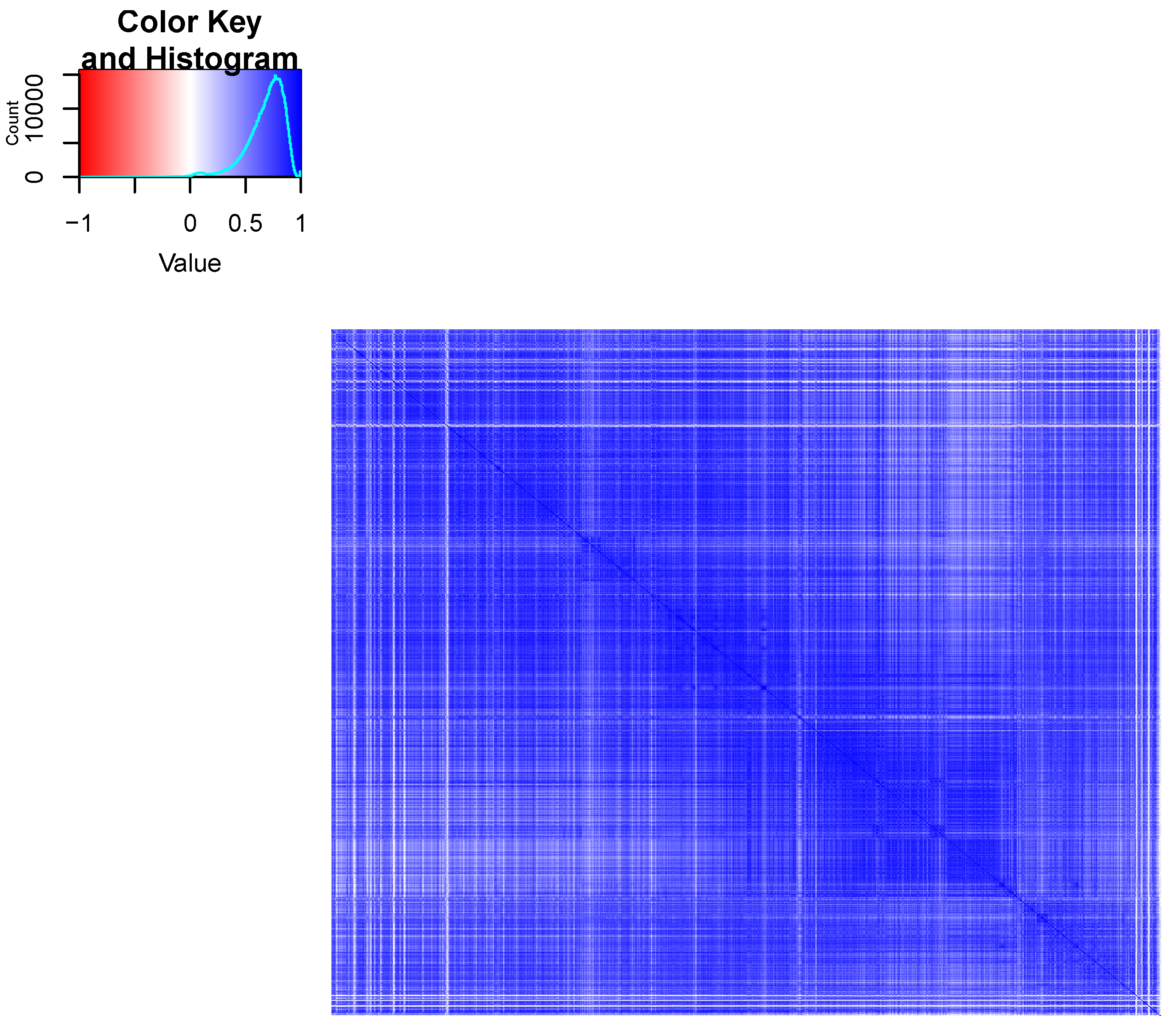
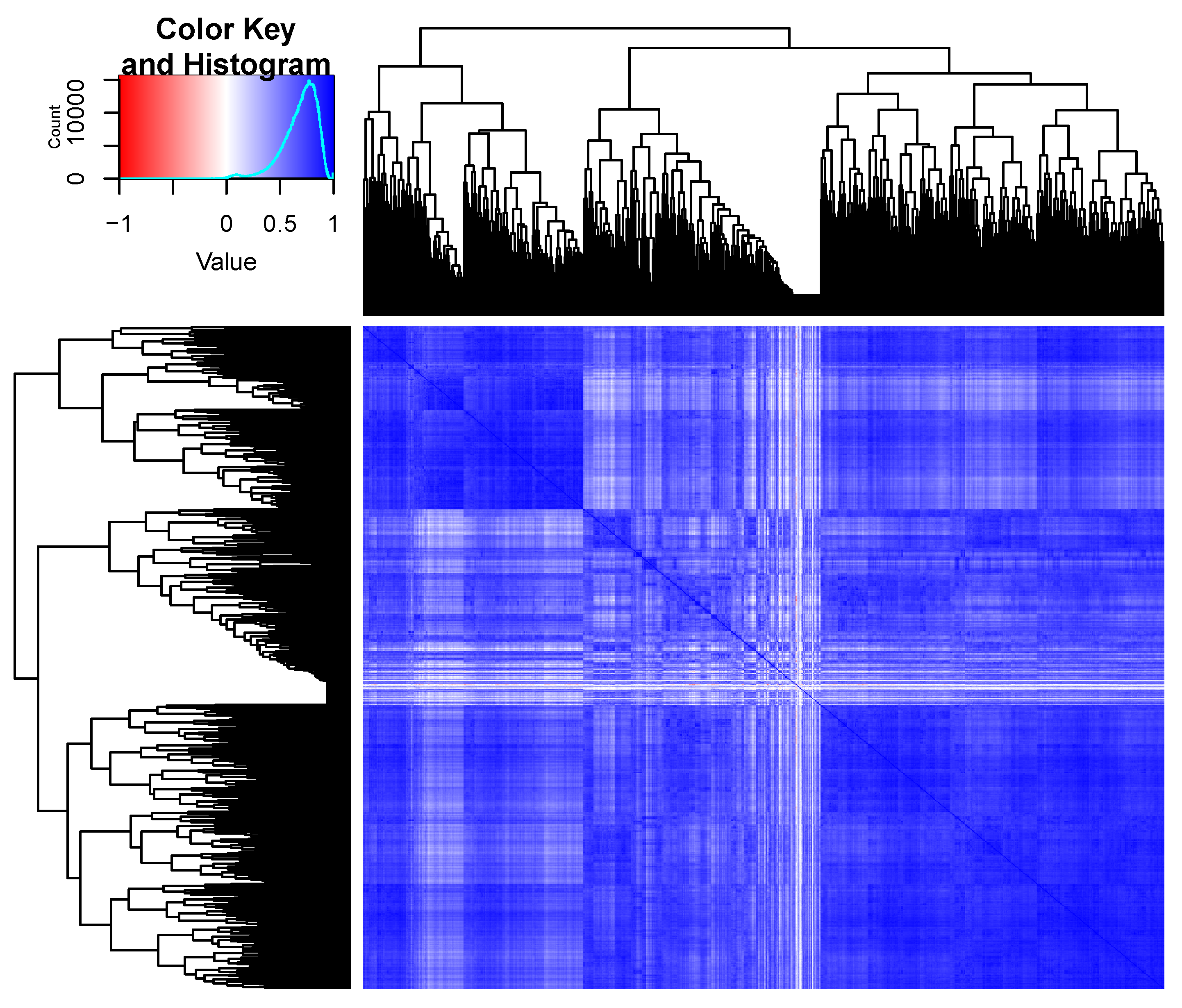
2.1.3. Visualizations
2.2. Statistical Background
2.2.1. Covariance Structures
- compound symmetry (CS)To ensure the positive definiteness of the matrix , we assume and ; cf. [19].
- banded symmetric Toeplitz structure ()where is an symmetric matrix with i-th superdiagonal and subdiagonal elements equal to 1 and all other elements equal to 0. In this paper, we consider the Toeplitz covariance matrix with and . The matrix is p.d. whenThe conditions for positive definiteness of the estimator of the matrix () is not expressed in the explicit form, cf. [20].
- autoregression of order one (AR(1))with . The matrix is p.d. when we assume and ; cf. [19]. The AR(1) structure is a special case of Toeplitz matrices with .
2.2.2. Identification Methods
- CS structurewith ,
- Toeplitz structure
- ○
- for
- ○
- forIn this case, the formulae for the estimator of the structure cannot be given in explicit form. To determine the estimator, the algorithm proposed by [20], p. 78, can be used.
- AR(1) structureTo determine the estimator of the AR(1) structure, the following system of equations should be solved:with . The above system of equations provides the local minimum of the discrepancy function; cf. [24].
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Winkelmüller, T.M.; Entila, F.; Anver, S.; Piasecka, A.; Song, B.; Dahms, E.; Sakakibara, H.; Gan, X.; Kułak, K.; Sawikowska, A.; et al. Gene expression evolution in pattern-triggered immunity within Arabidopsis thaliana and across Brassicaceae species. Plant Cell 2021, 33, 1863–1887. [Google Scholar] [CrossRef] [PubMed]
- Piasecka, A.; Sawikowska, A.; Kuczyńska, A.; Ogrodowicz, P.; Mikołajczak, K.; Krajewski, P.; Kachlicki, P. Phenolic metabolites from barley in contribution to phenome in soil moisture deficit. Int. J. Mol. Sci. 2020, 21, 6032. [Google Scholar] [CrossRef]
- Sawikowska, A.; Piasecka, A.; Kachlicki, P.; Krajewski, P. Separation of chromatographic co-eluted compounds by clustering and by functional data analysis. Metabolites 2021, 11, 214. [Google Scholar] [CrossRef]
- Kruszka, D.; Sawikowska, A.; Selvakesavan, R.K.; Krajewski, P.; Kachlicki, P.; Franklin, G. Silver nanoparticles affect phenolic and phytoalexin composition of Arabidopsis thaliana. Sci. Total Environ. 2020, 716, 135361. [Google Scholar] [CrossRef]
- Piasecka, A.; Sawikowska, A.; Kuczyńska, A.; Ogrodowicz, P.; Mikołajczak, K.; Krystowiak, K.; Gudyś, K.; Guzy-Wróbelska, J.; Krajewski, P.; Kachlicki, P. Drought related secondary metabolites of barley (Hordeum vulgare L.) leaves and their association with mQTLs. Plant J. 2017, 89, 898–913. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garibay-Hernández, A.; Kessler, N.; Józefowicz, A.M.; Türksoy, G.M.; Lohwasser, U.; Mock, H.-P. Untargeted metabotyping to study phenylpropanoid diversity in crop plants. Physiol. Plant. 2021, 173, 680–697. [Google Scholar] [CrossRef]
- Tracz, J.; Handschuh, L.; Lalowski, M.; Marczak, Ł.; Kostka-Jeziorny, K.; Perek, B.; Wanic-Kossowska, M.; Podkowińska, A.; Tykarski, A.; Formanowicz, D.; et al. Proteomic Profiling of Leukocytes Reveals Dysregulation of Adhesion and Integrin Proteins in Chronic Kidney Disease-Related Atherosclerosis. J. Proteome Res. 2021, 20, 3053–3067. [Google Scholar] [CrossRef]
- Thompson, R.M.; Dytfeld, D.; Reyes, L.; Robinson, R.M.; Smith, B.; Manevich, Y.; Jakubowiak, A.; Komarnicki, M.; Przybylowicz-Chalecka, A.; Szczepaniak, T.; et al. Glutaminase inhibitor CB-839 synergizes with carfilzomib in resistant multiple myeloma cells. Oncotarget 2017, 8, 35863–35876. [Google Scholar] [CrossRef] [Green Version]
- Luczak, M.; Suszynska-Zajczyk, J.; Marczak, L.; Formanowicz, D.; Pawliczak, E.; Wanic-Kossowska, M.; Stobiecki, M. Label-Free Quantitative Proteomics Reveals Differences in Molecular Mechanism of Atherosclerosis Related and Non-Related to Chronic Kidney Disease. Int. J. Mol. Sci. 2016, 17, 631. [Google Scholar] [CrossRef] [PubMed]
- Mieldzioc, A.; Mokrzycka, M.; Sawikowska, A. Covariance regularization for metabolomic data on the drought resistance of barley. Biom. Lett. 2020, 56, 165–181. [Google Scholar] [CrossRef] [Green Version]
- Filipiak, K.; Klein, D. Estimation and testing the covariance structure of doubly multivariate data. In Multivariate, Multilinear and Mixed Linear Models; Filipiak, K., Markiewicz, A., von Rosen, D., Eds.; Springer: Berlin/Heidelberg, Germany, 2021. [Google Scholar]
- Janiszewska, M.; Markiewicz, A.; Mokrzycka, M. Block matrix approximation via entropy loss function. Appl. Math. 2020, 65, 829–844. [Google Scholar] [CrossRef]
- Szczepańska-Álvarez, A.; Hao, C.; Liang, Y.; von Rosen, D. Estimation equations for multivariate linear models with Kronecker structured covariance matrices. Commun. Stat. Theory Methods 2017, 46, 7902–7915. [Google Scholar] [CrossRef]
- Swarcewicz, B.; Sawikowska, A.; Marczak, Ł.; Łuczak, M.; Ciesiołka, D.; Krystkowiak, K.; Kuczyńska, A.; Piślewska-Bednarek, M.; Krajewski, P.; Stobiecki, M. Effect of drought stress on metabolite contents in barley recombinant inbred line population revealed by untargeted GC–MS profiling. Acta Physiol. Plant 2017, 39, 158. [Google Scholar] [CrossRef]
- Chmielewska, K.; Rodziewicz, P.; Swarcewicz, B.; Sawikowska, A.; Krajewski, P.; Marczak, Ł.; Ciesiołka, D.; Kuczyńska, A.; Mikołajczak, K.; Ogrodowicz, P.; et al. Analysis of drought-induced proteomic and metabolomic changes in barley (Hordeum vulgare L.) leaves and roots unravels some aspects of biochemical mechanisms involved in drought tolerance. Front. Plant Sci. 2016, 7, 1108. [Google Scholar] [CrossRef] [PubMed]
- Pvclust: Hierarchical Clustering with P-Values via Multiscale Bootstrap Resampling. Available online: https://CRAN.R-project.org/package=pvclust (accessed on 11 June 2021).
- Murtagh, F.; Legendre, P. Ward’s Hierarchical Agglomerative Clustering Method: Which Algorithms Implement Ward’s Criterion? J. Classif. 2014, 31, 274–295. [Google Scholar] [CrossRef] [Green Version]
- Legendre, P.; Legendre, L. Numerical Ecology, 3rd ed.; Elsevier: Amsterdam, The Netherlands, 2012; Volume 24. [Google Scholar]
- Lin, L.; Higham, N.J.; Pan, J. Covariance structure regularization via entropy loss function. Comput. Stat. Data Anal. 2014, 72, 315–327. [Google Scholar] [CrossRef]
- Filipiak, K.; Markiewicz, A.; Mieldzioc, A.; Sawikowska, A. On projection of a positive definite matrix on a cone of nonnegative definite Toeplitz matrices. Electron. J. Linear Algebra 2018, 33, 74–82. [Google Scholar] [CrossRef] [Green Version]
- Filipiak, K.; Klein, D.; Mokrzycka, M. Separable covariance structure identification for doubly multivariate data. In Multivariate, Multilinear and Mixed Linear Models; Filipiak, K., Markiewicz, A., von Rosen, D., Eds.; Springer: Berlin/Heidelberg, Germany, 2021. [Google Scholar]
- Filipiak, K.; Klein, D.; Mokrzycka, M. Estimators comparison of separable covariance structure with one component as compound symmetry matrix. Electron. J. Linear Algebra 2018, 33, 83–98. [Google Scholar] [CrossRef] [Green Version]
- Filipiak, K.; Klein, D.; Markiewicz, A.; Mokrzycka, M. Approximation with a Kronecker product structure with one component as compound symmetry or autoregression via entropy loss function. Linear Algebra Appl. 2021, 610, 625–646. [Google Scholar] [CrossRef]
- Cui, X.; Li, X.; Zhao, J.; Zeng, L.; Zhang, D.; Pan, J. Covariance structure regularization via Frobenius norm discrepancy. Linear Algebra Appl. 2016, 510, 124–145. [Google Scholar] [CrossRef] [Green Version]
- Filipiak, K.; Klein, D. Approximation with Kronecker product structure with one component as compound symmetry or autoregression. Linear Algebra Appl. 2018, 559, 11–33. [Google Scholar] [CrossRef]
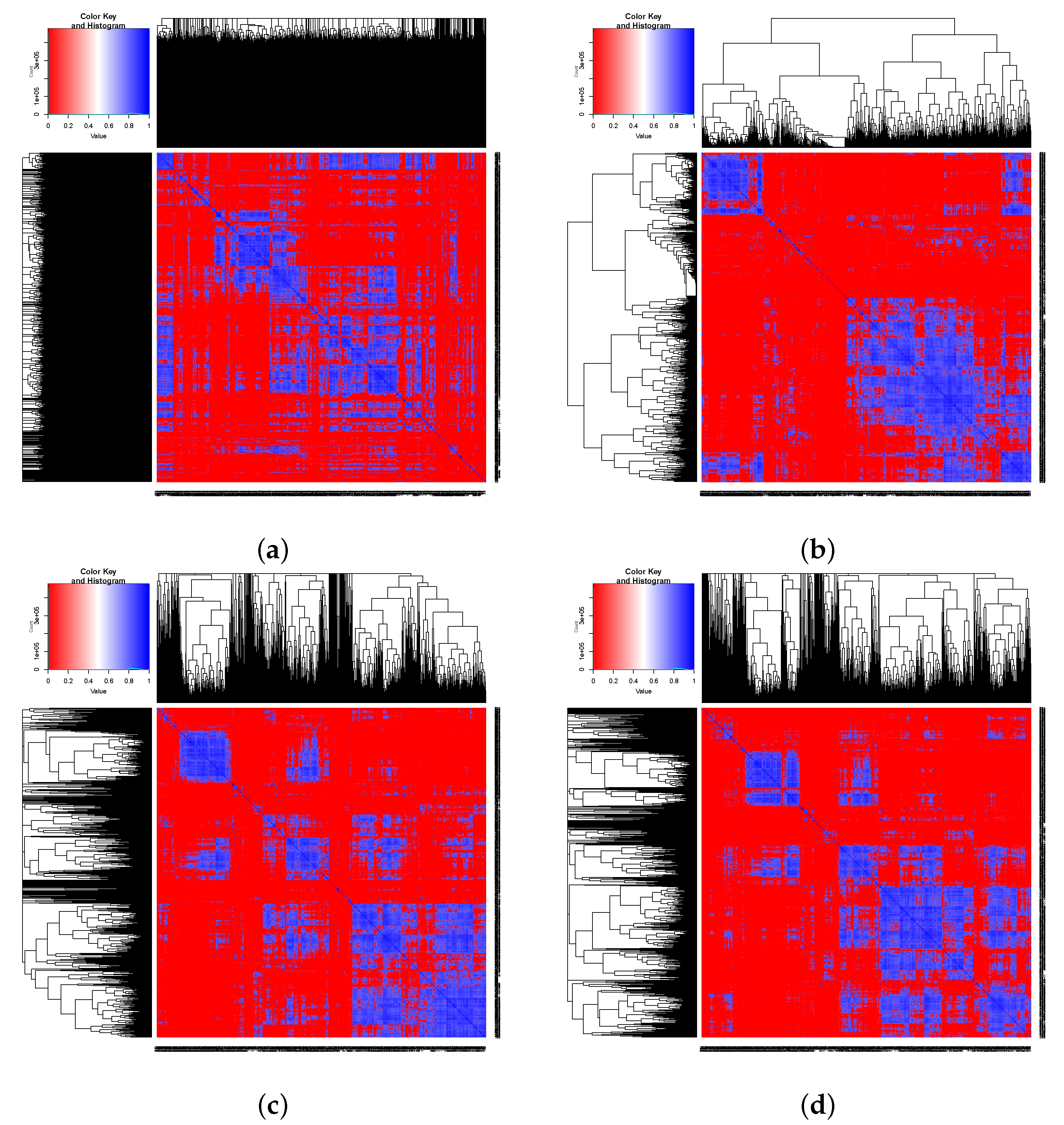
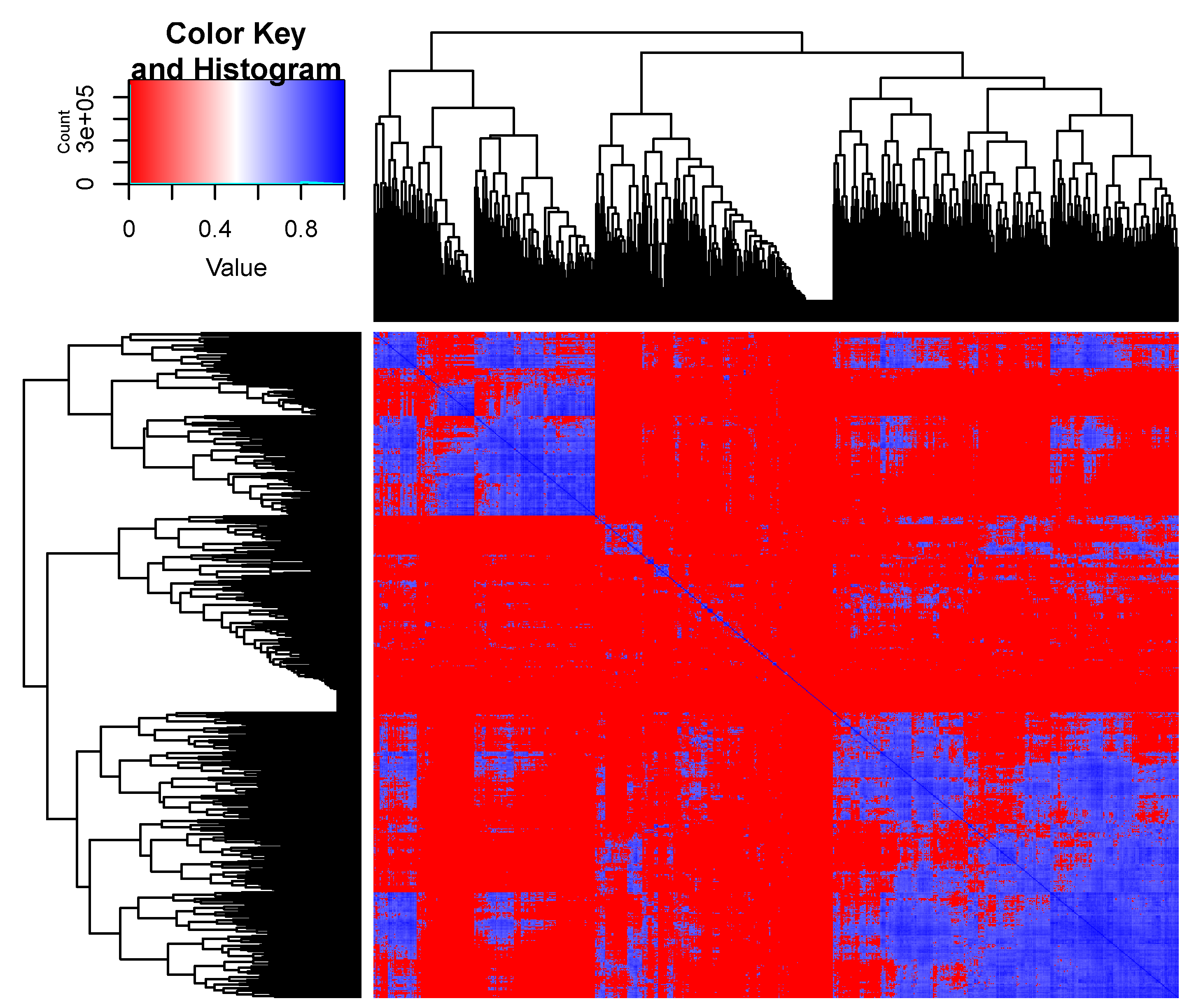
| 0.81877 | 0.81892 | 0.93560 | 0.93492 | ||
| 0.81882 | 0.81898 | 0.93565 | 0.93497 | ||
| 0.88871 | 0.88885 | 0.99738 | 0.99674 | ||
| 0.88805 | 0.88820 | 0.99680 | 0.99616 | ||
| 31.4724 | 29.14777 | 26.4708 | |
| 0.7920 | – | 0.7897 |
| 0.79225 | 0.79249 | 0.81832 | 0.81794 |
| 14.7186 | 18.5793 | 16.7440 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mieldzioc, A.; Mokrzycka, M.; Sawikowska, A. Identification of Block-Structured Covariance Matrix on an Example of Metabolomic Data. Separations 2021, 8, 205. https://doi.org/10.3390/separations8110205
Mieldzioc A, Mokrzycka M, Sawikowska A. Identification of Block-Structured Covariance Matrix on an Example of Metabolomic Data. Separations. 2021; 8(11):205. https://doi.org/10.3390/separations8110205
Chicago/Turabian StyleMieldzioc, Adam, Monika Mokrzycka, and Aneta Sawikowska. 2021. "Identification of Block-Structured Covariance Matrix on an Example of Metabolomic Data" Separations 8, no. 11: 205. https://doi.org/10.3390/separations8110205
APA StyleMieldzioc, A., Mokrzycka, M., & Sawikowska, A. (2021). Identification of Block-Structured Covariance Matrix on an Example of Metabolomic Data. Separations, 8(11), 205. https://doi.org/10.3390/separations8110205






