The Molecular Identification and Comprehensive Analysis of Klebsiella pneumoniae Isolated from Industrial Wastewater
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation and Culture of Bacteria
2.2. Genome and Plasmid DNA Extraction
2.3. Phylogenetic Tree Generation
2.4. Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry (MALDI-TOF MS) Identification
2.5. Biochemical Analysis and Identification of Microorganisms
2.6. Antibiotic Resistance Analysis
2.7. Bacterial Growth Curve Analysis
2.8. Antimicrobial Resistance Gene Detection
2.9. Proteomic Analysis
3. Results
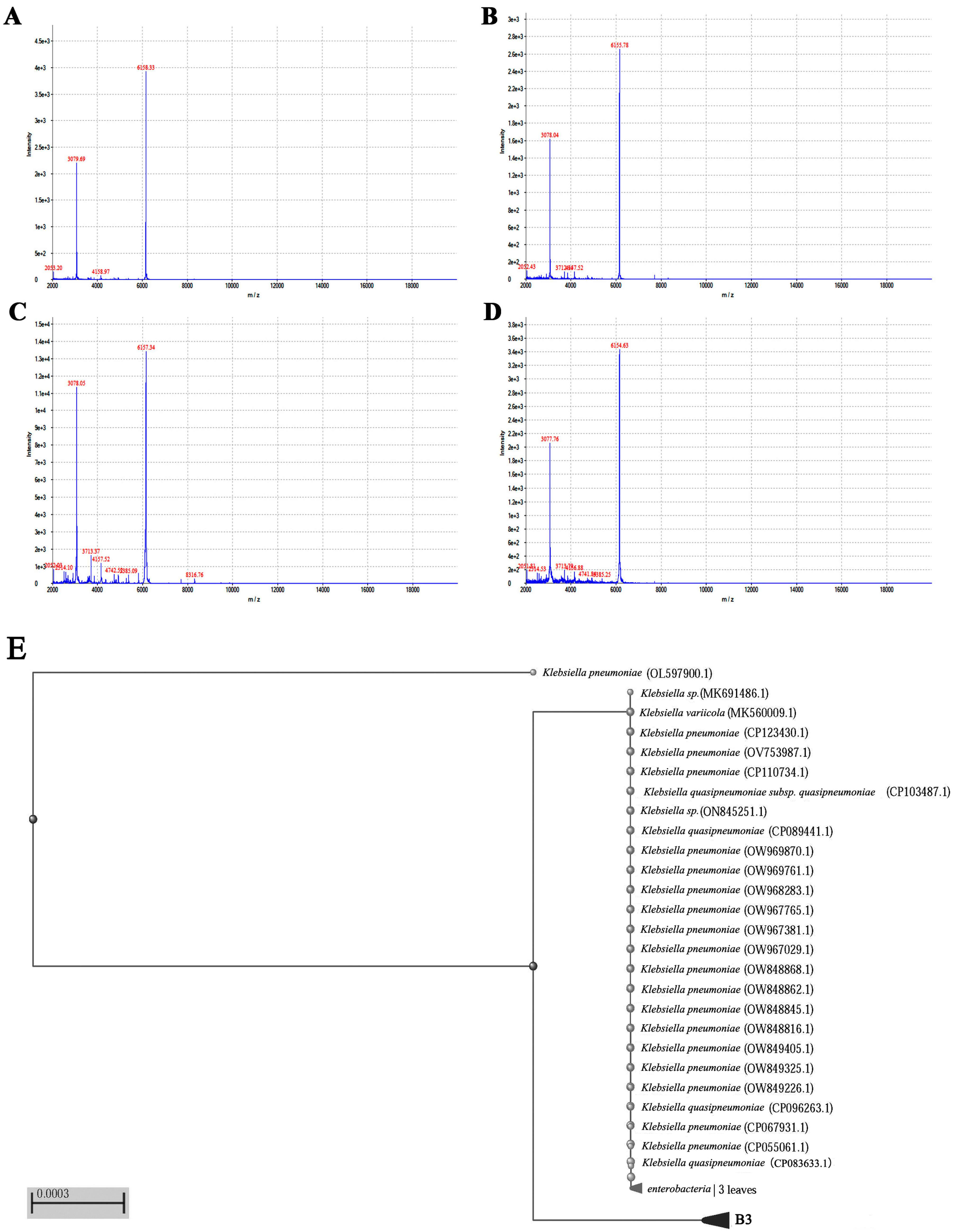
3.1. MALDI-TOF MS Enables the Rapid Identification of Klebsiella Pneumoniae
3.2. Molecular Identification of Klebsiella pneumoniae
3.3. Biochemical Identification of Klebsiella pneumoniae
3.4. Antibiotic Resistance Analysis of Klebsiella pneumoniae
3.5. Growth Experiment of Klebsiella Pneumoniae
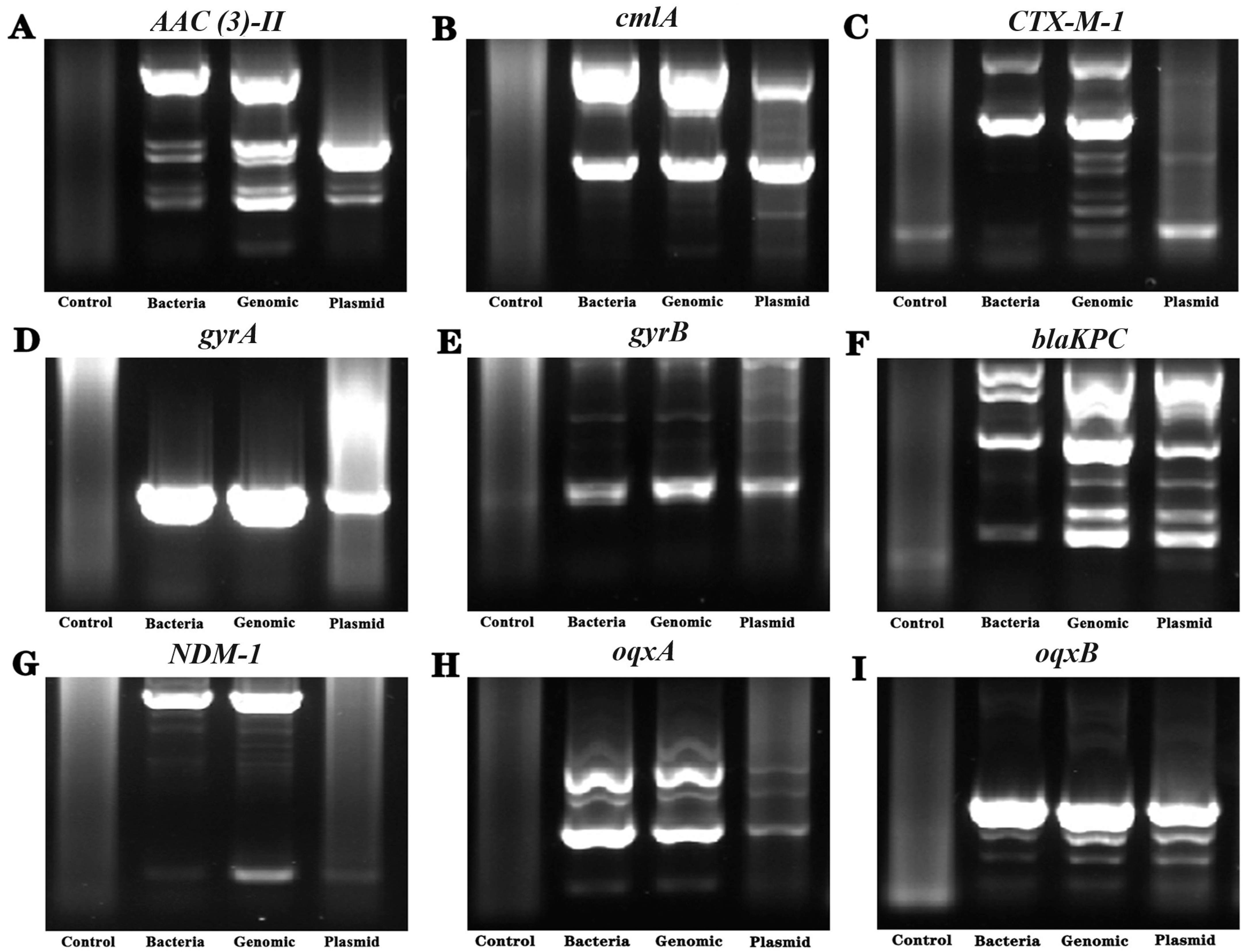
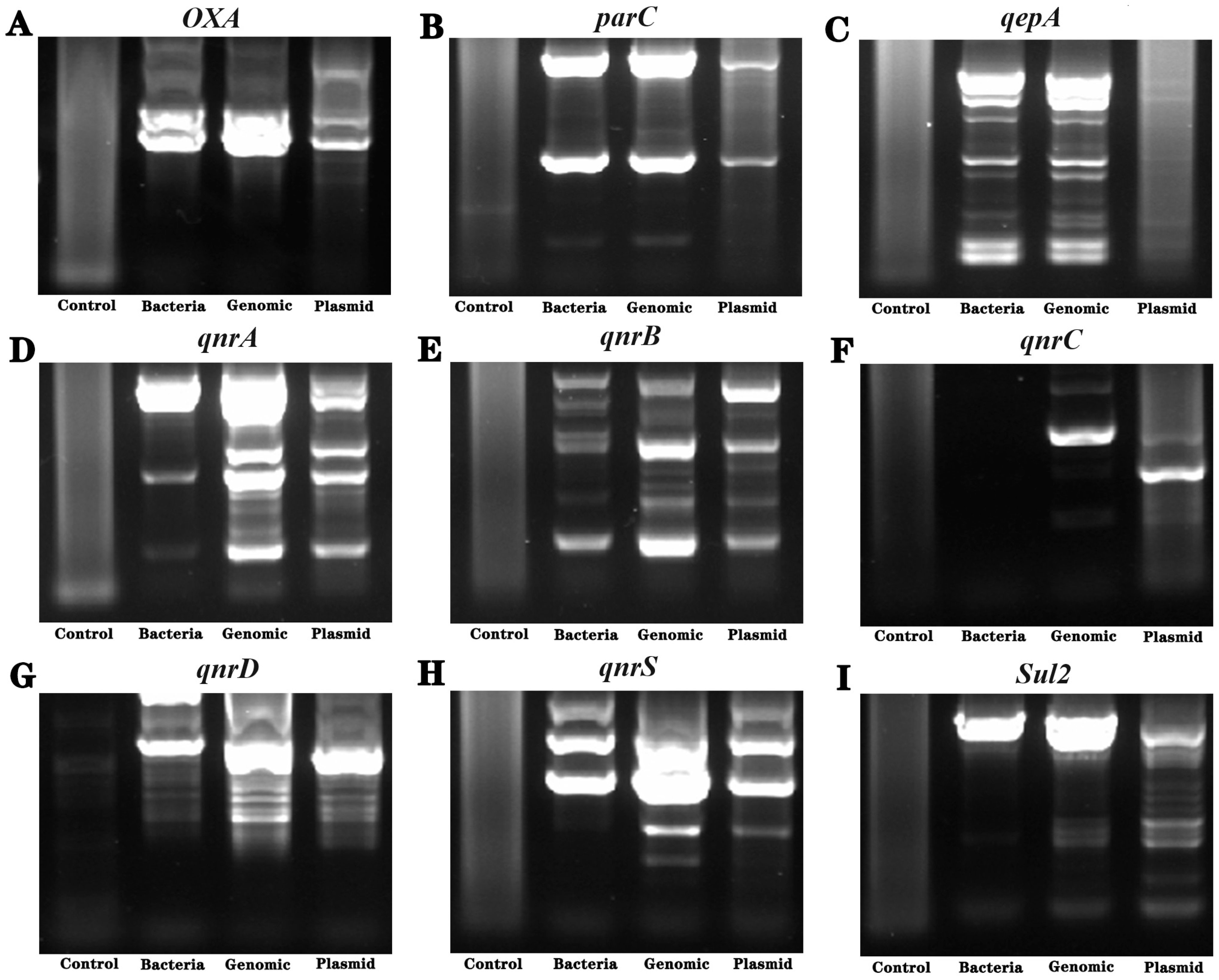
3.6. Detection of Drug Resistance-Related Genes
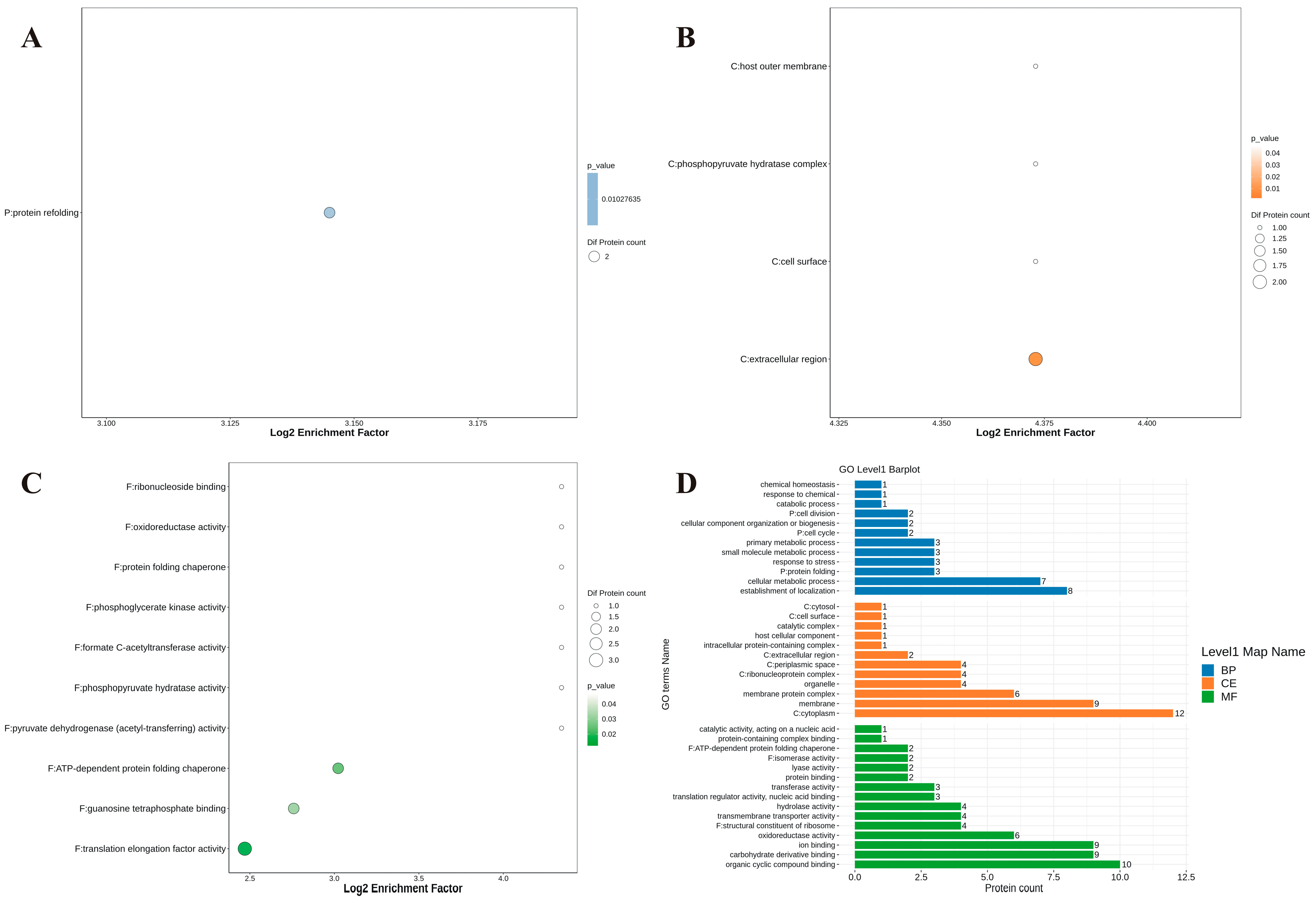
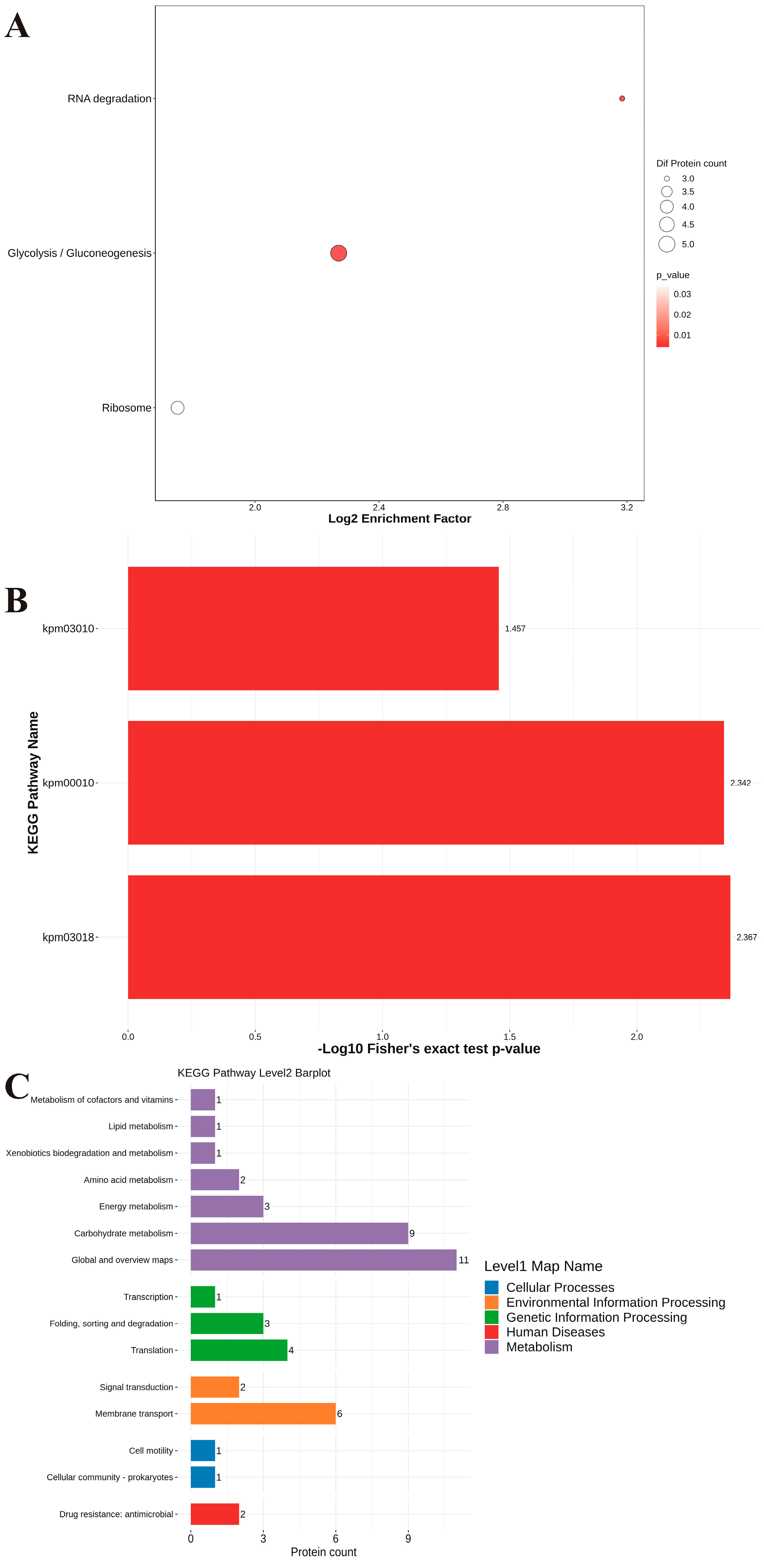
3.7. Proteomic Analysis
4. Discussion
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Behera, S.; Tanuku, N.R.S.; Moturi, S.R.K.; Gudapati, G.; Tadi, S.R.; Modali, S. Anthropogenic impact and antibiotic resistance among the indicator and pathogenic bacteria from several industrial and sewage discharge points along the coast from Pydibhimavaram to Tuni, East Coast of India. Environ. Monit. Assess. 2023, 195, 546. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Zou, H.Y.; Zhao, L.; Li, Q.; Meng, M.; Li, X.W.; Berglund, B. High prevalence of Escherichia coli co-harboring conjugative plasmids with colistin- and carbapenem resistance genes in a wastewater treatment plant in China. Int. J. Hyg. Environ. Health 2023, 250, 114159. [Google Scholar] [CrossRef] [PubMed]
- Cahill, N.; Hooban, B.; Fitzhenry, K.; Joyce, A.; O’Connor, L.; Miliotis, G.; McDonagh, F.; Burke, L.; Chueiri, A.; Farrell, M.L.; et al. First reported detection of the mobile colistin resistance genes, mcr-8 and mcr-9, in the Irish environment. Sci. Total Environ. 2023, 876, 162649. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.C.; Jian, Z.; Liu, W.; Li, J.; Pei, N. One Health Analysis of mcr-Carrying Plasmids and Emergence of mcr-10.1 in Three Species of Klebsiella Recovered from Humans in China. Microbiol. Spectr. 2022, 10, e0230622. [Google Scholar] [CrossRef] [PubMed]
- Mutuku, C.; Melegh, S.; Kovacs, K.; Urban, P.; Virag, E.; Heninger, R.; Herczeg, R.; Sonnevend, A.; Gyenesei, A.; Fekete, C.; et al. Characterization of beta-Lactamases and Multidrug Resistance Mechanisms in Enterobacterales from Hospital Effluents and Wastewater Treatment Plant. Antibiotics 2022, 11, 776. [Google Scholar] [CrossRef] [PubMed]
- Bong, C.W.; Low, K.Y.; Chai, L.C.; Lee, C.W. Prevalence and Diversity of Antibiotic Resistant Escherichia coli From Anthropogenic-Impacted Larut River. Front. Public Health 2022, 10, 794513. [Google Scholar] [CrossRef] [PubMed]
- Savin, M.; Bierbaum, G.; Mutters, N.T.; Schmithausen, R.M.; Kreyenschmidt, J.; Garcia-Menino, I.; Schmoger, S.; Kasbohrer, A.; Hammerl, J.A. Genetic Characterization of Carbapenem-Resistant Klebsiella spp. from Municipal and Slaughterhouse Wastewater. Antibiotics 2022, 11, 435. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.F.; Du, L.Y.; Luo, J.; He, H.X. Isolation, identification and characterization of Morganella morganii from Naja naja atra in Beijing, China. Cell Mol. Biol. 2017, 63, 52–58. [Google Scholar] [CrossRef] [PubMed]
- Yigit, H.; Queenan, A.M.; Anderson, G.J.; Domenech-Sanchez, A.; Biddle, J.W.; Steward, C.D.; Alberti, S.; Bush, K.; Tenover, F.C. Novel carbapenem-hydrolyzing beta-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 2001, 45, 1151–1161. [Google Scholar] [CrossRef]
- Yu, Y.; Ji, S.; Chen, Y.; Zhou, W.; Wei, Z.; Li, L.; Ma, Y. Resistance of strains producing extended-spectrum beta-lactamases and genotype distribution in China. J. Infect. 2007, 54, 53–57. [Google Scholar] [CrossRef]
- Kim, H.B.; Wang, M.; Park, C.H.; Kim, E.C.; Jacoby, G.A.; Hooper, D.C. oqxAB encoding a multidrug efflux pump in human clinical isolates of Enterobacteriaceae. Antimicrob. Agents Chemother. 2009, 53, 3582–3584. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.H.; Deng, Y.T.; Zeng, Z.L.; Gao, J.H.; Chen, L.; Arakawa, Y.; Chen, Z.L. Coprevalence of plasmid-mediated quinolone resistance determinants QepA, Qnr, and AAC(6′)-Ib-cr among 16S rRNA methylase RmtB-producing Escherichia coli isolates from pigs. Antimicrob. Agents Chemother. 2008, 52, 2992–2993. [Google Scholar] [CrossRef] [PubMed]
- Obasi, A.; Nwachukwu, S.; Ugoji, E.; Kohler, C.; Gohler, A.; Balau, V.; Pfeifer, Y.; Steinmetz, I. Extended-Spectrum -Lactamase-Producing Klebsiella pneumoniae from Pharmaceutical Wastewaters in South-Western Nigeria. Microb. Drug Resist. 2017, 23, 1013–1018. [Google Scholar] [CrossRef] [PubMed]
- Aristizabal-Hoyos, A.M.; Rodriguez, E.A.; Torres-Palma, R.A.; Jimenez, J.N. Concern levels of beta-lactamase-producing Gram-negative bacilli in hospital wastewater: Hotspot of antimicrobial resistance in Latin-America. Diagn. Microbiol. Infect. Dis. 2023, 105, 115819. [Google Scholar] [CrossRef] [PubMed]
- Loudermilk, E.M.; Kotay, S.M.; Barry, K.E.; Parikh, H.I.; Colosi, L.M.; Mathers, A.J. Tracking Klebsiella pneumoniae carbapenemase gene as an indicator of antimicrobial resistance dissemination from a hospital to surface water via a municipal wastewater treatment plant. Water Res. 2022, 213, 118151. [Google Scholar] [CrossRef] [PubMed]
- Dolejska, M.; Frolkova, P.; Florek, M.; Jamborova, I.; Purgertova, M.; Kutilova, I.; Cizek, A.; Guenther, S.; Literak, I. CTX-M-15-producing Escherichia coli clone B2-O25b-ST131 and Klebsiella spp. isolates in municipal wastewater treatment plant effluents. J. Antimicrob. Chemother. 2011, 66, 2784–2790. [Google Scholar] [CrossRef]
- Mahgoub, S.A.; Qattan, S.Y.A.; Salem, S.S.; Abdelbasit, H.M.; Raafat, M.; Ashkan, M.F.; Al-Quwaie, D.A.; Motwali, E.A.; Alqahtani, F.S.; Abd El-Fattah, H.I. Characterization and Biodegradation of Phenol by Pseudomonas aeruginosa and Klebsiella variicola Strains Isolated from Sewage Sludge and Their Effect on Soybean Seeds Germination. Molecules 2023, 28, 1203. [Google Scholar] [CrossRef]
- Shaker, R.A.E.; Nagy, Y.I.; Adly, M.E.; Khattab, R.A.; Ragab, Y.M. Acinetobacter baumannii, Klebsiella pneumoniae and Elizabethkingia miricola isolated from wastewater have biodegradable activity against fluoroquinolone. World J. Microbiol. Biotechnol. 2022, 38, 187. [Google Scholar] [CrossRef]
- Wang, Y.; Wan, S.; Yu, W.; Yuan, D.; Sun, L. Newly isolated Enterobacter cloacae sp. HN01 and Klebsiella pneumoniae sp. HN02 collaborate with self-secreted biosurfactant to improve solubility and bioavailability for the biodegradation of hydrophobic and toxic gaseous para-xylene. Chemosphere 2022, 304, 135328. [Google Scholar] [CrossRef]
- Nie, M.; Yin, X.; Jia, J.; Wang, Y.; Liu, S.; Shen, Q.; Li, P.; Wang, Z. Production of a novel bioflocculant MNXY1 by Klebsiella pneumoniae strain NY1 and application in precipitation of cyanobacteria and municipal wastewater treatment. J. Appl. Microbiol. 2011, 111, 547–558. [Google Scholar] [CrossRef]
- Soda, S.; Otsuki, H.; Inoue, D.; Tsutsui, H.; Sei, K.; Ike, M. Transfer of antibiotic multiresistant plasmid RP4 from Escherichia coli to activated sludge bacteria. J. Biosci. Bioeng. 2008, 106, 292–296. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Li, Y.; Duan, J. Biodegradation of 2-methylquinoline by Klebsiella pneumoniae TJ-A isolated from acclimated activated sludge. J. Environ. Sci. Health A Tox Hazard. Subst. Environ. Eng. 2014, 49, 27–38. [Google Scholar] [CrossRef] [PubMed]
- Abbas, S.Z.; Riaz, M.; Ramzan, N.; Zahid, M.T.; Shakoori, F.R.; Rafatullah, M. Isolation and characterization of arsenic resistant bacteria from wastewater. Braz. J. Microbiol. 2014, 45, 1309–1315. [Google Scholar] [CrossRef] [PubMed]
- Zulfiqar, S.; Shakoori, A.R. Molecular characterization, metal uptake and copper induced transcriptional activation of efflux determinants in copper resistant isolates of Klebsiella pneumoniae. Gene 2012, 510, 32–38. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Liang, J.; Wang, S.; Yang, B.; Chen, M.; Liu, Y. Production of a bioflocculant from Klebsiella sp. OS-1 using brewery wastewater as a source. Environ. Technol. 2019, 40, 44–52. [Google Scholar] [CrossRef]
- Awasthi, S.; Srivastava, P.; Singh, P.; Tiwary, D.; Mishra, P.K. Biodegradation of thermally treated high-density polyethylene (HDPE) by Klebsiella pneumoniae CH001. 3 Biotech 2017, 7, 332. [Google Scholar] [CrossRef]
- Rajkumari, J.; Paikhomba Singha, L.; Pandey, P. Genomic insights of aromatic hydrocarbon degrading Klebsiella pneumoniae AWD5 with plant growth promoting attributes: A paradigm of soil isolate with elements of biodegradation. 3 Biotech 2018, 8, 118. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Guo, Z.; Zhu, G.; Li, N.; Bai, G.; Jiang, M. Genomic Characteristics and Phylogenetic Analyses of a Multiple Drug-Resistant Klebsiella pneumoniae Harboring Plasmid-Mediated MCR-1 Isolated from Tai’an City, China. Pathogens 2023, 12, 221. [Google Scholar] [CrossRef]
- Yu, M.; Wei, Q.; Song, W.; Yuan, J. Phenotypic and Genetic Analysis of KPC-49, a KPC-2 Variant Conferring Resistance to Ceftazidime-Avibactam and Maintaining Resistance to Imipenem and Meropenem. Infect. Drug Resist. 2023, 16, 2477–2485. [Google Scholar] [CrossRef]
- Singh, V.; Dhankhar, P.; Dalal, V.; Tomar, S.; Kumar, P. In-silico functional and structural annotation of hypothetical protein from Klebsiella pneumonia: A potential drug target. J. Mol. Graph. Model. 2022, 116, 108262. [Google Scholar] [CrossRef]

| Name | Abbreviation | Results |
|---|---|---|
| Amino acid control | C | Negative |
| Arginine dihydrolase | ADH | Negative |
| Urease | URE | Positive |
| Lysine decarboxylase | LDC | Positive |
| Aescin hydrolysis | ESC | Positive |
| Ornithine decarboxylase | ODC | Negative |
| Rhamnose fermentation | RHAf | Positive |
| Melibiose fermentation | MELf | Positive |
| Galactosidase | ONPG | Positive |
| Glucose fermentation | GLUf | Positive |
| Hydrogen sulfide production | H2S | Negative |
| Utilization of citrate | CIT | Positive |
| Gelatin hydrolysis | GEL | Negative |
| Lactose fermentation | LACf | Negative |
| Tryptophan deaminase | TDA | Negative |
| Cellulose disaccharide fermentation | CELf | Positive |
| Production of indole | IND | Negative |
| Arabinose fermentation | ARAf | Positive |
| VP Experiment | VP | Positive |
| Bitter almond glycoside fermentation | AMYf | Negative |
| Mannitol fermentation | MANf | Positive |
| Inositol fermentation | INOf | Positive |
| Sucrose fermentation | SACf | Positive |
| Sorbitol fermentation | SORf | Positive |
| Drug Name | Abbreviation | MIC Value |
|---|---|---|
| Piperacillin/tazobactam | P/T | =4/4 |
| Tobramycin | TOB | ≤1 |
| Furantoin | FD | ≥128 |
| Cefazolin | CFZ | ≤2 |
| Ceftazidime | CAZ | ≤0.5 |
| Ampicillin | AMP | >32 |
| Moxifloxacin | MXF | ≤0.5 |
| Minocycline | MIN | ≤4 |
| Gentamicin | GEN | ≤1 |
| MeropeneM | MRP | ≤0.06 |
| Cefoperazone/sulbactam | CPS | ≤16/8 |
| Ceftazidime/clavulanic acid | CAZ/C | ≤1/4 |
| Imipenem | IPM | ≤0.25 |
| Cefuroxime | CXM | ≤8 |
| Ticarcillin/clavulanic acid | TIM | ≤16/2 |
| Cefoxitin | FOX | ≤8 |
| Levofloxacin | LEV | =0.12 |
| Polymyxin B | PB | ≤1 |
| Cefepime | FEP | ≤0.12 |
| Cefotaxime/clavulanic acid | CTX/C | ≤1/4 |
| Amoxicillin/clavulanate | AMC | ≤8/4 |
| Aztreonam | ATM | ≤0.25 |
| Ertapenem | ETP | ≤0.5 |
| Amikacin | AMK | ≤4 |
| Compound xinnuomin | SXT | ≤2/38 |
| Cefotaxime | CTX | ≤0.12 |
| Ciprofloxacin | CIP | ≤0.015 |
| Chloramphenicol | CHL | ≥32 |
| Tigecycline | TGC | ≤0.25 |
| Ampicillin/sulbactam | AMS | =16/8 |
| Test Group | Time (h) | Peak Point (OD) |
|---|---|---|
| Klebsiella pneumoniae-1 μL | 13 | 5 |
| Klebsiella pneumoniae-10 μL | 13 | 5.08 |
| Klebsiella pneumoniae-20 μL | 13 | 5.07 |
| Klebsiella pneumoniae-40 μL | 12 | 4.9 |
| Klebsiella pneumoniae-60 μL | 13 | 4.82 |
| Klebsiella pneumoniae-80 μL | 11 | 4.73 |
| Klebsiella pneumoniae-100 μL | 11 | 4.66 |
| Item | Proteomics Analysis |
|---|---|
| 1 | Multidrug-resistant outer membrane protein (MdtQ) |
| 2 | Multidrug-resistant secretion protein |
| 3 | Modulator of drug activity B |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, K.; Li, C.; Wang, W.; Guo, J.; Wang, H. The Molecular Identification and Comprehensive Analysis of Klebsiella pneumoniae Isolated from Industrial Wastewater. Separations 2024, 11, 121. https://doi.org/10.3390/separations11040121
Yan K, Li C, Wang W, Guo J, Wang H. The Molecular Identification and Comprehensive Analysis of Klebsiella pneumoniae Isolated from Industrial Wastewater. Separations. 2024; 11(4):121. https://doi.org/10.3390/separations11040121
Chicago/Turabian StyleYan, Kai, Changfu Li, Weiyu Wang, Juan Guo, and Haifeng Wang. 2024. "The Molecular Identification and Comprehensive Analysis of Klebsiella pneumoniae Isolated from Industrial Wastewater" Separations 11, no. 4: 121. https://doi.org/10.3390/separations11040121
APA StyleYan, K., Li, C., Wang, W., Guo, J., & Wang, H. (2024). The Molecular Identification and Comprehensive Analysis of Klebsiella pneumoniae Isolated from Industrial Wastewater. Separations, 11(4), 121. https://doi.org/10.3390/separations11040121







