Mass Spectrometric Determination of the Effect of Surface Deactivation on Membranes Used for In-Situ Sampling of Cerebrospinal Fluid (CSF)
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Cerebrospinal Fluid Collection
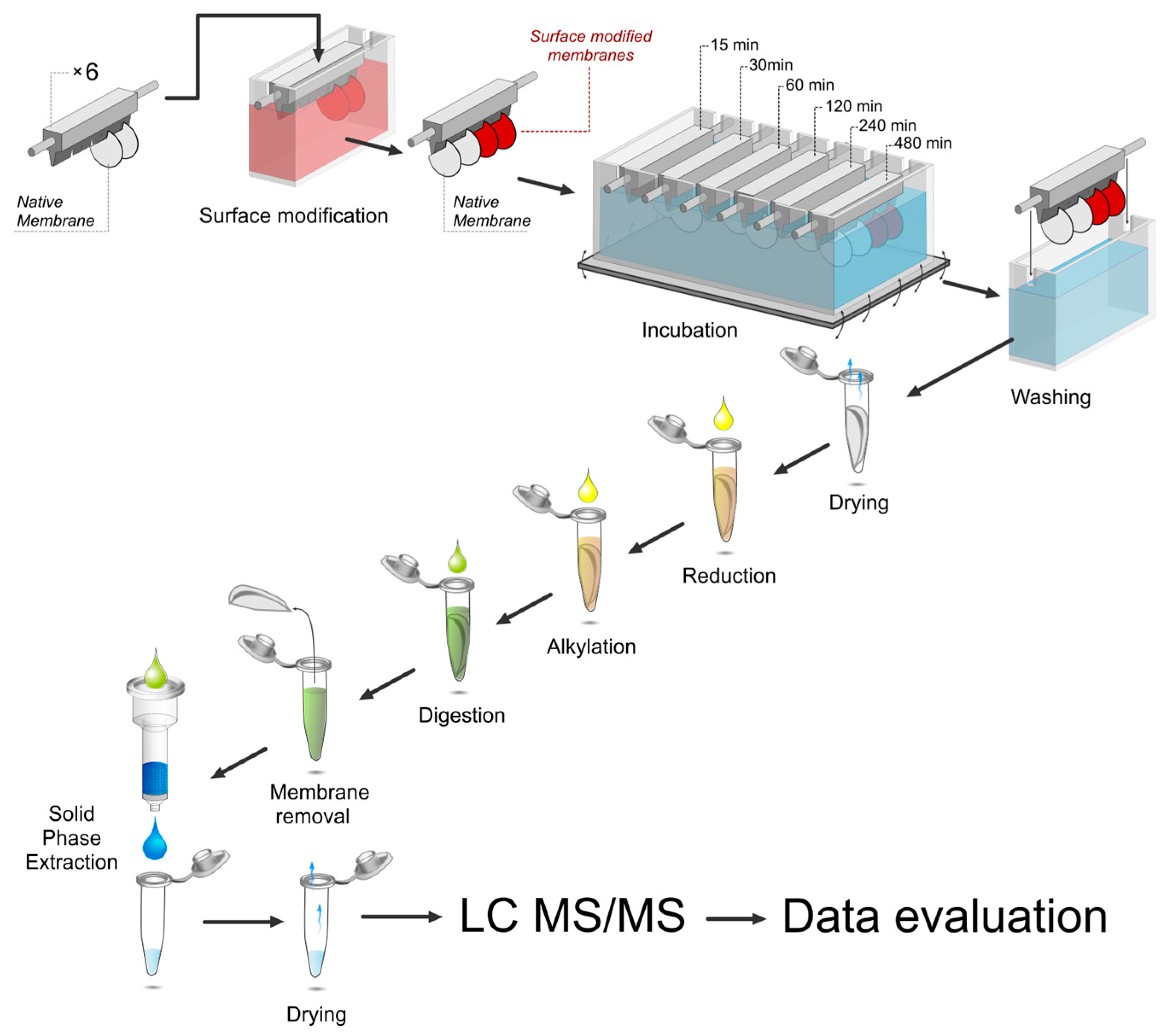
2.3. Surface Modification Procedure
2.4. Protein Adsorption in vCSF
2.5. On-Surface Enzymatic Digestion (oSED)
2.6. Nano-LC-MS/MS
2.7. Data Evaluation
3. Results
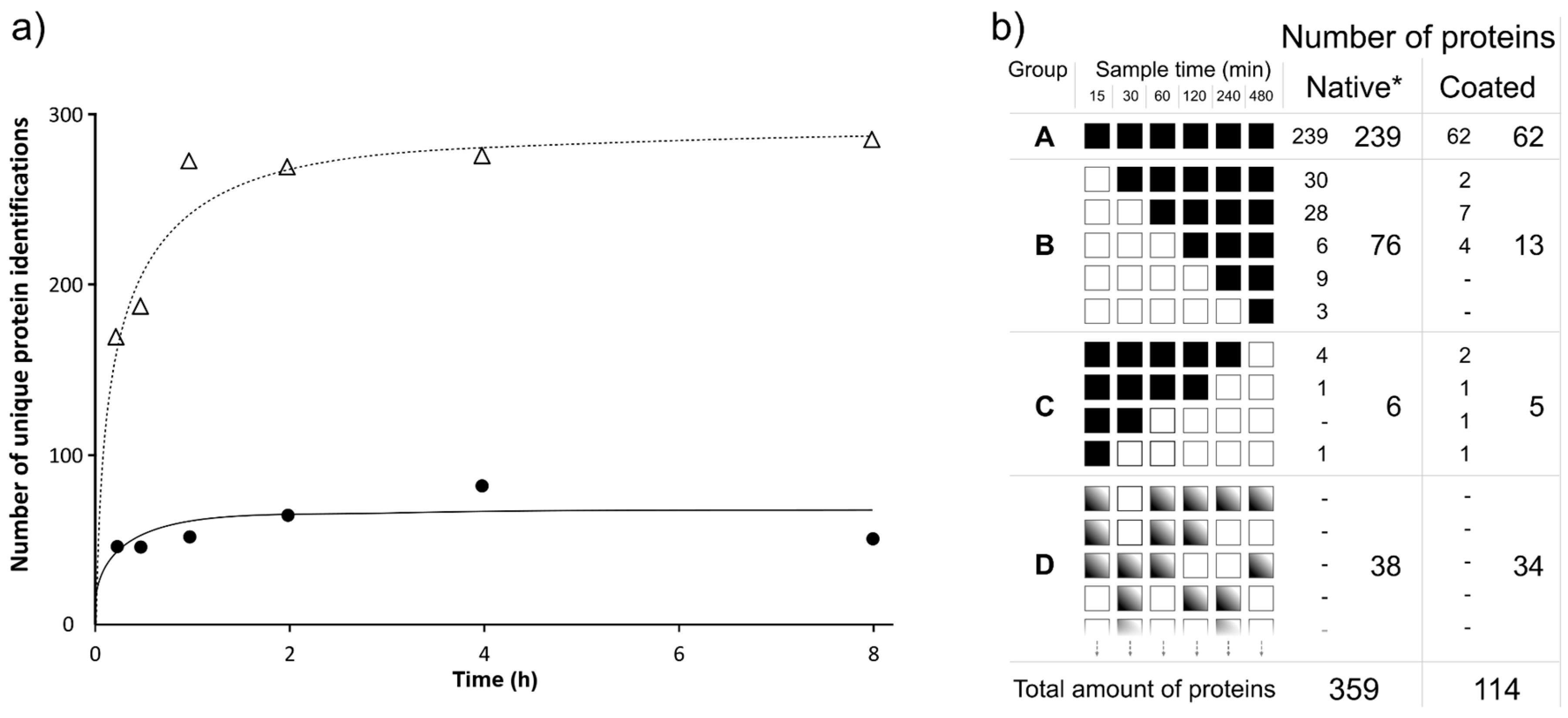
3.1. Protein Adsorption Over Time
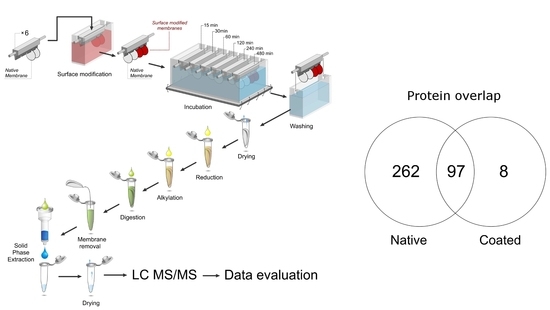
3.2. Competitive Protein Adsorption Analysis
3.3. Individual Protein Adsorption Behavior
3.4. Proteins Uniquely Adsorbed to Coated Membranes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Anderson, J.M.; Rodriguez, A.; Chang, D.T. Foreign body reaction to biomaterials. Semin. Immunol. 2008, 20, 86–100. [Google Scholar] [CrossRef] [PubMed]
- Rabe, M.; Verdes, D.; Seeger, S. Understanding protein adsorption phenomena at solid surfaces. Adv. Colloid Interface Sci. 2011, 162, 87–106. [Google Scholar] [CrossRef] [PubMed]
- Sides, C.R.; Stenken, J.A. Microdialysis sampling techniques applied to studies of the foreign body reaction. Eur. J. Pharm. Sci. 2014, 57, 74–86. [Google Scholar] [CrossRef] [PubMed]
- Wisniewski, N.; Klitzman, B.; Miller, B.; Reichert, W.M. Decreased analyte transport through implanted membranes: differentiation of biofouling from tissue effects. J. Biomed. Mater. Res. 2001, 57, 513–521. [Google Scholar] [CrossRef]
- Wei, Q.; Becherer, T.; Angioletti-Uberti, S.; Dzubiella, J.; Wischke, C.; Neffe, A.T.; Lendlein, A.; Ballauff, M.; Haag, R. Protein interactions with polymer coatings and biomaterials. Angew. Chem. Int. Ed. Engl. 2014, 53, 8004–8031. [Google Scholar] [CrossRef] [PubMed]
- Dahlin, A.P.; Hjort, K.; Hillered, L.; Sjodin, M.O.; Bergquist, J.; Wetterhall, M. Multiplexed quantification of proteins adsorbed to surface-modified and non-modified microdialysis membranes. Anal. Bioanal. Chem. 2012, 402, 2057–2067. [Google Scholar] [CrossRef] [PubMed]
- Undin, T.; Bergstrom, L.; Dahlin, A.P. A mass spectrometry based method for investigating time dependent protein adsorption on surfaces in contact with complex biological samples. Future Sci. OA 2015, 1, FSO32. [Google Scholar] [CrossRef] [PubMed]
- Undin, T.; Dahlin, A.; Hörnaeus, K.; Bergquist, J.; Bergstrom Lind, S. Mechanistic investigation of the on-surface enzymatic digestion (oSED) protein adsorption detection method using targeted mass spectrometry. Analyst 2016, 141, 1714–1720. [Google Scholar] [CrossRef] [PubMed]
- Lundgren, D.H.; Hwang, S.I.; Wu, L.; Han, D.K. Role of spectral counting in quantitative proteomics. Expert Rev. Proteomics 2010, 7, 39–53. [Google Scholar] [CrossRef] [PubMed]
- Higuchi, A.; Sugiyama, K.; Yoon, B.O.; Sakurai, M.; Hara, M.; Sumita, M.; Sugawara, S.; Shirai, T. Serum protein adsorption and platelet adhesion on pluronic (TM)-adsorbed polysulfone membranes. Biomaterials 2003, 24, 3235–3245. [Google Scholar] [CrossRef]


© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Undin, T.; P. Dahlin, A.; Bergquist, J.; Bergström Lind, S. Mass Spectrometric Determination of the Effect of Surface Deactivation on Membranes Used for In-Situ Sampling of Cerebrospinal Fluid (CSF). Separations 2018, 5, 27. https://doi.org/10.3390/separations5020027
Undin T, P. Dahlin A, Bergquist J, Bergström Lind S. Mass Spectrometric Determination of the Effect of Surface Deactivation on Membranes Used for In-Situ Sampling of Cerebrospinal Fluid (CSF). Separations. 2018; 5(2):27. https://doi.org/10.3390/separations5020027
Chicago/Turabian StyleUndin, Torgny, Andreas P. Dahlin, Jonas Bergquist, and Sara Bergström Lind. 2018. "Mass Spectrometric Determination of the Effect of Surface Deactivation on Membranes Used for In-Situ Sampling of Cerebrospinal Fluid (CSF)" Separations 5, no. 2: 27. https://doi.org/10.3390/separations5020027
APA StyleUndin, T., P. Dahlin, A., Bergquist, J., & Bergström Lind, S. (2018). Mass Spectrometric Determination of the Effect of Surface Deactivation on Membranes Used for In-Situ Sampling of Cerebrospinal Fluid (CSF). Separations, 5(2), 27. https://doi.org/10.3390/separations5020027