Chromatographic Profiling with Machine Learning Discriminates the Maturity Grades of Nicotiana tabacum L. Leaves
Abstract
1. Introduction
2. Materials and Methods
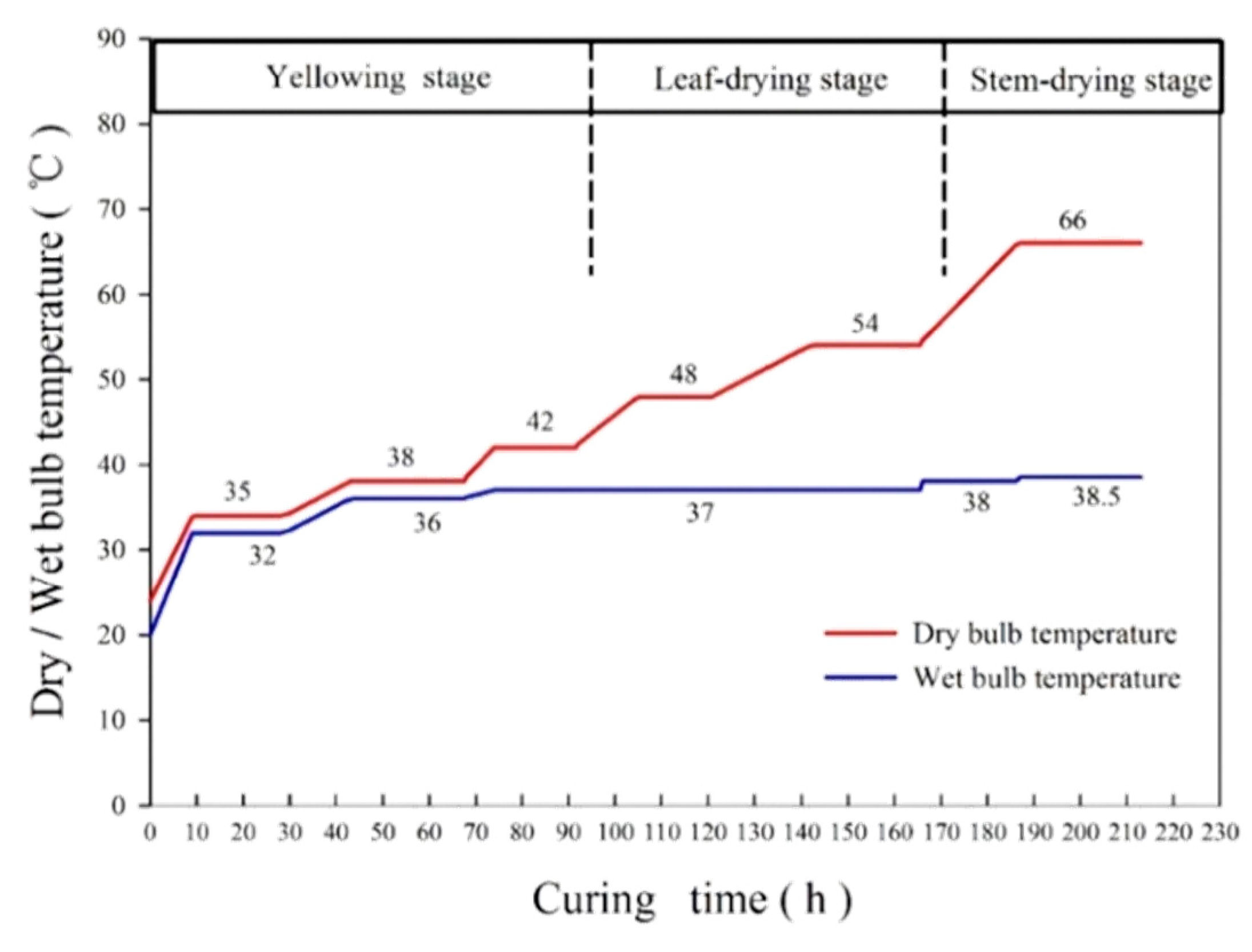
2.1. Materials and Reagents
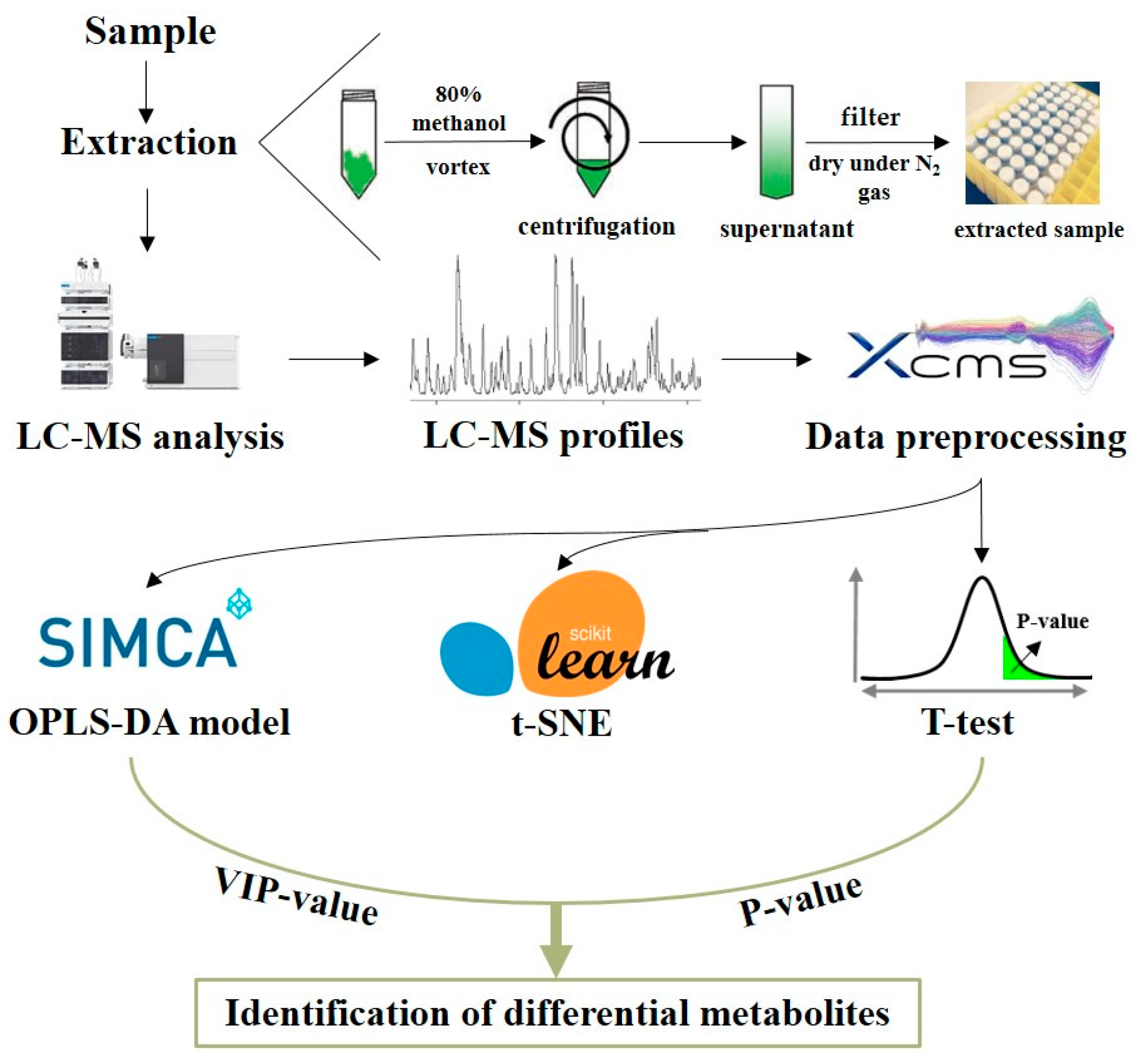
2.2. Sample Preparation
2.3. LC-MS Analysis
2.4. Data Preprocessing
2.5. Machine Learning Models of Maturity Grades
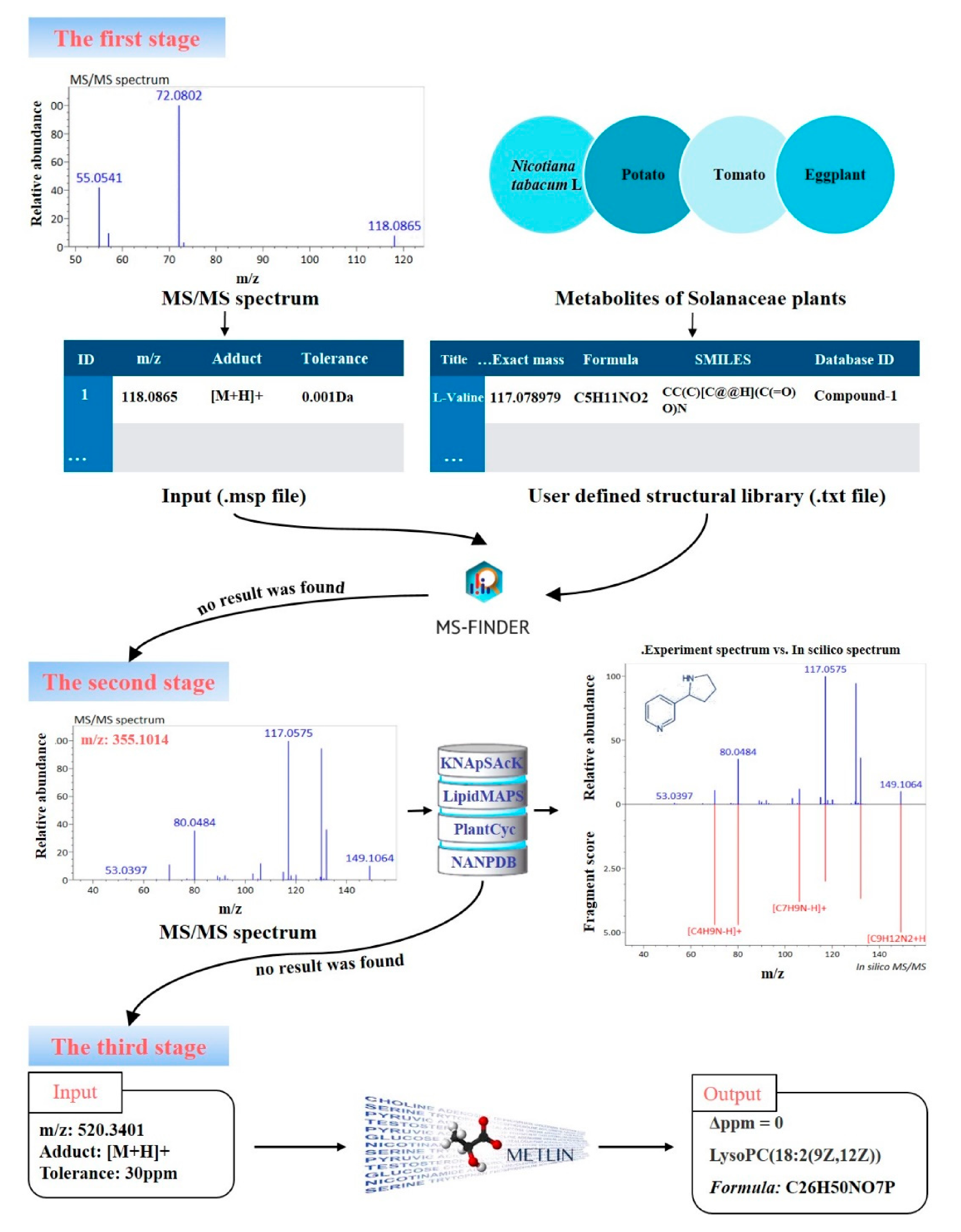
2.6. Identification of Metabolites
3. Results
3.1. Development of the Analytical Method
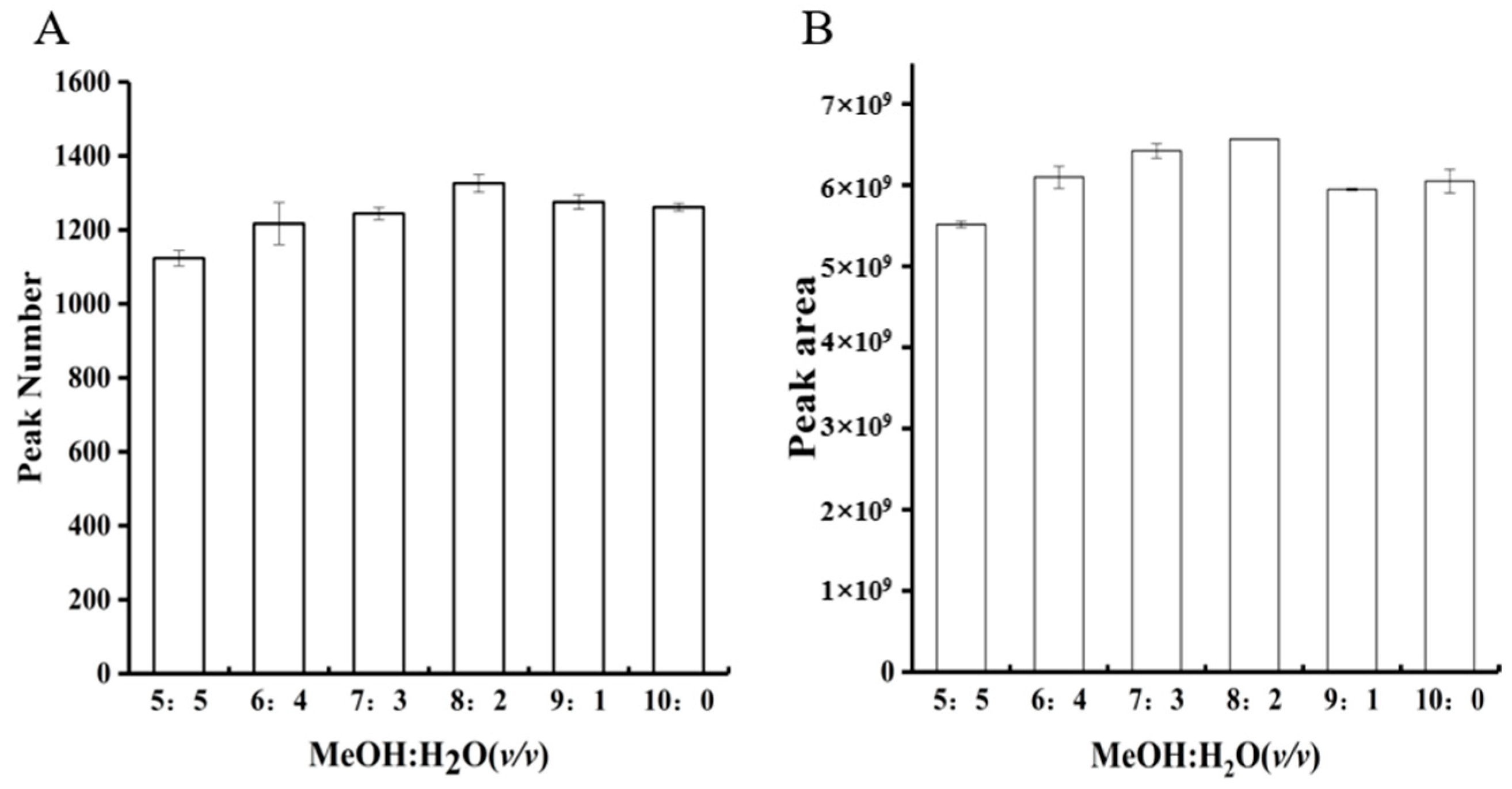
3.1.1. Extraction Solvent Optimization
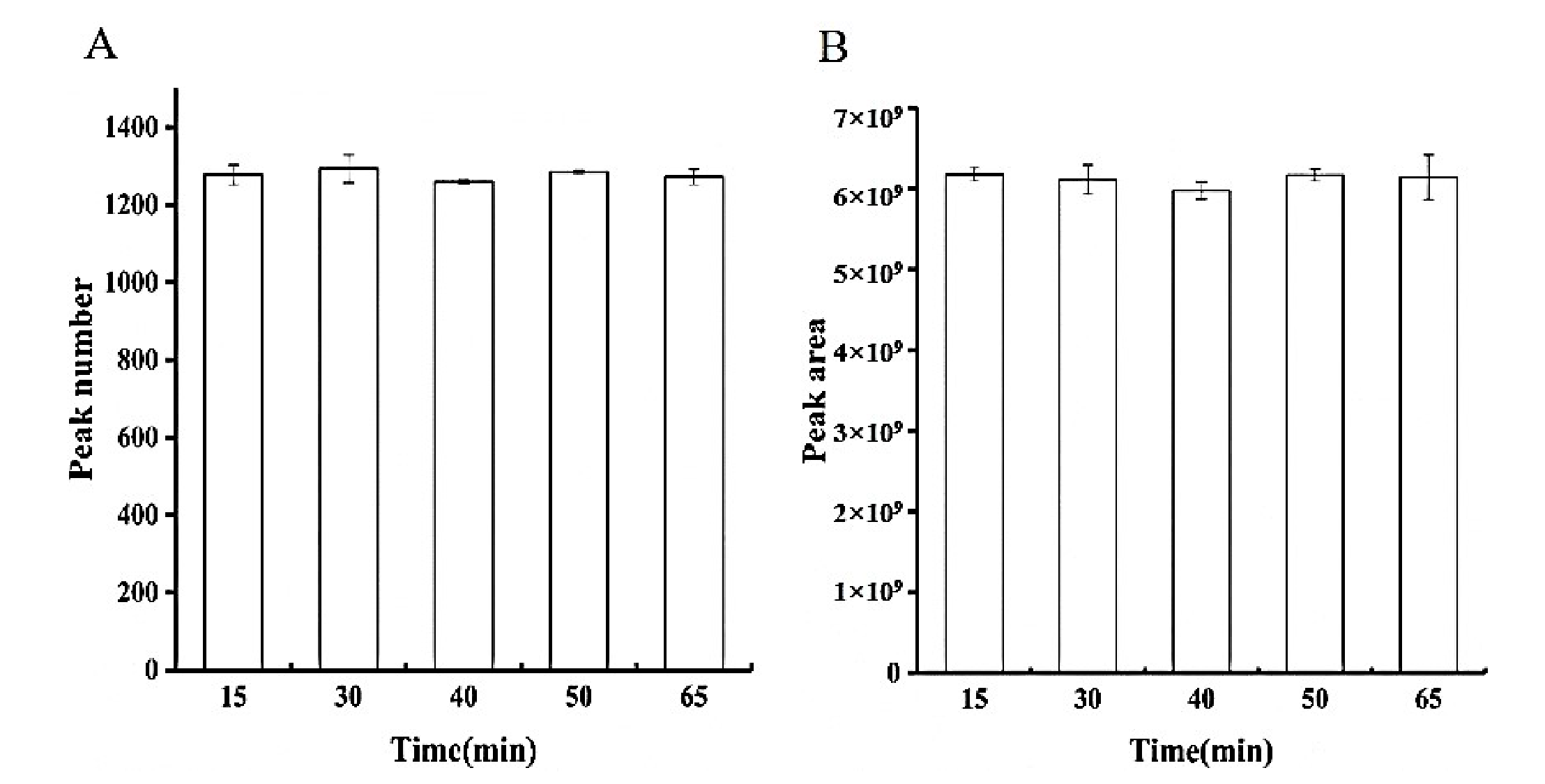
3.1.2. Extraction Time Selection
3.1.3. Investigation of UPLC/IT-TOF MS Parameters
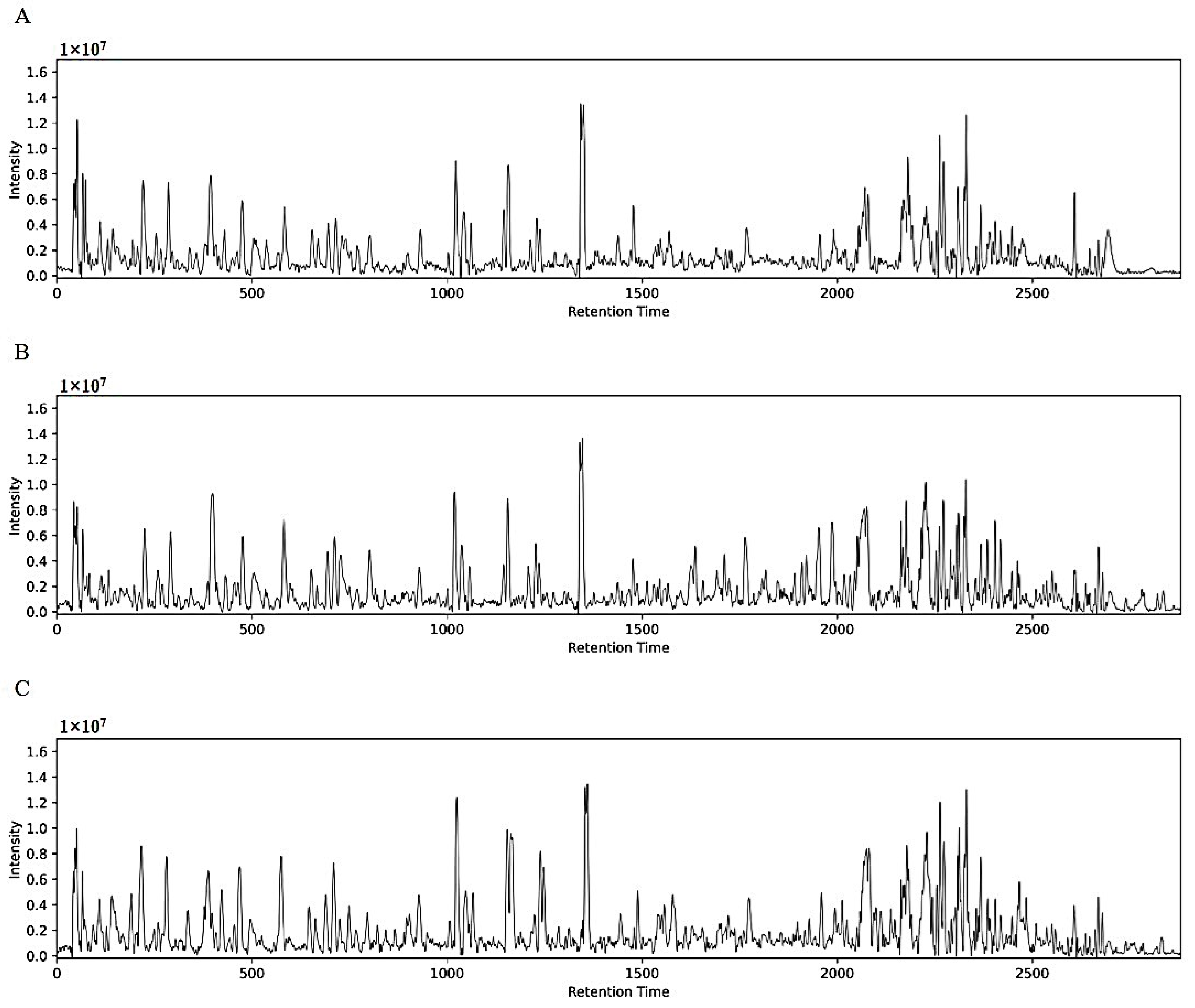
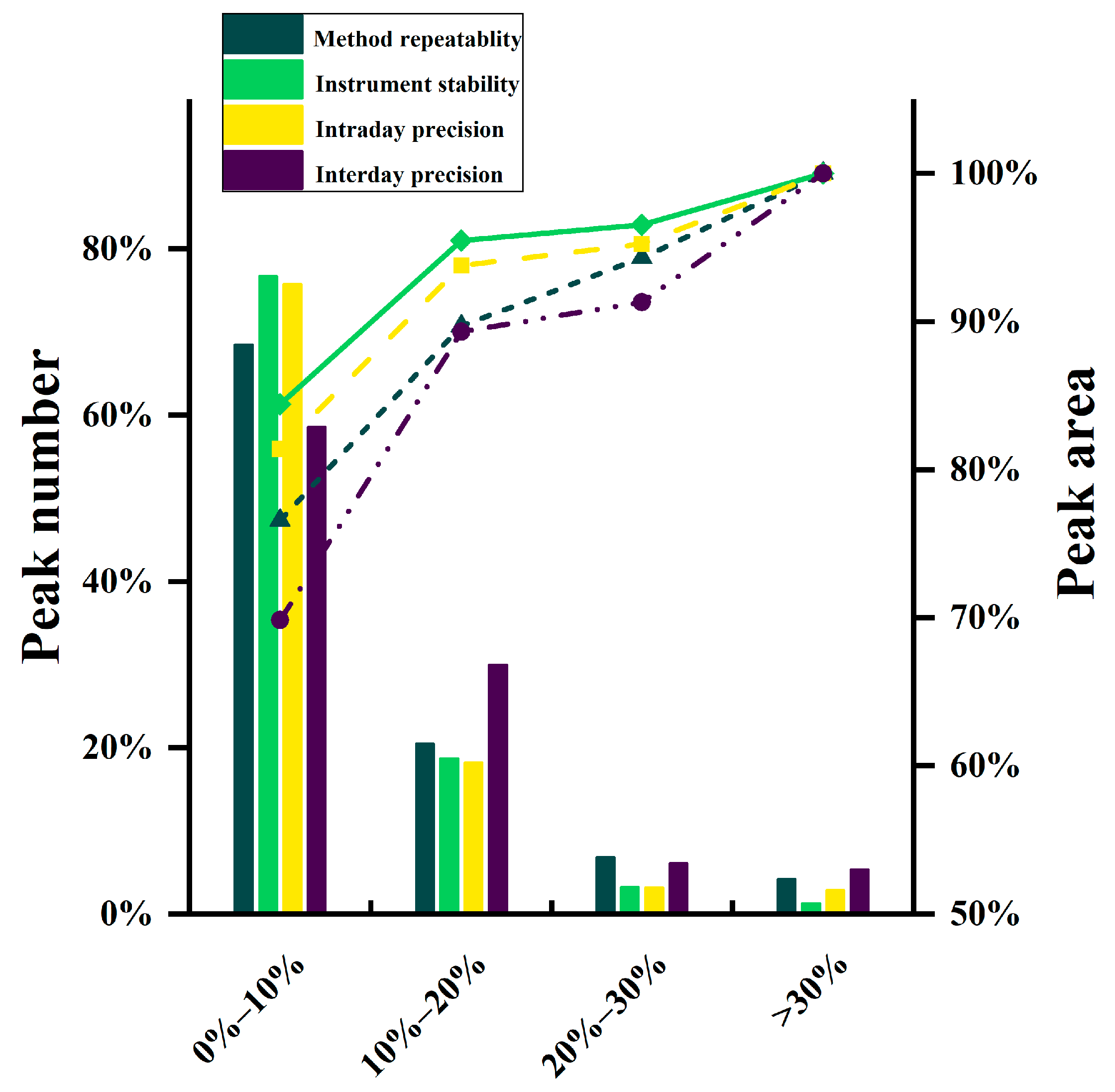
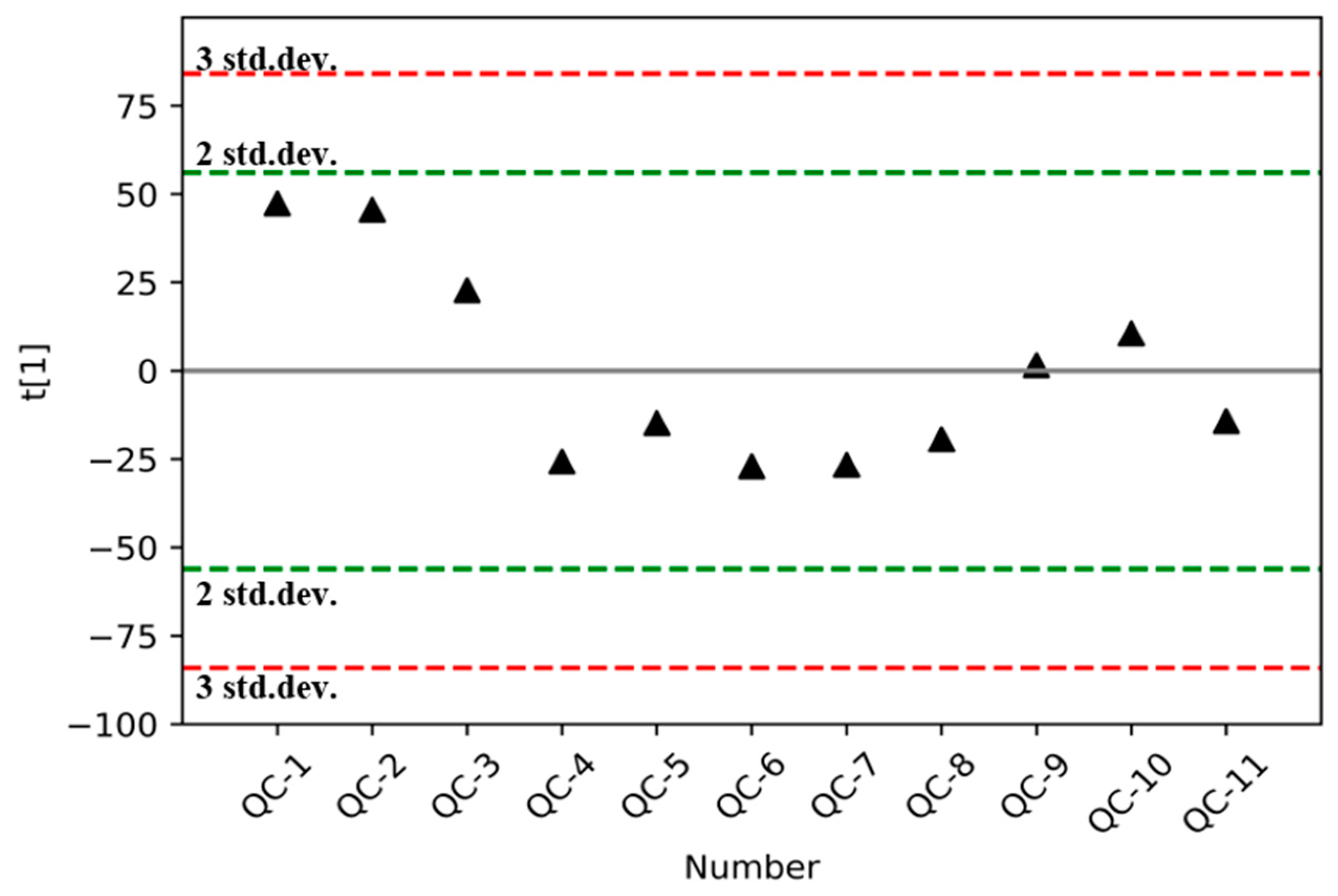
3.2. Validation of the Analytical Method
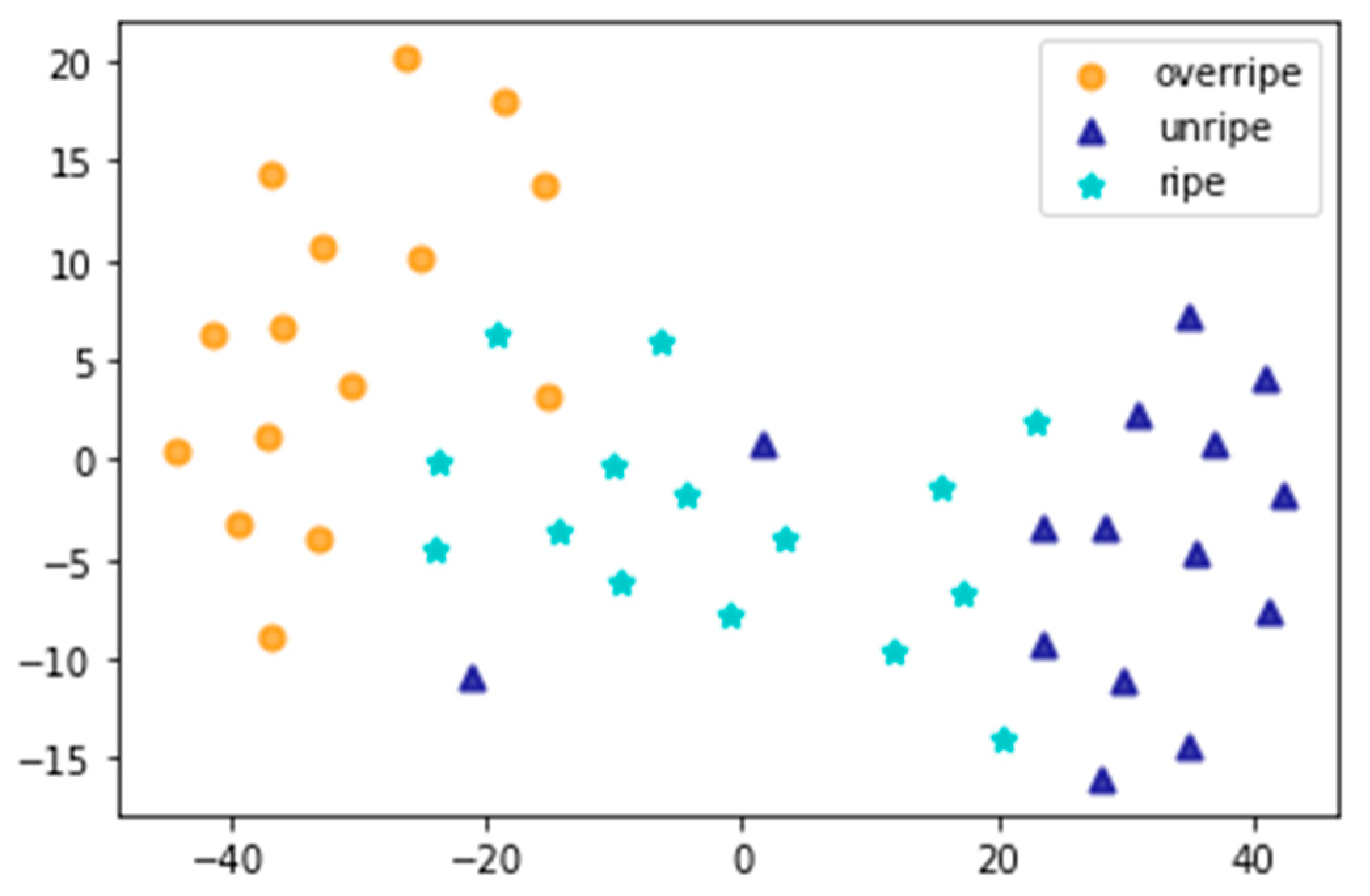
3.3. Classification of the Leaves from Different Maturity Grades
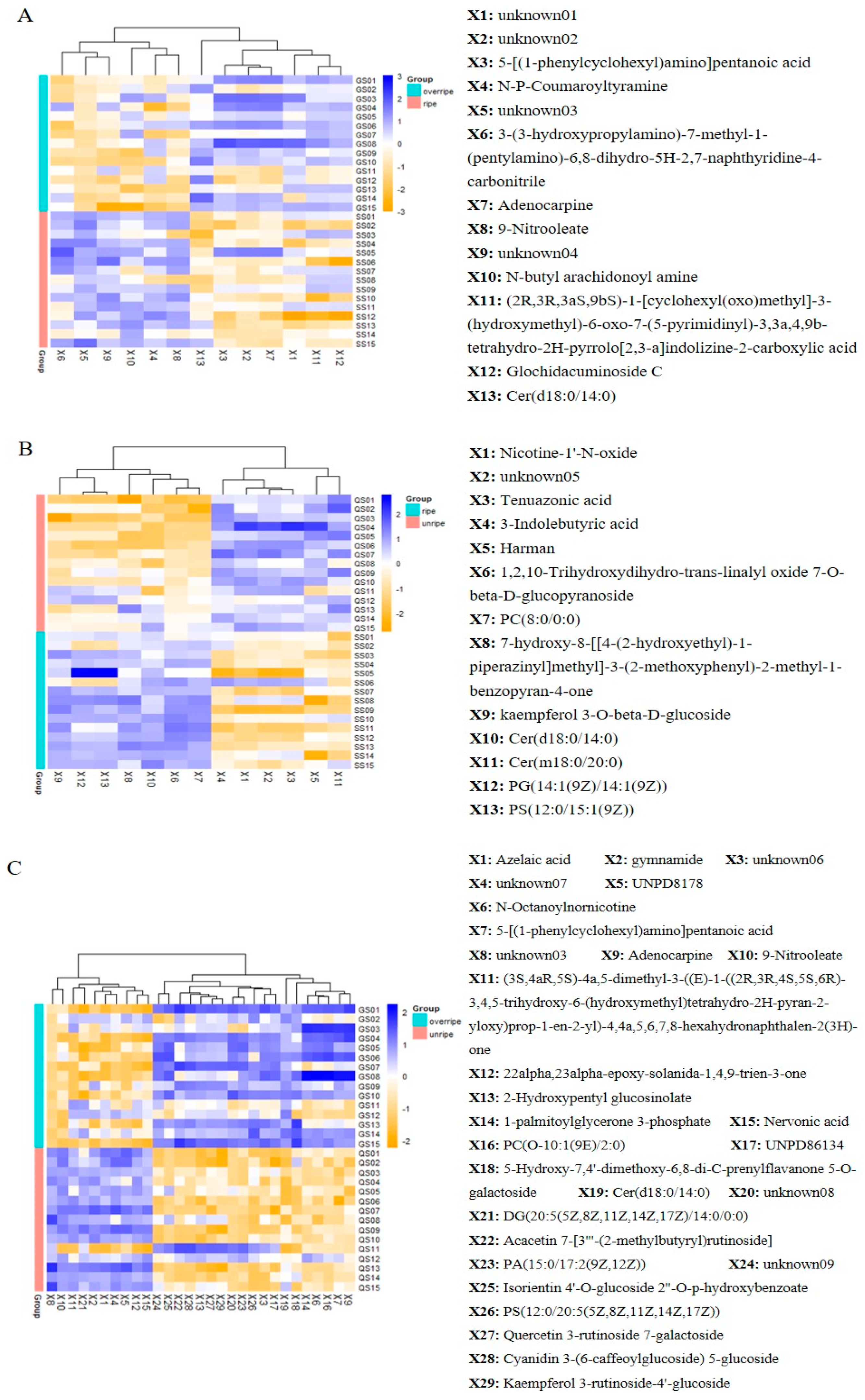
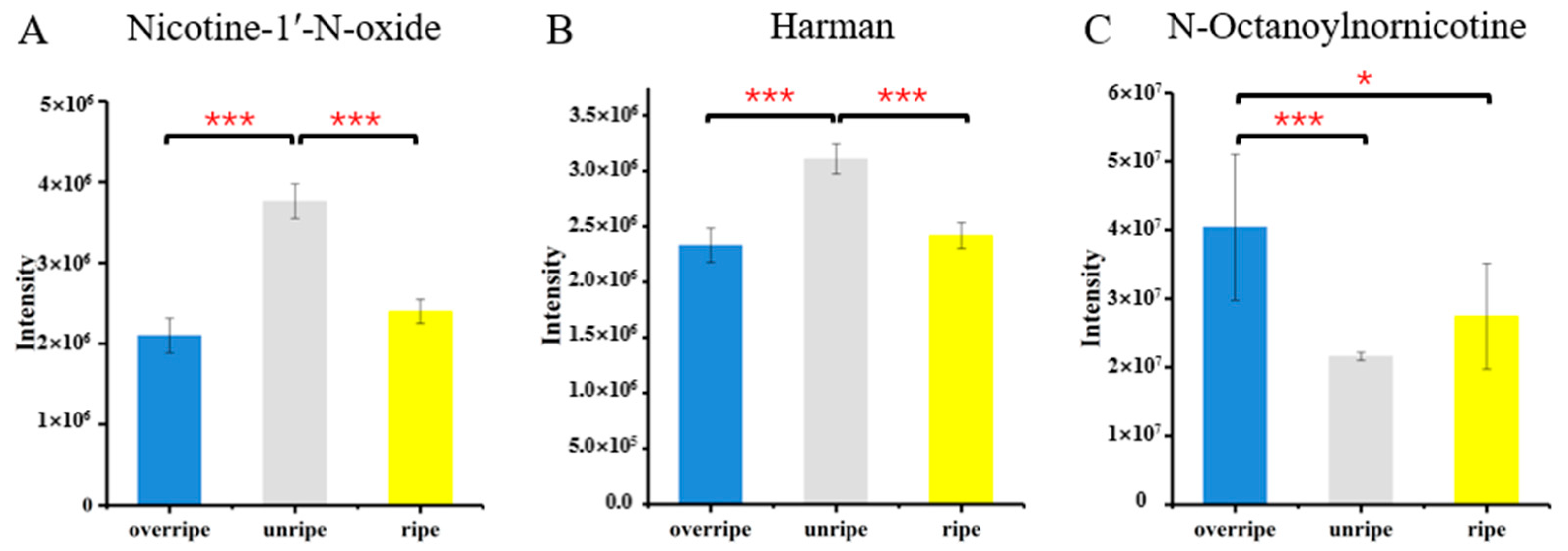
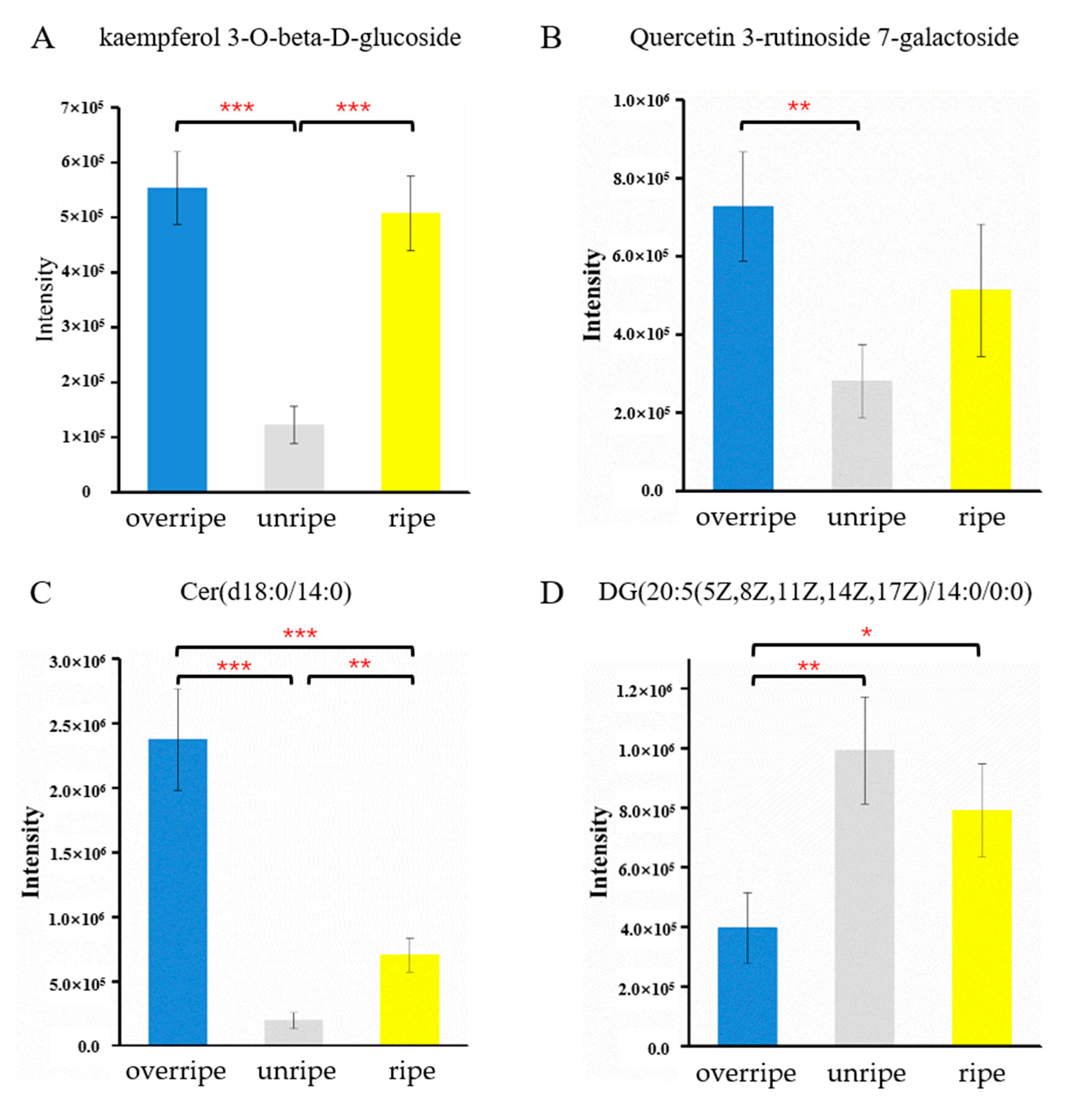
3.4. Identification of Metabolites from Different Maturity Grades
4. Discussions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chen, Y.; Ren, K.; He, X.; Gong, J.; Hu, X.; Su, J.; Jin, Y.; Zhao, Z.; Zhu, Y.; Zou, C. Dynamic changes in physiological and biochemical properties of flue-cured tobacco of different leaf ages during flue-curing and their effects on yield and quality. BMC Plant Biol. 2019, 19, 555. [Google Scholar] [CrossRef] [PubMed]
- Cui, G.; Huang, W.; Zhao, G.; Han, S. Effects of different maturity on appearance grade quality and key chemical components of baked tobacco leaves. J. Anhui Agric. Sci. 2013, 41, 10819–10822. [Google Scholar]
- Musilová, J.; Glatz, Z. Metabolomics-Basic concepts, Strategies and Methodologies. Chem. Listy 2011, 105, 745–751. [Google Scholar]
- Lisec, J.; Schauer, N.; Kopka, J.; Willmitzer, L.; Fernie, A.R. Gas chromatography mass spectrometry-based metabolite profiling in plants. Nat. Protoc. 2006, 1, 387–396. [Google Scholar] [CrossRef] [PubMed]
- Ramautar, R.; Busnel, J.-M.; Deelder, A.M.; Mayboroda, O.A. Enhancing the coverage of the urinary metabolome by sheathless capillary electrophoresis-mass spectrometry. Anal. Chem. 2012, 84, 885–892. [Google Scholar] [CrossRef] [PubMed]
- Núñez, N.; Vidal-Casanella, O.; Sentellas, S.; Saurina, J.; Núñez, O. Non-Targeted Ultra-High Performance Liquid Chromatography-High-Resolution Mass Spectrometry (UHPLC-HRMS) Fingerprints for the Chemometric Characterization and Classification of Turmeric and Curry Samples. Separations 2020, 7, 32. [Google Scholar] [CrossRef]
- De Vos, R.C.; Moco, S.; Lommen, A.; Keurentjes, J.J.; Bino, R.J.; Hall, R.D. Untargeted large-scale plant metabolomics using liquid chromatography coupled to mass spectrometry. Nat. Protoc. 2007, 2, 778–791. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhao, J.; Zhao, C.; Zhou, H.; Li, Y.; Zhang, J.; Li, L.; Hu, C.; Li, W.; Peng, X. A metabolomics study delineating geographical location-associated primary metabolic changes in the leaves of growing tobacco plants by GC-MS and CE-MS. Sci. Rep. 2015, 5, 16346. [Google Scholar] [CrossRef]
- Li, L.; Zhao, J.; Zhao, Y.; Lu, X.; Zhou, Z.; Zhao, C.; Xu, G. Comprehensive investigation of tobacco leaves during natural early senescence via multi-platform metabolomics analyses. Sci. Rep. 2016, 6, 37976. [Google Scholar] [CrossRef]
- Wang, X.; Sun, H.; Zhang, A.; Wang, P.; Han, Y. Ultra-performance liquid chromatography coupled to mass spectrometry as a sensitive and powerful technology for metabolomic studies. J. Sep. Sci. 2011, 34, 3451–3459. [Google Scholar] [CrossRef]
- Patti, G.J. Separation strategies for untargeted metabolomics. J. Sep. Sci. 2011, 34, 3460–3469. [Google Scholar] [CrossRef] [PubMed]
- Monga, G.K.; Ghosal, A.; Ramanathan, D. To Develop the Method for UHPLC-HRMS to Determine the Antibacterial Potential of a Central American Medicinal Plant. Separations 2019, 6, 37. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhao, C.; Lu, X.; Zhou, H.; Li, Y.; Zhou, J.; Chang, Y.; Zhang, J.; Jin, L.; Lin, F. Investigation of the relationship between the metabolic profile of tobacco leaves in different planting regions and climate factors using a pseudotargeted method based on gas chromatography/mass spectrometry. J. Proteome Res. 2013, 12, 5072–5083. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Zhao, Y.; Hu, C.; Zhao, C.; Zhang, J.; Li, L.; Zeng, J.; Peng, X.; Lu, X.; Xu, G. Metabolic profiling with gas chromatography–mass spectrometry and capillary electrophoresis–mass spectrometry reveals the carbon–nitrogen status of tobacco leaves across different planting areas. J. Proteome Res. 2016, 15, 468–476. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Lu, X.; Zhao, J.; Zhang, J.; Zhao, Y.; Zhao, C.; Xu, G. Lipidome and metabolome analysis of fresh tobacco leaves in different geographical regions using liquid chromatography–mass spectrometry. Anal. Bioanal. Chem. 2015, 407, 5009–5020. [Google Scholar] [CrossRef]
- Li, L.; Zhao, C.; Chang, Y.; Lu, X.; Zhang, J.; Zhao, Y.; Zhao, J.; Xu, G. Metabolomics study of cured tobacco using liquid chromatography with mass spectrometry: Method development and its application in investigating the chemical differences of tobacco from three growing regions. J. Sep. Sci. 2014, 37, 1067–1074. [Google Scholar] [CrossRef]
- Li, Q.; Zhao, C.; Li, Y.; Chang, Y.; Wu, Z.; Pang, T.; Lu, X.; Wu, Y.; Xu, G. Liquid chromatography/mass spectrometry-based metabolic profiling to elucidate chemical differences of tobacco leaves between Zimbabwe and China. J. Sep. Sci. 2011, 34, 119–126. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, X.; Guo, J.; Xia, Q.; Zhao, G.; Zhou, H.; Xie, F. Metabolic profiling of Chinese tobacco leaf of different geographical origins by GC-MS. J. Agric. Food Chem. 2013, 61, 2597–2605. [Google Scholar] [CrossRef]
- Lommen, A.; Kools, H.J. MetAlign 3.0: Performance enhancement by efficient use of advances in computer hardware. Metabolomics 2012, 8, 719–726. [Google Scholar] [CrossRef]
- Pluskal, T.; Castillo, S.; Villar-Briones, A.; Orešič, M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinform. 2010, 11, 395. [Google Scholar] [CrossRef]
- Smith, C.A.; Want, E.J.; O’Maille, G.; Abagyan, R.; Siuzdak, G. XCMS: Processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem. 2006, 78, 779–787. [Google Scholar] [CrossRef] [PubMed]
- Coble, J.B.; Fraga, C.G. Comparative evaluation of preprocessing freeware on chromatography/mass spectrometry data for signature discovery. J. Chromatogr. A 2014, 1358, 155–164. [Google Scholar] [CrossRef] [PubMed]
- Treviño, V.; Yañez-Garza, I.L.; Rodriguez-López, C.E.; Urrea-López, R.; Garza-Rodriguez, M.-L.; Barrera-Saldaña, H.-A.; Tamez-Peña, J.G.; Winkler, R.; Díaz de-la-Garza, R.I. GridMass: A fast two-dimensional feature detection method for LC/MS. J. Mass Spectrom. 2015, 50, 165–174. [Google Scholar] [CrossRef] [PubMed]
- Tautenhahn, R.; Boettcher, C.; Neumann, S. Highly sensitive feature detection for high resolution LC/MS. BMC Bioinform. 2008, 9, 504. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Zhao, J.; Wang, C.; Yan, C. Comprehensive evaluation of robotic global performance based on modified principal component analysis. Int. J. Adv. Robot. Syst. 2020, 17, 1729881419896881. [Google Scholar] [CrossRef]
- Maaten, L.v.d.; Hinton, G. Visualizing data using t-SNE. J. Mach. Learn. Res. 2008, 9, 2579–2605. [Google Scholar]
- Cloarec, O.; Dumas, M.-E.; Craig, A.; Barton, R.H.; Trygg, J.; Hudson, J.; Blancher, C.; Gauguier, D.; Lindon, J.C.; Holmes, E. Statistical total correlation spectroscopy: An exploratory approach for latent biomarker identification from metabolic 1H NMR data sets. Anal. Chem. 2005, 77, 1282–1289. [Google Scholar] [CrossRef]
- Bylesjo, M.; Rantalainen, M.; Cloarec, O.; Nicholson, J.K.; Holmes, E.; Trygg, J. OPLS discriminant analysis: Combining the strengths of PLS-DA and SIMCA classification. J. Chemom. 2006, 20, 341–351. [Google Scholar] [CrossRef]
- Guijas, C.; Montenegro-Burke, J.R.; Domingo-Almenara, X.; Palermo, A.; Warth, B.; Hermann, G.; Koellensperger, G.; Huan, T.; Uritboonthai, W.; Aisporna, A.E. METLIN: A technology platform for identifying knowns and unknowns. Anal. Chem. 2018, 90, 3156–3164. [Google Scholar] [CrossRef]
- Ivanova, P.T.; Milne, S.B.; Myers, D.S.; Brown, H.A. Lipidomics: A mass spectrometry based systems level analysis of cellular lipids. Curr. Opin. Chem. Biol. 2009, 13, 526–531. [Google Scholar] [CrossRef]
- Horai, H.; Arita, M.; Kanaya, S.; Nihei, Y.; Ikeda, T.; Suwa, K.; Ojima, Y.; Tanaka, K.; Tanaka, S.; Aoshima, K. MassBank: A public repository for sharing mass spectral data for life sciences. J. Mass Spectrom. 2010, 45, 703–714. [Google Scholar] [CrossRef] [PubMed]
- Matsuda, F.; Yonekura-Sakakibara, K.; Niida, R.; Kuromori, T.; Shinozaki, K.; Saito, K. MS/MS spectral tag-based annotation of non-targeted profile of plant secondary metabolites. Plant J. 2009, 57, 555–577. [Google Scholar] [CrossRef] [PubMed]
- Röst, H.L.; Sachsenberg, T.; Aiche, S.; Bielow, C.; Weisser, H.; Aicheler, F.; Andreotti, S.; Ehrlich, H.-C.; Gutenbrunner, P.; Kenar, E. OpenMS: A flexible open-source software platform for mass spectrometry data analysis. Nat. Methods 2016, 13, 741–748. [Google Scholar] [CrossRef] [PubMed]
- Bijlsma, S.; Bobeldijk, I.; Verheij, E.R.; Ramaker, R.; Kochhar, S.; Macdonald, I.A.; Van Ommen, B.; Smilde, A.K. Large-scale human metabolomics studies: A strategy for data (pre-) processing and validation. Anal. Chem. 2006, 78, 567–574. [Google Scholar] [CrossRef]
- Chong, J.; Xia, J. MetaboAnalystR: An R package for flexible and reproducible analysis of metabolomics data. Bioinformatics 2018, 34, 4313–4314. [Google Scholar] [CrossRef]
- Triba, M.N.; Le Moyec, L.; Amathieu, R.; Goossens, C.; Bouchemal, N.; Nahon, P.; Rutledge, D.N.; Savarin, P. PLS/OPLS models in metabolomics: The impact of permutation of dataset rows on the K-fold cross-validation quality parameters. Mol. Biosyst. 2015, 11, 13–19. [Google Scholar] [CrossRef]
- Creydt, M.; Arndt, M.; Hudzik, D.; Fischer, M. Plant metabolomics: Evaluation of different extraction parameters for nontargeted UPLC-ESI-QTOF-Mass spectrometry at the example of white Asparagus officinalis. J. Agric. Food Chem. 2018, 66, 12876–12887. [Google Scholar] [CrossRef]
- Hsu, P.C.; Lan, R.S.; Brasky, T.M.; Marian, C.; Cheema, A.K.; Ressom, H.W.; Loffredo, C.A.; Pickworth, W.B.; Shields, P.G. Metabolomic profiles of current cigarette smokers. Mol. Carcinog. 2017, 56, 594–606. [Google Scholar] [CrossRef]
- Rabara, R.C.; Tripathi, P.; Rushton, P.J. Comparative metabolome profile between tobacco and soybean grown under water-stressed conditions. Biomed Res. Int. 2017, 2017, 3065251. [Google Scholar] [CrossRef]
- Zhao, J.; Li, L.; Zhao, Y.; Zhao, C.; Chen, X.; Liu, P.; Zhou, H.; Zhang, J.; Hu, C.; Chen, A. Metabolic changes in primary, secondary, and lipid metabolism in tobacco leaf in response to topping. Anal. Bioanal. Chem. 2018, 410, 839–851. [Google Scholar] [CrossRef]
- Ishida, N. A Simultaneous analytical method to profile non-volatile components with low polarity elucidating differences between tobacco leaves using atmospheric pressure chemical ionization mass spectrometry detection. Beiträge Zur Tab. Int./Contrib. Tob. Res. 2016, 27, 60–74. [Google Scholar] [CrossRef][Green Version]
- Madala, N.E.; Piater, L.A.; Steenkamp, P.A.; Dubery, I.A. Multivariate statistical models of metabolomic data reveals different metabolite distribution patterns in isonitrosoacetophenone-elicited Nicotiana tabacum and Sorghum bicolor cells. SpringerPlus 2014, 3, 254. [Google Scholar] [CrossRef] [PubMed]
- Leffingwell, J.C. Chemical constituents of tobacco leaf and differences among tobacco types. Chem. Prepr. Arch. 2001, 2001, 173–232. [Google Scholar]
- Rizzato, G.; Scalabrin, E.; Radaelli, M.; Capodaglio, G.; Piccolo, O. A new exploration of licorice metabolome. Food Chem. 2017, 221, 959–968. [Google Scholar] [CrossRef]
- Tugizimana, F.; Steenkamp, P.A.; Piater, L.A.; Dubery, I.A. Multi-platform metabolomic analyses of ergosterol-induced dynamic changes in Nicotiana tabacum cells. PLoS ONE 2014, 9, e87846. [Google Scholar] [CrossRef]
- Cho, K.; Kim, Y.; Wi, S.J.; Seo, J.B.; Kwon, J.; Chung, J.H.; Park, K.Y.; Nam, M.H. Nontargeted metabolite profiling in compatible pathogen-inoculated tobacco (Nicotiana tabacum L. cv. Wisconsin 38) using UPLC-Q-TOF/MS. J. Agric. Food Chem. 2012, 60, 11015–11028. [Google Scholar] [CrossRef]
- Beato, V.M.; Navarro-Gochicoa, M.T.; Rexach, J.; Herrera-Rodríguez, M.B.; Camacho-Cristóbal, J.J.; Kempa, S.; Weckwerth, W.; González-Fontes, A. Expression of root glutamate dehydrogenase genes in tobacco plants subjected to boron deprivation. Plant Physiol. Biochem. 2011, 49, 1350–1354. [Google Scholar] [CrossRef]
- Xu, J.; Chen, Z.; Wang, F.; Jia, W.; Xu, Z. combined transcriptomic and metabolomic analyses uncover rearranged gene expression and metabolite metabolism in tobacco during cold acclimation. Sci. Rep. 2020, 10, 1–13. [Google Scholar] [CrossRef]
- Scalabrin, E.; Radaelli, M.; Rizzato, G.; Bogani, P.; Buiatti, M.; Gambaro, A.; Capodaglio, G. Metabolomic analysis of wild and transgenic Nicotiana langsdorffii plants exposed to abiotic stresses: Unraveling metabolic responses. Anal. Bioanal. Chem. 2015, 407, 6357–6368. [Google Scholar] [CrossRef]
- Arndt, D.; Wachsmuth, C.; Buchholz, C.; Bentley, M. A complex matrix characterization approach, applied to cigarette smoke, that integrates multiple analytical methods and compound identification strategies for non-targeted liquid chromatography with high-resolution mass spectrometry. Rapid Commun. Mass Spectrom. 2020, 34, e8571. [Google Scholar] [CrossRef]
- Moco, S.; Bino, R.J.; Vorst, O.; Verhoeven, H.A.; de Groot, J.; van Beek, T.A.; Vervoort, J.; De Vos, C.R. A liquid chromatography-mass spectrometry-based metabolome database for tomato. Plant Physiol. 2006, 141, 1205–1218. [Google Scholar] [CrossRef] [PubMed]
- Torras-Claveria, L.; Jáuregui, O.; Codina, C.; Tiburcio, A.F.; Bastida, J.; Viladomat, F. Analysis of phenolic compounds by high-performance liquid chromatography coupled to electrospray ionization tandem mass spectrometry in senescent and water-stressed tobacco. Plant Sci. 2012, 182, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Claassen, C.; Kuballa, J.r.; Rohn, S. Metabolomics-Based approach for the discrimination of potato varieties (Solanum tuberosum) using UPLC-IMS-QToF. J. Agric. Food Chem. 2019, 67, 5700–5709. [Google Scholar] [CrossRef] [PubMed]
- Tsaballa, A.; Sarrou, E.; Xanthopoulou, A.; Tsaliki, E.; Kissoudis, C.; Karagiannis, E.; Michailidis, M.; Martens, S.; Sperdouli, E.; Hilioti, Z. Comprehensive approaches reveal key transcripts and metabolites highlighting metabolic diversity among three oriental tobacco varieties. Ind. Crop. Prod. 2020, 143, 111933. [Google Scholar] [CrossRef]
- Popova, V.; Ivanova, T.; Stoyanova, A.; Nikolova, V.; Hristeva, T.; Gochev, V.; Yonchev, Y.; Nikolov, N.; Zheljazkov, V.D. Terpenoids in the essential oil and concentrated aromatic products obtained from Nicotiana glutinosa L. Leaves. Molecules 2020, 25, 30. [Google Scholar] [CrossRef]
- Jassbi, A.R.; Zare, S.; Asadollahi, M.; Schuman, M.C. Ecological roles and biological activities of specialized metabolites from the genus Nicotiana. Chem. Rev. 2017, 117, 12227–12280. [Google Scholar] [CrossRef]
- Tsugawa, H.; Kind, T.; Nakabayashi, R.; Yukihira, D.; Tanaka, W.; Cajka, T.; Saito, K.; Fiehn, O.; Arita, M. Hydrogen rearrangement rules: Computational MS/MS fragmentation and structure elucidation using MS-FINDER software. Anal. Chem. 2016, 88, 7946–7958. [Google Scholar] [CrossRef]
- Chang, Y.; Zhao, C.; Zhu, Z.; Wu, Z.; Zhou, J.; Zhao, Y.; Lu, X.; Xu, G. Metabolic profiling based on LC/MS to evaluate unintended effects of transgenic rice with cry1Ac and sck genes. Plant Mol. Biol. 2012, 78, 477–487. [Google Scholar] [CrossRef]
- Vinatoru, M. An overview of the ultrasonically assisted extraction of bioactive principles from herbs. Ultrason. Sonochem. 2001, 8, 303–313. [Google Scholar] [CrossRef]
- Lisuma, J.; Mbega, E.; Ndakidemi, P. Influence of Tobacco Plant on Macronutrient Levels in Sandy Soils. Agronomy 2020, 10, 418. [Google Scholar] [CrossRef]
- Morita, M.; Shitan, N.; Sawada, K.; Van Montagu, M.C.; Inzé, D.; Rischer, H.; Goossens, A.; Oksman-Caldentey, K.-M.; Moriyama, Y.; Yazaki, K. Vacuolar transport of nicotine is mediated by a multidrug and toxic compound extrusion (MATE) transporter in Nicotiana tabacum. Proc. Natl. Acad. Sci. USA 2009, 106, 2447–2452. [Google Scholar] [CrossRef] [PubMed]
- Abo-Zaid, G.A.; Matar, S.M.; Abdelkhalek, A. Induction of Plant Resistance against Tobacco Mosaic Virus Using the Biocontrol Agent Streptomyces cellulosae Isolate Actino 48. Agronomy 2020, 10, 1620. [Google Scholar] [CrossRef]
- Kachlicki, P.; Piasecka, A.; Stobiecki, M.; Marczak, Ł. Structural characterization of flavonoid glycoconjugates and their derivatives with mass spectrometric techniques. Molecules 2016, 21, 1494. [Google Scholar] [CrossRef] [PubMed]

| Maturity Grade | Producing Area | Variety | Growth Period | HARVEST PERIOD | Number of Samples |
|---|---|---|---|---|---|
| Unripe sample | Yunnan, China | K326 | April to August, 2020 | Transplanted for 70 days | 15 |
| Ripe sample | Yunnan, China | K326 | April to August, 2020 | Transplanted for 80 days | 15 |
| Overripe sample | Yunnan, China | K326 | April to August, 2020 | Transplanted for 90 days | 15 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Y.; Tian, M.; Zhao, G.; Lu, H.; Zhang, Z.; Zou, C. Chromatographic Profiling with Machine Learning Discriminates the Maturity Grades of Nicotiana tabacum L. Leaves. Separations 2021, 8, 9. https://doi.org/10.3390/separations8010009
Chen Y, Tian M, Zhao G, Lu H, Zhang Z, Zou C. Chromatographic Profiling with Machine Learning Discriminates the Maturity Grades of Nicotiana tabacum L. Leaves. Separations. 2021; 8(1):9. https://doi.org/10.3390/separations8010009
Chicago/Turabian StyleChen, Yi, Miao Tian, Gaokun Zhao, Hongmei Lu, Zhimin Zhang, and Congming Zou. 2021. "Chromatographic Profiling with Machine Learning Discriminates the Maturity Grades of Nicotiana tabacum L. Leaves" Separations 8, no. 1: 9. https://doi.org/10.3390/separations8010009
APA StyleChen, Y., Tian, M., Zhao, G., Lu, H., Zhang, Z., & Zou, C. (2021). Chromatographic Profiling with Machine Learning Discriminates the Maturity Grades of Nicotiana tabacum L. Leaves. Separations, 8(1), 9. https://doi.org/10.3390/separations8010009





