Multivariate Analysis of UPLC-MS/MS Metabolomic Profiles in Four Hiraea Species (Malpighiaceae)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant
2.2. Preparation of Extracts
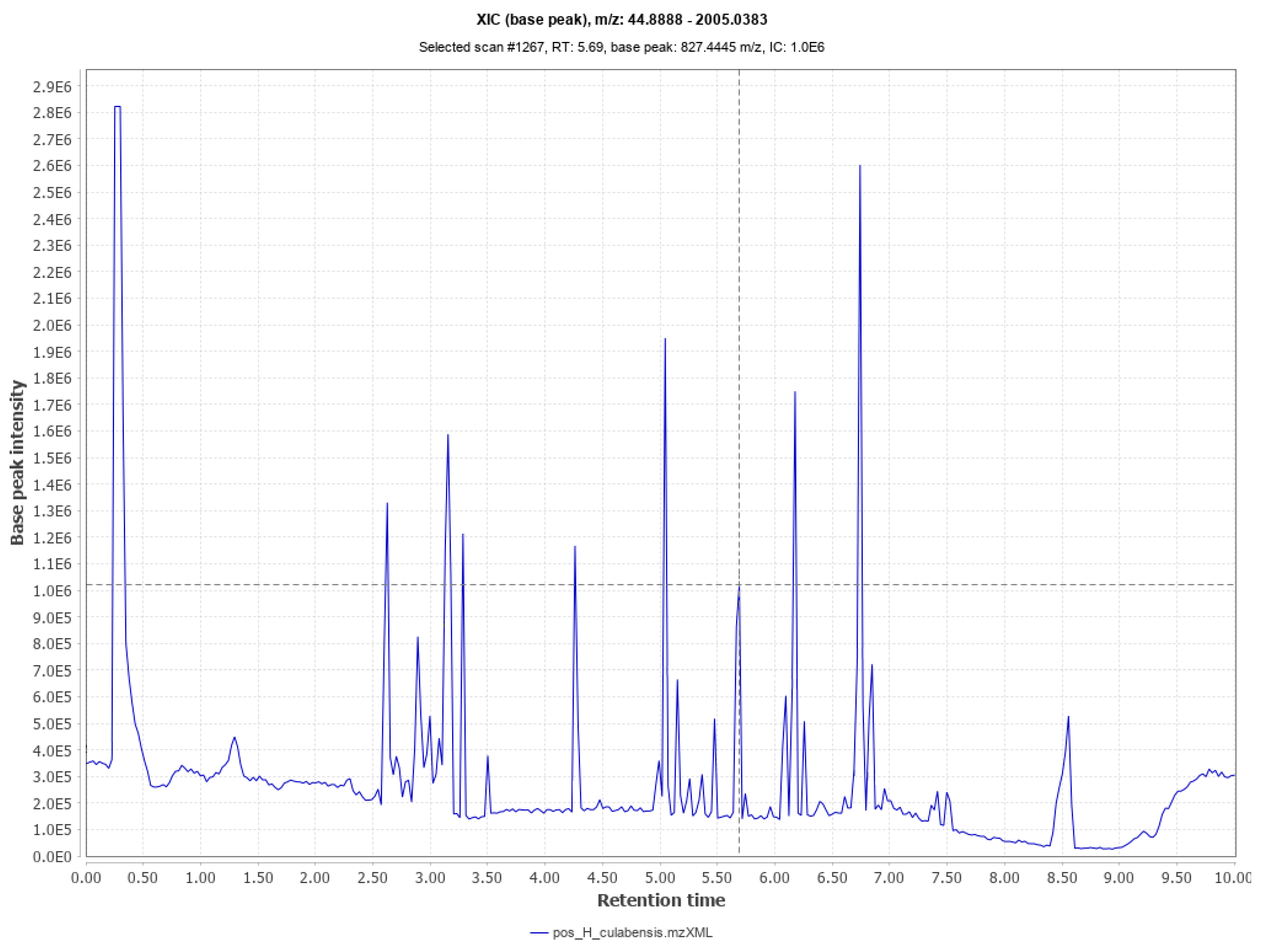
2.3. UPLC-MS/MS Analyses
2.4. VIP Score (PLS-DA)
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| H. | Hiraea |
| PLS-DA | Discriminant Analysis-Partial Least Squares |
| UPLC-MS/MS | Ultra-performance Liquid Chromatography coupled to Mass Spectrometry |
| VIP | Variable Importance in Projection |
References
- Powo Malpighiaceae Juss. Plants of the World Online. 2025. Available online: https://powo.science.kew.org/taxon/urn:lsid:ipni.org:names:30000017-2 (accessed on 15 March 2025).
- Anderson, C.E. Resolution of the Hiraea cephalotes Complex (Malpighiaceae). Edinb. J. Bot. 2013, 70, 413–432. [Google Scholar] [CrossRef]
- Menard, K.L.; Schwartz, M.D. Four New Species of Phytocoris Fallén (Hemiptera, Miridae) from the Davis Mountains in Texas and Further Documentation of Known Species of Jeff Davis County. Zookeys 2023, 2023, 97–139. [Google Scholar] [CrossRef] [PubMed]
- Guimarães da Silva, J.M.; de Almeida, R.F.; Zeraik, M.L. Comparative Metabolite Profiling of Three Savannic Species of Banisteriopsis (Malpighiaceae) via UPLC-MS/MS and Chemometric Tools. Chem. Biodivers. 2024, 21, e202400679. [Google Scholar] [CrossRef]
- Mannochio-Russo, H.; de Almeida, R.F.; Nunes, W.D.G.; Bueno, P.C.P.; Caraballo-Rodríguez, A.M.; Bauermeister, A.; Dorrestein, P.C.; Bolzani, V.S. Untargeted Metabolomics Sheds Light on the Diversity of Major Classes of Secondary Metabolites in the Malpighiaceae Botanical Family. Front. Plant Sci. 2022, 13, 854842. [Google Scholar] [CrossRef]
- Abbas, H.A.; Tadros, S.H.; El-Toumy, S.A.; Salama, A.M.; El Gedaily, R.A. A Review on Traditional Uses, Phytochemistry and Pharmacological Potential of Family Malpighiaceae. Egypt. J. Chem. 2022, 65, 235–274. [Google Scholar] [CrossRef]
- Barônio, G.J.; Haleem, M.A.; Marsaioli, A.J.; Torezan-Silingardi, H.M. Characterization of Malpighiaceae Flower-Visitor Interactions in a Brazilian Savannah: How Do Floral Resources and Visitor Abundance Change over Time. Flora 2017, 234, 126–134. [Google Scholar] [CrossRef]
- Mannochio-Russo, H.; Bueno, P.C.P.; Bauermeister, A.; De Almeida, R.F.; Dorrestein, P.C.; Cavalheiro, A.J.; Bolzani, V.S. Can Statistical Evaluation Tools for Chromatographic Method Development Assist in the Natural Products Workflow? A Case Study on Selected Species of the Plant Family Malpighiaceae. J. Nat. Prod. 2020, 83, 3239–3249. [Google Scholar] [CrossRef]
- Mannochio Russo, H.; Ferreira Queiroz, E.; Marcourt, L.; Rutz, A.; Allard, P.M.; de Almeida, R.F.; Marques Carvalho, N.; Wolfender, J.L.; da Silva Bolzani, V. Phytochemical Analysis of the Methanolic Leaves Extract of Niedenzuella multiglandulosa (Malpighiaceae), a Plant Species Toxic to Cattle in Brazil. Phytochem. Lett. 2020, 37, 10–16. [Google Scholar] [CrossRef]
- Alexandre, G.P.; Simão, J.L.S.; Tavares, M.O.A.; Zuffo, I.M.S.; Prado, S.V.; De Paiva, J.A.; Mustapha, A.N.; De Oliveira, A.E.; Kato, L.; Severino, V.G.P. Dereplication by HPLC-ESI-MS and Antioxidant Activity of Phenolic Compounds from Banisteriopsis laevifolia (Malpighiaceae). An. Acad. Bras. Cienc. 2022, 94, e20201844. [Google Scholar] [CrossRef]
- Nieto Camacho, A.; Baca Ibarra, I.I.; Huerta-Reyes, M. Antioxidant and Anti-Inflammatory Profiles of Two Mexican Heteropterys Species and Their Relevance for the Treatment of Mental Diseases: H. brachiata (L.) DC. and H. cotinifolia A. Juss. (Malpighiaceae). Molecules 2024, 29, 3053. [Google Scholar] [CrossRef]
- Allwood, J.W.; Williams, A.; Uthe, H.; van Dam, N.M.; Luis, L.A.J.; Grant, M.R.; Pétriacq, P. Unravelling Plant Responses to Stress—The Importance of Targeted and Untargeted Metabolomics. Metabolites 2021, 11, 558. [Google Scholar] [CrossRef] [PubMed]
- Schymanski, E.L.; Jeon, J.; Gulde, R.; Fenner, K.; Ruff, M.; Singer, H.P.; Hollender, J. Identifying Small Molecules via High Resolution Mass Spectrometry: Communicating Confidence. Environ. Sci. Technol. 2014, 48, 2097–2098. [Google Scholar] [CrossRef]
- Alazem, M.; Lin, N.S. Antiviral Roles of Abscisic Acid in Plants. Front. Plant Sci. 2017, 8, 1760. [Google Scholar] [CrossRef] [PubMed]
- Shomali, A.; Das, S.; Arif, N.; Sarraf, M.; Zahra, N.; Yadav, V.; Aliniaeifard, S.; Chauhan, D.K.; Hasanuzzaman, M. Diverse Physiological Roles of Flavonoids in Plant Environmental Stress Responses and Tolerance. Plants 2022, 11, 3158. [Google Scholar] [CrossRef]
- Yuan, X.; He, C.; Xue, X.; Liang, Z.; Chen, Y. Optimisation Study on the Flavonoid Extraction Process from Abrus precatorius Leaves and the Comparison of Total Flavonoid Content by HPLC and UV. J. Holist. Integr. Pharm. 2023, 4, 119–126. [Google Scholar] [CrossRef]
- Vukovic, N.; Mladenović, M.; Stankovic, M.; Sukdolak, S.; Solujic, S.; Mihailovic, V.; Mladenovic, M.; Stojanovic, J.; Stankovic, M.S. Chemical Composition and Antimicrobial Activity of Teucrium Arduini Essential Oil and Cirsimarin from Montenegro. Artic. J. Med. Plants Res. 2011, 5, 1244–1250. [Google Scholar]
- Chalker-Scott, L. Environmental Significance of Anthocyanins in Plant Stress Responses. Photochem. Photobiol. 1999, 70, 1–9. [Google Scholar] [CrossRef]
- Samanta, A.; Das, S.K. Roles of Flavonoids in Plants. Int. J. Pharm. Sci. Tech. 2011, 6, 1–22. [Google Scholar]
- Hidalgo, M.; Sánchez-Moreno, C.; de Pascual-Teresa, S. Flavonoid-Flavonoid Interaction and Its Effect on Their Antioxidant Activity. Food Chem. 2010, 121, 691–696. [Google Scholar] [CrossRef]
- Islam, M.S.; Yoshimoto, M.; Yamakawa, O. Distribution and Physiological Functions of Caffeoylquinic Acid Derivatives in Leaves of Sweetpotato Genotypes. J. Food Sci. 2003, 68, 111–116. [Google Scholar] [CrossRef]
- Meng, S.; Cao, J.; Feng, Q.; Peng, J.; Hu, Y. Roles of Chlorogenic Acid on Regulating Glucose and Lipids Metabolism: A Review. Evid. Based Complement. Altern. Med. 2013, 2013, 801457. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Shen, Q.; Chen, Y.; Pan, R.; Kuang, S.; Liu, G.; Sun, G.; Sun, X. Myricitrin Alleviates Oxidative Stress-Induced Inflammation and Apoptosis and Protects Mice against Diabetic Cardiomyopathy. Sci. Rep. 2017, 7, 44239. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.T.; Cheng, C.; Shi, J.X.; Zhang, W.T.; Sun, H.; Liu, C.M. Avicularin Attenuated Lead-Induced Ferroptosis, Neuroinflammation, and Memory Impairment in Mice. Antioxidants 2024, 13, 1024. [Google Scholar] [CrossRef] [PubMed]
- Noor, S.; Mohammad, T.; Rub, M.A.; Raza, A.; Azum, N.; Yadav, D.K.; Hassan, M.I.; Asiri, A.M. Biomedical Features and Therapeutic Potential of Rosmarinic Acid. Arch. Pharmacal Res. 2022, 45, 205–228. [Google Scholar] [CrossRef]


| Hiraea Leaves | Collection Herbarium (Voucher Number) | Local | Date (dd/mm/yyyy) |
|---|---|---|---|
| H. cuiabensis | Francener (1218-SP) | Araguainha/MT-Brazil | 29 December 2012 |
| H. hatschbachii | Almeida (HUEFS) | Foz do Iguaçu/PR-Brazil | 8 June 2013 |
| H. reclinata | Pace (518-SPF) | Iquitos/Loreto-Peru | 18 September 2014 |
| H. restingae | Almeida (SP-518) | Soretano/ES-Brazil | 20 January 2012 |
| N° | Ion (m/z) | Exact Mass | Adduct | Error (ppm) | Fragment | Metabolites (Class) | Molecular Formula | Rt ** (min) | Leaves | VIP Score Values |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 265.1452 | 265.1439 | [M + H]+ | 4.90 | 155.1085, 142.1008 | Abscisic acid * (hormone) | C15H20O4 | 2.34 | H. cuiabensis; H. hatschbachii; H. reclinata | 1.2780 |
| 2 | 268.1055 | 268.1045 | [M + H]+ | 3.72 | 136.0627, 119.0358, 108.0425 | Adenosine (nitrogenous) | C10H13N5O4 | 0.38 | All | 0.4441 |
| 3 | 433.2312 | 433.2331 | [M + Na]+ | −4.38 | 158.0090 | 1-Hexadecanoyl-2-glycero-3-phosphate * (glycerol) | C19H39O7P | 5.26 | All | 1.2552 |
| 4 | 435.0916 | 435.0927 | [M + H]+ | −2.52 | 303.0491 | Quercetin-3-O-pentoside (flavonoid) | C20H18O11 | 3.12 | H. cuiabensis; H. hatschbachii | 1.2161 |
| 5 | 463.1221 | 463.1240 | [M]+ | −4.10 | 317.0641 | Peonidin-3-O-glucoside * (anthocyanin) | C22H23O11 | 3.04 | H. hatschbachii; H. reclinata; H. restingae | 1.0280 |
| 6 | 465.1041 | 465.1033 | [M + H]+ | 1.72 | 303.0521 | Isoquercitrin * (flavonoid) | C21H20O12 | 2.82 | All | 0.2580 |
| 7 | 477.1366 | 477.1396 | [M + H]+ | −6.28 | 315.0838 | Cirsimarin * (flavonoid) | C23H24O11 | 3.42 | H. restingae | 1.2780 |
| 8 | 480.1259 | 480.1267 | [M + H]+ | −2.85 | 318.0731, 301.0704 | Petunidin-3- galactoside * (anthocyanin) | C22H23O12 | 3.02 | All | - *** |
| 9 | 480.1268 | 480.1267 | [M + H]+ | 0.35 | 318.0740, 317.0661 | Petunidin-3-O-glucoside * (anthocyanin) | C22H23O12 | 3.31 | H. reclinata | 1.1070 |
| 10 | 491.1156 | 491.1189 | [M + H]+ | −6.71 | 295.0574 | Kaempferol-O-acetylhexoside * (flavonoid) | C23H22O12 | 3.30 | H. cuiabensis | 0.2621 |
| 11 | 493.1312 | 493.1346 | [M]+ | −6.89 | 329.0628 | Malvidin-3-O-galactoside * (anthocyanin) | C23H25O12 | 3.19 | H. hatschbachii | 1.2161 |
| 12 | 551.1017 | 551.1037 | [M + H]+ | −3.62 | 303.0485 | Flavonol (flavonoid) | C24H22O15 | 3.05 | All | 1.0077 |
| 13 | 565.1550 | 565.1557 | [M + H]+ | 1.23 | 325.1134, 287.0558 | Isoschaftoside * (flavonoid) | C26H28O14 | 2.78 | H. reclinata; H. restingae | 0.4825 |
| 14 | 573.0847 | 573.0856 | [M + Na]+ | −1.57 | 308.1107 | Quercetin-3-O-malonylglucoside * (flavonoid) | C24H22O15 | 3.04 | H. hatschbachii | 1.2161 |
| 15 | 595.1654 | 595.1663 | [M + H]+ | −1.51 | 287.0557 | Kaempferol-3-O-rutinoside (flavonoid) * | C27H30O15 | 3.14 | H. reclinata | 1.0281 |
| 16 | 595.1672 | 595.1663 | [M]+ | 1.51 | 287.0564 | Cyanidin-3-rutinoside * (anthocyanin) | C27H31O15 | 3.26 | H. hatschbachii | - *** |
| 17 | 597.1461 | 597.1455 | [M]+ | 1.00 | 301.0354 | Delphinidin-3-O-sambubioside * (anthocyanin) | C26H29O16 | 3.92 | H. cuiabensis | 0.2621 |
| 18 | 609.1822 | 609.1819 | [M + H]+ | 0.49 | 303.0793, 301.0715 | Diosmetin-7-O-rutinoside * (flavonoid) | C28H32O15 | 3.11 | H. cuiabensis | - *** |
| 19 | 611.1606 | 611.1612 | [M + H]+ | 0.98 | 303.0509 | Rutin (flavonoid) | C27H30O16 | 2.93 | All | 1.2944 |
| 20 | 627.1570 | 627.1561 | [M + H]+ | 1.43 | 301.0352 | Myricetin-3-rutinoside * (flavonoid) | C27H30O17 | 2.69 | H. cuiabensis; H. restingae | - *** |
| 21 | 649.2180 | 649.2191 | [M + H–H2O]+ | −1.69 | 149.0450, 148.0371 | Maltotetraose * (carbohydrate) | C24H42O21 | 0.38 | H. cuiabensis | 1.0281 |
| 22 | 675.6782 | 675.6766 | [2M + H]+ | 2.36 | 343.4278, 329.4122 | Docosenamide * (amide) | C22H43NO | 6.82 | H. cuiabensis; H. hatschbachii; H. restingae | - *** |
| N° | Ion (m/z) [M–H]− | Exact Mass | Error (ppm) | Fragment | Metabolites (Class) | Molecular Formula | Rt *** (min) | Leaves | VIP Score Values |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 153.0193 | 153.0187 | 3.92 | 109.0284, 108.0206 | Pyrocatechuic acid (phenolic * compound) | C7H6O4 | 1.24 | All | - **** |
| 2 | 177.0195 | 177.0187 | 4.51 | 176.0109, 106.0054 | Aesculetin * (flavonoid) | C9H6O4 | 2.69 | All | - **** |
| 3 | 201.1138 | 201.1126 | 5.92 | 184.1111, 139.0759, 137.0602 | Decanedioic acid * (acids) | C10H18O4 | 3.43 | H. hatschbachii; H. reclinata | 1.0431 |
| 4 | 204.0309 | 204.0296 | 6.37 | 161.0476, 160.0398, 159.0320 | Xanthurenic acid * (acids) | C10H7NO4 | 1.26 | H. hatschbachii | 1.0431 |
| 5 | 210.0777 | 210.0766 | 5.23 | 148.0524, 124.0524, 118.0054 | Enicoflavine * (flavonoid) | C10H13NO4 | 2.98 | H. hatschbachii | 1.0431 |
| 6 | 269.0463 | 269.0450 | 4.83 | 245.1595 | Apigenin * (flavonoid) | C15H10O5 | 4.19 | H. cuiabensis; H. reclinata; H. restingae | 1.0431 |
| 7 | 282.0852 | 282.0838 | 4.96 | 150.0430 | Guanosine * (flavonoid) | C10H13N5O5 | 0.74 | H. reclinata | 0.9781 |
| 8 | 285.0410 | 285.0399 | 3.85 | 284.0332 | Luteolin (flavonoid) | C15H10O6 | 3.49 | H. reclinata | 0.6566 |
| 9 | 289.0726 | 289.0712 | 4.84 | 125.0226 | Catechin * (flavonoid) | C15H14O6 | 3.92 | All | 1.0431 |
| 10 | 325.0937 | 325.0923 | 4.30 | 146.0316 | Coumaroyl hexoside * (phenolic glycoside) | C15H18O8 | 2.59 | H. reclinata; H. restingae | 1.0431 |
| 11 | 351.0880 | 351.0868 | 3.41 | 191.0539 | Pinastric acid * (acids) | C20H16O6 | 1.61 | H. cuiabensis; H. hatschbachii | 1.0431 |
| 12 | 353.0883 | 353.0872 | 3.11 | 191.0518, 135.0418 | Caffeoyl quinic acid * (phenolic compound) | C16H18O9 | 1.46 | H. hatschbachii; H. reclinata; H. restingae | 1.0431 |
| 13 | 353.0886 | 353.0872 | 3.96 | 191.0530, 179.1326, 135.0426 | Chlorogenic acid * (phenolic compound) | C16H18O9 | 3.08 | H. cuiabensis; H. reclinata; H. restingae | 1.0436 |
| 14 | 359.0787 | 359.0766 | 5.84 | 198.0892, 177.0187, 164.0109 | Rosmarinic acid * (phenolic compound) | C18H16O8 | 3.53 | H. hatschbachii; H. reclinata | 1.2542 |
| 15 | 367.1045 | 367.1029 | 4.33 | 196.0735, 179.0708 | 3-O-feruloylquinic * acid (phenolic compound) | C17H20O9 | 2.77 | H. hatschbachii | 1.0431 |
| 16 | 431.0996 | 431.0978 | 4.17 | 283.0187 | Isovitexin (flavonoid) | C21H20O10 | 3.74 | H. cuiabensis | 1.1601 |
| 17 | 433.0788 | 433.0770 | 4.15 | 303.0234, 302.0156, 301.0079 | Avicularin * (flavonoid) | C20H18O11 | 4.02 | H. cuiabensis | 1.5502 |
| 18 | 433.0792 | 433.0770 | 5.07 | 301.0080 | Quercetin-3-O-beta-D-xylopyranoside * (flavonoid) | C20H18O11 | 3.55 | All | 1.0431 |
| 19 | 459.0944 | 459.0927 | 3.70 | 177.0400 | Oroxindin * (flavonoid) | C22H20O11 | 3.53 | H. hatschbachii | 0.1471 |
| 20 | 463.0889 | 463.0876 | 2.80 | 301.0264, 178.9916 | Myricitrin * (flavonoid) | C21H20O12 | 2.98 | H. cuiabensis; H. reclinata | 1.6217 |
| 21 | 463.0893 | 463.0876 | 3.67 | 175.0256 | Hyperoside * (flavonoid) | C21H20O12 | 3.69 | All | 0.6566 |
| 22 | 477.1048 | 477.1033 | 3.14 | 313.0364, 301.0364 | Isorhamnetin-3-O-glucoside * (flavonoid) | C22H22O12 | 3.60 | H. cuiabensis; H. reclinata; H. restingae | 0.7729 |
| 23 | 479.0826 | 479.0825 | 0.20 | 478.0747 | Myricetin-3-O-beta-D-galactopyranoside * (flavonoid) | C21H20O13 | 6.42 | H. restingae | 1.0431 |
| 24 | 515.1213 | 515.1189 | 4.65 | 190.0472, 174.0522 | Dicaffeoyl quinic acid * (acids) | C25H24O12 | 3.18 | H. hatschbachii | 0.1248 |
| 25 | 579.1463 ** | 579.1502 | −6.73 | 124.0312 | Epicatechin (flavonoid) | C15H14O6 | 2.21 | H. reclinata | - **** |
| 26 | 623.1586 | 623.1612 | −4.17 | 317.0293, 301.0348, 300.0270 | Isorhamnetin-3-galactoside-6″-rhamnoside * (flavonoid) | C28H32O16 | 3.46 | H. cuiabensis; H. hatschbachii | 0.5496 |
| 27 | 739.2062 | 739.2085 | −3.11 | 597.9844, 596.5766 | Kaempferol-3-glucoside-2″-rhamnoside-7-rhamnoside * (flavonoid) | C33H40O19 | 3.14 | H. cuiabensis | 0.1179 |
| 28 | 895.1954 ** | 895.1932 | 2.45 | 447.0928 | Quercetin-3-O-rhamnoside * (flavonoid) | C21H20O11 | 3.51 | H. cuiabensis | 1.0692 |
| Measure | Positive Ionization Mode | Negative Ionization Mode |
|---|---|---|
| Q2 | 0.9987 | 0.9998 |
| R2 | 0.9991 | 0.9997 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Silva, J.M.G.d.; Almeida, R.F.d.; Zeraik, M.L. Multivariate Analysis of UPLC-MS/MS Metabolomic Profiles in Four Hiraea Species (Malpighiaceae). Separations 2025, 12, 159. https://doi.org/10.3390/separations12060159
Silva JMGd, Almeida RFd, Zeraik ML. Multivariate Analysis of UPLC-MS/MS Metabolomic Profiles in Four Hiraea Species (Malpighiaceae). Separations. 2025; 12(6):159. https://doi.org/10.3390/separations12060159
Chicago/Turabian StyleSilva, Jaqueline Munise Guimarães da, Rafael Felipe de Almeida, and Maria Luiza Zeraik. 2025. "Multivariate Analysis of UPLC-MS/MS Metabolomic Profiles in Four Hiraea Species (Malpighiaceae)" Separations 12, no. 6: 159. https://doi.org/10.3390/separations12060159
APA StyleSilva, J. M. G. d., Almeida, R. F. d., & Zeraik, M. L. (2025). Multivariate Analysis of UPLC-MS/MS Metabolomic Profiles in Four Hiraea Species (Malpighiaceae). Separations, 12(6), 159. https://doi.org/10.3390/separations12060159








