Comparison of Pre-Processing and Variable Selection Strategies in Group-Based GC×GC-TOFMS Analysis
Abstract
1. Introduction
2. Materials and Methods
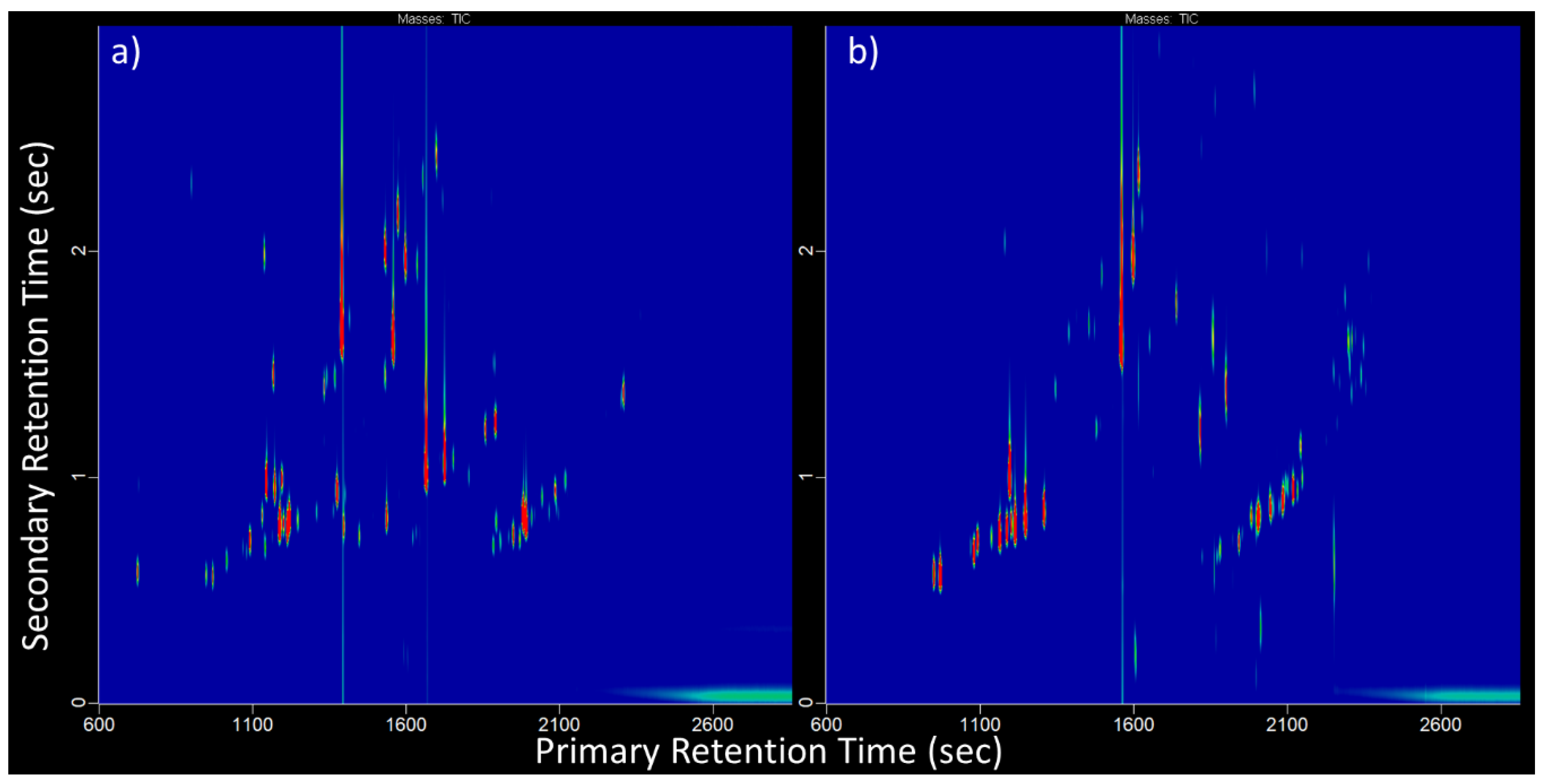
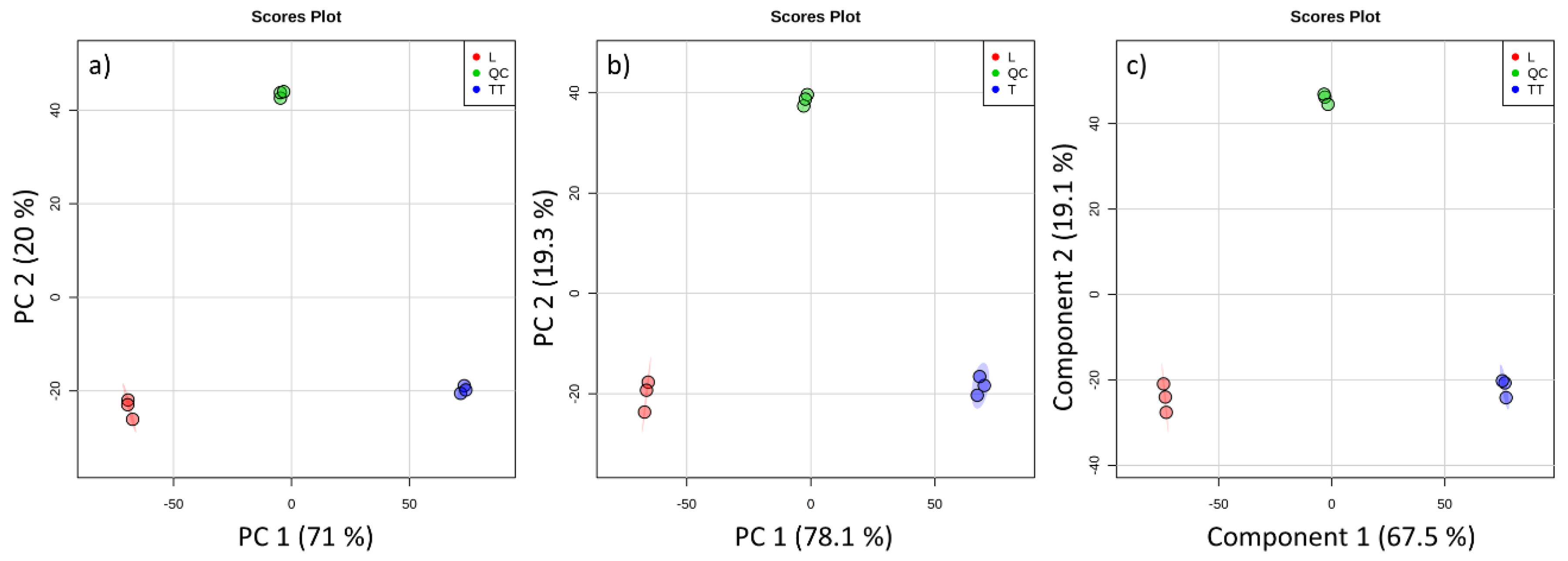
3. Results and Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gruber, B.; Weggler, B.A.; Jaramillo, R.; Murrell, K.A.; Piotrowski, P.K.; Dorman, L.F. Comprehensive two-dimensional gas chromatography in forensic science: A critical review of recent trends. TrAC Trends Anal. Chem. 2018, 105, 292–301. [Google Scholar] [CrossRef]
- Megson, D.; Reiner, E.J.; Jobst, K.J.; Dorman, F.L.; Robson, M.; Focant, J.F. A review of the determination of persistent organic pollutants for environmental forensics investigations. Anal. Chim. Acta 2016, 941, 10–25. [Google Scholar] [CrossRef] [PubMed]
- Keppler, E.A.H.; Jenkins, C.L.; Davis, T.J.; Bean, H.D. Advances in the application of comprehensive two-dimensional gas chromatography in metabolomics. TrAC Trends Anal. Chem. 2018, 109, 275–286. [Google Scholar] [CrossRef] [PubMed]
- Reichenbach, S.E.; Tian, X.; Tao, Q.; Ledford, E.B., Jr.; Wu, Z.; Fiehn, O. Informatics for cross-sample analysis with comprehensive two-dimensional gas chromatography and high-resolution mass spectrometry (GC×GC–HRMS). Talanta 2011, 83, 1279–1288. [Google Scholar] [CrossRef] [PubMed]
- De Carvalho Rocha, W.F.; Schantz, M.M.; Sheen, D.A.; Chu, P.M.; Lippa, K.A. Unsupervised classification of petroleum Certified Reference Materials and other fuels by chemometric analysis of gas chromatography-mass spectrometry data. Fuel 2017, 197, 248–258. [Google Scholar] [CrossRef] [PubMed]
- Jennerwein, M.K.; Eschner, M.; Gröger, T.; Wilharm, T.; Zimmermann, R. Complete group-type quantification of petroleum middle distillates based on comprehensive two-dimensional gas chromatography time-of-flight mass spectrometry (GC×GC-TOFMS) and visual basic scripting. Energy Fuels 2014, 28, 5670–5681. [Google Scholar] [CrossRef]
- Weggler, B.A.; Gröger, T.; Zimmermann, R. Advanced scripting for the automated profiling of two-dimensional gas chromatography-time-of-flight mass spectrometry data from combustion aerosol. J. Chromatogr. A 2014, 1364, 241–248. [Google Scholar] [CrossRef]
- Pena-Abaurrea, M.; Jobst, K.J.; Ruffolo, R.; Shen, L.; McCrindle, R.; Helm, P.A.; Reiner, E.J. Identification of potential novel bioaccumulative and persistent chemicals in sediments from Ontario (Canada) using scripting approaches with GC×GC-TOF MS analysis. Environ. Sci. Technol. 2014, 48, 9591–9599. [Google Scholar] [CrossRef] [PubMed]
- Hilton, D.C.; Jones, R.S.; Sjödin, A. A method for rapid, non-targeted screening for environmental contaminants in household dust. J. Chromatogr. A 2010, 1217, 6851–6856. [Google Scholar] [CrossRef]
- Piotrowski, P.K.; Weggler, B.A.; Barth-Naftilan, E.; Kelly, C.N.; Zimmermann, R.; Saiers, J.E.; Dorman, F.L. Non-Targeted chemical characterization of a Marcellus shale gas well through GC×GC with scripting algorithms and high-resolution time-of-flight mass spectrometry. Fuel 2018, 215, 363–369. [Google Scholar] [CrossRef]
- Piotrowski, P.K.; Weggler, B.A.; Yoxtheimer, D.A.; Kelly, C.N.; Barth-Naftilan, E.; Saiers, J.E.; Dorman, F.L. Elucidating Environmental Fingerprinting Mechanisms of Unconventional Gas Development through Hydrocarbon Analysis. Anal. Chem. 2018, 90, 5466–5473. [Google Scholar] [CrossRef] [PubMed]
- Bowman, D.T.; Warren, L.A.; McCarry, B.E.; Slater, G.F. Profiling of individual naphthenic acids at a composite tailings reclamation fen by comprehensive two-dimensional gas chromatography-mass spectrometry. Sci. Total Environ. 2019, 649, 1522–1531. [Google Scholar] [CrossRef] [PubMed]
- Perrault, K.; Stefanuto, P.H.; Dubois, L.; Cnuts, D.; Rots, V.; Focant, J.F. A new approach for the characterization of organic residues from stone tools using GC×GC-TOFMS. Separations 2016, 3, 16. [Google Scholar] [CrossRef]
- Stefanuto, P.H.; Perrault, K.A.; Stadler, S.; Pesesse, R.; LeBlanc, H.N.; Forbes, S.L.; Focant, J.F. GC×GC–TOFMS and supervised multivariate approaches to study human cadaveric decomposition olfactive signatures. Anal. Bioanal. Chem. 2015, 407, 4767–4778. [Google Scholar] [CrossRef] [PubMed]
- Weggler, B.A.; Ly-Verdu, S.; Jennerwein, M.; Sippula, O.; Reda, A.A.; Orasche, J.; Gröger, T.; Jokiniemi, J.; Zimmermann, R. Untargeted identification of wood type-specific markers in particulate matter from wood combustion. Environ. Sci. Technol. 2016, 50, 10073–10081. [Google Scholar] [CrossRef]
- Pesesse, R.; Stefanuto, P.H.; Schleich, F.; Louis, R.; Focant, J.F. Multimodal chemometric approach for the analysis of human exhaled breath in lung cancer patients by TD-GC×GC-TOFMS. J. Chromatogr. B. 2019, 1114, 146–153. [Google Scholar] [CrossRef]
- Berrueta, L.A.; Alonso-Salces, R.M.; Héberger, K. Supervised pattern recognition in food analysis. J. Chromatogr. A 2007, 1158, 196–214. [Google Scholar] [CrossRef]
- Escandar, G.M.; Olivieri, A.C. Multi-way chromatographic calibration—A review. J. Chromatogr. A 2019, 1587, 2–13. [Google Scholar] [CrossRef]
- Pierce, K.M.; Kehimkar, B.; Marney, L.C.; Hoggard, J.C.; Synovec, R.E. Review of chemometric analysis techniques for comprehensive two dimensional separations data. J. Chromatogr. A 2012, 1255, 3–11. [Google Scholar] [CrossRef]
- Gomez-Caravaca, A.M.; Maggio, R.M.; Cerretani, L. Chemometric applications to assess quality and critical parameters of virgin and extra-virgin olive oil. A review. Anal. Chim. Acta 2016, 913, 1–21. [Google Scholar] [CrossRef]
- Marriott, P.J.; Haglund, P.; Ong, R.C. A review of environmental toxicant analysis by using multidimensional gas chromatography and comprehensive GC. Clin. Chim. Acta 2003, 328, 1–19. [Google Scholar] [CrossRef]
- Martín-Alberca, C.; Ortega-Ojeda, F.E.; García-Ruiz, C. Analytical tools for the analysis of fire debris. A review: 2008–2015. Anal. Chim. Acta 2016, 928, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Patterson, R.E.; Kirpich, A.S.; Koelmel, J.P.; Kalavalapalli, S.; Morse, A.M.; Cusi, K.; Sunny, N.E.; McIntyre, L.M.; Garrett, T.J.; Yost, R.A. Improved experimental data processing for UHPLC–HRMS/MS lipidomics applied to nonalcoholic fatty liver disease. Metabolomics 2017, 13, 142. [Google Scholar] [CrossRef]
- Da Silva, M.D.R.G.; Cardeal, Z.; Marriott, P.J. Comprehensive two-dimensional gas chromatography: Application to aroma and essential oil analysis. ACS Symp. Ser. 2008, 988, 3–24. [Google Scholar]
- Shellie, R.A. Volatile components of plants, essential oils, and fragrances. Compr. Anal. Chem. 2009, 55, 189–213. [Google Scholar]
- Tranchida, P.Q.; Shellie, R.A.; Purcaro, G.; Conte, L.S.; Dugo, P.; Dugo, G.; Mondello, L. Analysis of fresh and aged tea tree essential oils by using GC×GC-QMS. J. Chromatogr. Sci. 2010, 48, 262–266. [Google Scholar] [CrossRef]
- Smelcerovic, A.; Djordjevic, A.; Lazarevic, J.; Stojanovic, G. Recent advances in analysis of essential oils. Curr. Anal. Chem. 2013, 9, 61–70. [Google Scholar] [CrossRef]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Piotrowski, P.; Place, B. Comparison of Pre-Processing and Variable Selection Strategies in Group-Based GC×GC-TOFMS Analysis. Separations 2019, 6, 41. https://doi.org/10.3390/separations6030041
Piotrowski P, Place B. Comparison of Pre-Processing and Variable Selection Strategies in Group-Based GC×GC-TOFMS Analysis. Separations. 2019; 6(3):41. https://doi.org/10.3390/separations6030041
Chicago/Turabian StylePiotrowski, Paulina, and Benjamin Place. 2019. "Comparison of Pre-Processing and Variable Selection Strategies in Group-Based GC×GC-TOFMS Analysis" Separations 6, no. 3: 41. https://doi.org/10.3390/separations6030041
APA StylePiotrowski, P., & Place, B. (2019). Comparison of Pre-Processing and Variable Selection Strategies in Group-Based GC×GC-TOFMS Analysis. Separations, 6(3), 41. https://doi.org/10.3390/separations6030041




