Identification of Ginseng Radix et Rhizoma, Panacis Quinquefolii Radix, Notoginseng Radix et Rhizoma, and Platycodonis Radix Based on UHPLC-QTOF-MS and “Matrix Characteristics”
Abstract
1. Introduction
2. Materials and Methods
2.1. Herbs and Reagent Materials
2.2. Sample Pretreatment
2.3. UHPLC Analysis Conditions
2.4. Mass Spectrometry Conditions
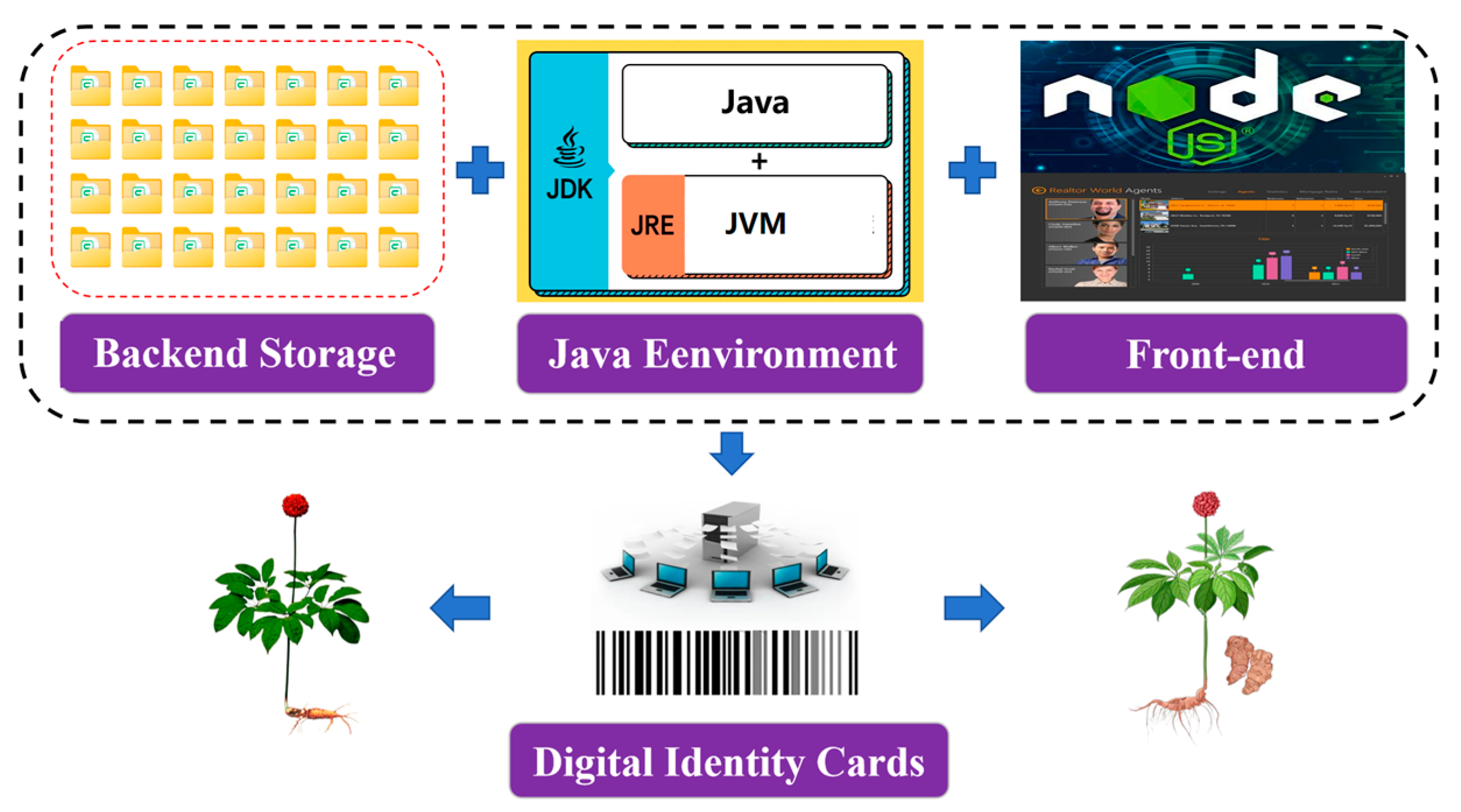
2.5. Data Processing and Analysis
2.5.1. Data Processing
2.5.2. Identification Algorithm Flow
3. Results
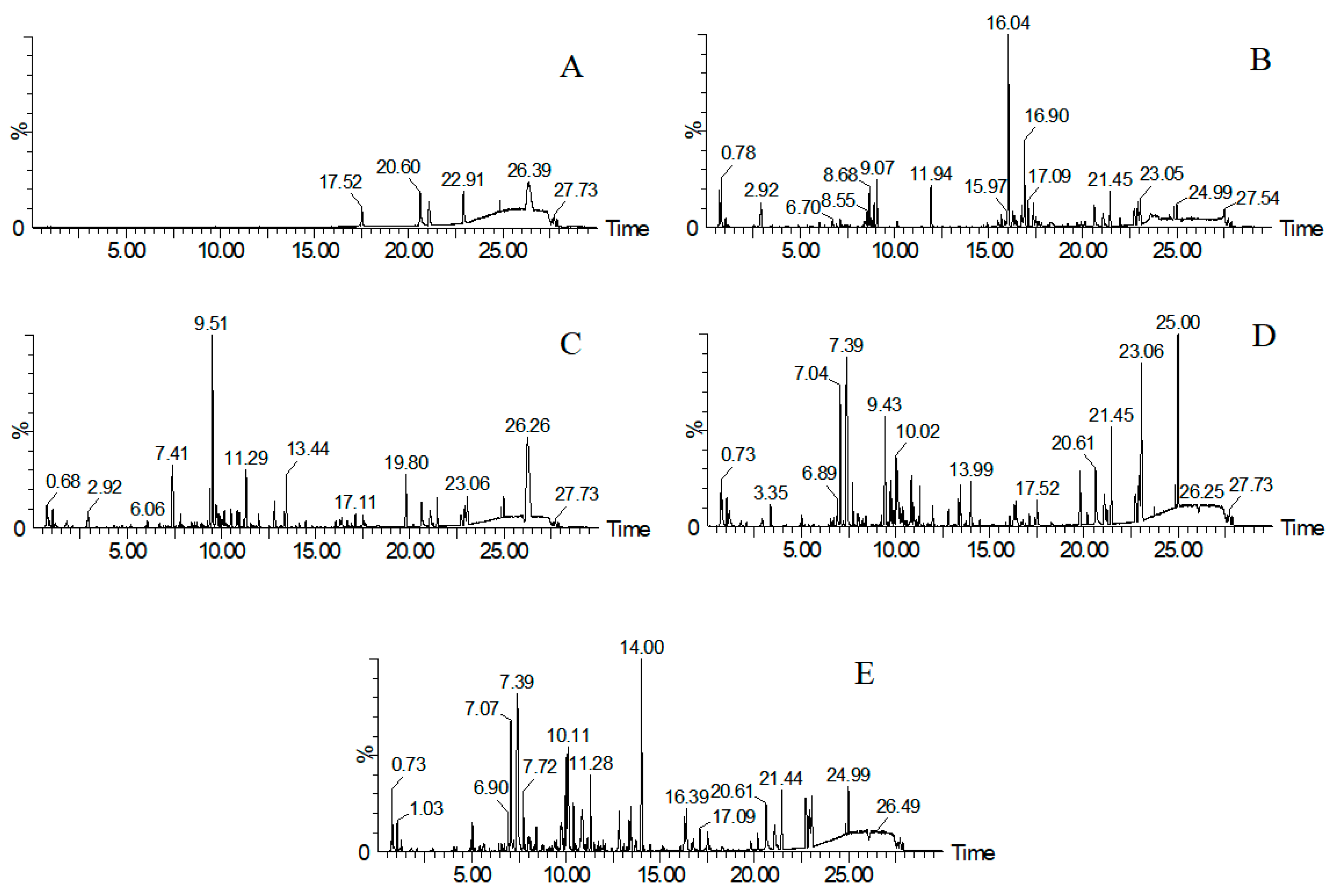
3.1. Results of UHPLC-QTOF-MS Analysis
3.2. The Results of Digital Quantization Processing
3.3. Results of Algorithm Programming
3.4. Results of Identification Analysis and Verification
4. Discussion
4.1. Present Situation and Digitization of TCM Identification
4.2. Discussion of Research Ideas and Strengths
4.3. Optimization of UHPLC-QTOF-MS Analysis
4.4. Sample Selection Principles
4.5. Parameter Selection, Setting and Suggestion
4.6. Research Shortcomings and Prospects
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhou, X.; Zhang, K.; Liu, L.; Zhao, Q.; Huang, M.; Shao, R.; Wang, Y.; Qu, B.; Wang, Y. Anti-fatigue effect from Ginseng Radix et Rhizoma: A suggestive and promising treatment for long COVID. Acupunct. Herb. Med. 2022, 2, 69–77. [Google Scholar] [PubMed]
- Xiang, Y.Z.; Shang, H.C.; Gao, X.M.; Zhang, B.L. A comparison of the ancient use of ginseng in traditional Chinese medicine with modern pharmacological experiments and clinical trials. Phytother. Res. PTR 2008, 22, 851–858. [Google Scholar] [CrossRef] [PubMed]
- Arring, N.M.; Millstine, D.; Marks, L.A.; Nail, L.M. Ginseng as a Treatment for Fatigue: A Systematic Review. J. Altern. Complement. Med. 2018, 24, 624–633. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, H.; Dai, X.; Zhu, R.; Chen, B.; Xia, B.; Ye, Z.; Zhao, D.; Gao, S.; Orekhov, A.N.; et al. A comprehensive review on the phytochemistry, pharmacokinetics, and antidiabetic effect of Ginseng. Phytomedicine 2021, 92, 153717. [Google Scholar] [CrossRef]
- Jang, W.Y.; Hwang, J.Y.; Cho, J.Y. Ginsenosides from Panax ginseng as Key Modulators of NF-κB Signaling Are Powerful Anti-Inflammatory and Anticancer Agents. Int. J. Mol. Sci. 2023, 24, 6119. [Google Scholar] [CrossRef]
- Huang, J.; Liu, D.; Wang, Y.; Liu, L.; Li, J.; Yuan, J.; Jiang, Z.; Jiang, Z.; Hsiao, W.W.; Liu, H.; et al. Ginseng polysaccharides alter the gut microbiota and kynurenine/tryptophan ratio, potentiating the antitumour effect of antiprogrammed cell death 1/programmed cell death ligand 1 (anti-PD-1/PD-L1) immunotherapy. Gut 2020, 71, 734–745. [Google Scholar]
- Yang, L.; Hou, A.; Zhang, J.; Wang, S.; Man, W.; Yu, H.; Zheng, S.; Wang, X.; Liu, S.; Jiang, H. Panacis Quinquefolii Radix: A Review of the Botany, Phytochemistry, Quality Control, Pharmacology, Toxicology and Industrial Applications Research Progress. Front. Pharmacol. 2020, 11, 602092. [Google Scholar] [CrossRef]
- Wang, X.X.; Zou, H.Y.; Cao, Y.N.; Zhang, X.M.; Sun, M.; Tu, P.F.; Liu, K.C.; Zhang, Y. Radix Panacis quinquefolii Extract Ameliorates Inflammatory Bowel Disease through Inhibiting Inflammation. Chin. J. Integr. Med. 2023, 29, 825–831. [Google Scholar] [CrossRef]
- Wang, T.; Guo, R.; Zhou, G.; Zhou, X.; Kou, Z.; Sui, F.; Li, C.; Tang, L.; Wang, Z. Traditional uses, botany, phytochemistry, pharmacology and toxicology of Panax notoginseng (Burk.) F.H. Chen: A review. J. Ethnopharmacol. 2016, 188, 234–258. [Google Scholar]
- Wang, X.; Liu, J.; Tian, R.; Zheng, B.; Li, C.; Huang, L.; Lu, Z.; Zhang, J.; Mao, W.; Liu, B.; et al. Sanqi Oral Solution Mitigates Proteinuria in Rat Passive Heymann Nephritis and Blocks Podocyte Apoptosis via Nrf2/HO-1 Pathway. Front. Pharmacol. 2021, 12, 727874. [Google Scholar] [CrossRef]
- Zhang, L.L.; Huang, M.Y.; Yang, Y.; Huang, M.Q.; Shi, J.J.; Zou, L.; Lu, J.J. Bioactive platycodins from Platycodonis Radix: Phytochemistry, pharmacological activities, toxicology and pharmacokinetics. Food Chem. 2020, 327, 127029. [Google Scholar] [PubMed]
- Li, J.J.; Liu, M.L.; Lv, J.N.; Chen, R.L.; Ding, K.; He, J.Q. Polysaccharides from Platycodonis Radix ameliorated respiratory syncytial virus-induced epithelial cell apoptosis and inflammation through activation of miR-181a-mediated Hippo and SIRT1 pathways. Int. Immunopharmacol. 2022, 104, 108510. [Google Scholar] [CrossRef]
- Zhang, C.Y.; Dong, L.; Wang, J.; Chen, S.L. Simultaneous determination of ten ginsenosides in panacis quinquefolii radix by ultra performance liquid chromatography and quality evaluation based on chemometric methods. Pharmazie 2011, 66, 553–559. [Google Scholar] [PubMed]
- Balkrishna, A.; Verma, S.; Tiwari, D.; Srivastava, J.; Varshney, A. UPLC-QToF-MS based fingerprinting of polyphenolic metabolites in the bark extract of Boehmeria rugulosa Wedd. J. Mass Spectrom. JMS 2022, 57, e4890. [Google Scholar] [CrossRef]
- Kou, T.Y. Terahertz spectroscopy for accurate identification of ginseng and panax quinquefolium. Opt. Instrum. 2020, 42, 27–32. [Google Scholar]
- Wang, X.L.; Lei, R.; Zhao, Z.X.; Su, J.; Cheng, X.L.; Liu, Y.L.; Ma, S.C. Analysis of supplementary testing methods and supervision strategy for illegal admixture of Panax quinquefolium in Panax ginseng and its preparations. China J. Chin. Mater. Medica 2024, 7, 1–14. [Google Scholar]
- Jiang, C.; Luo, Y.Q.; Yuan, Y.; Huang, L.Q.; Jin, Y.; Zhao, Y.Y. Identification of Panax ginseng, P. notoginseng and P. quinquefolius admixture by multiplex allele-specific polymerase chain reaction. China J. Chin. Mater. Medica 2017, 42, 1319–1323. [Google Scholar]
- National Pharmacopoeia Commission. Pharmacopoeia of the People’s Republic of China, 11th ed.; China Medical Science Press: Beijing, China, 2020; p. 289. [Google Scholar]
- Lin, F.; Kennelly, E.J.; Linington, R.G.; Long, C. Comprehensive Metabolite Profiling of Two Edible Garcinia Species Based on UPLC-ESI-QTOF-MSE Coupled with Bioactivity Assays. J. Agric. Food Chem. 2023, 71, 7604–7617. [Google Scholar] [CrossRef]
- GB 19298-2014; National Food Safety Standard—Packaged Water for Drinking. Standardization Administration of China: Beijing, China, 2014.
- Rui Wang, X.; Ting Zhang, J.; Guang Jing, W.; Hua Li, M.; Han Guo, X.; Long Cheng, X.; Wei, F. Digital identification and adulteration analysis of Pulsatilla Radix and Pulsatilla Cernua based on “matrix characteristics” and UHPLC-QTOF-MSE. J. Chromatogr. B 2024, 1244, 124257. [Google Scholar] [CrossRef]
- Wang, X.R.; Zhang, J.T.; He, F.; Fu, R.; Jing, W.G.; Guo, X.; Li, M.; Cheng, X.L.; Wei, F. Identification Analysis of Angelicae sinensis radix and Angelicae pubescentis radix Based on Quantized “matrix characteristics” and UHPLC-QTOF-MSE Analysis. J. Am. Soc. Mass Spectrom. 2024, 35, 2222–2229. [Google Scholar] [CrossRef]
- Hou, J.; Yao, C.; Li, Y.; Yang, L.; Chen, X.; Nie, M.; Qu, H.; Ji, S.; Guo, D.A. A MS-feature-based medicinal plant database-driven strategy for ingredient identification of Chinese medicine prescriptions. J. Pharm. Biomed. Anal. 2023, 234, 115482. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhang, J.; He, F.; Jing, W.; Li, M.; Guo, X.; Cheng, X.; Wei, F. Differential Chemical Components Analysis of Periplocae Cortex, Lycii Cortex, and Acanthopanacis Cortex Based on Mass Spectrometry Data and Chemometrics. Molecules 2024, 29, 3807. [Google Scholar] [CrossRef]
- Wu, C.S.; Chen, Y.H.; Chen, C.L.; Chien, S.K.; Syifa, N.; Hung, Y.C.; Cheng, K.J.; Hu, S.C.; Lo, P.T.; Lin, S.Y.; et al. Constructing a bilingual website with validated database for Herb and Western medicine interactions using Ginseng, Ginkgo and Dong Quai as examples. BMC Complement. Altern. Med. 2019, 19, 335. [Google Scholar]
- Nair, H.K. The dawn of the digital era. J. Wound Care 2022, 31 (Suppl. S4), S3. [Google Scholar] [CrossRef]
- Mann, D.M.; Lawrence, K. Reimagining Connected Care in the Era of Digital Medicine. JMIR Mhealth Uhealth 2022, 10, e34483. [Google Scholar] [CrossRef] [PubMed]
- Sloop, J.T.; Chao, A.; Gundersen, J.; Phillips, A.L.; Sobus, J.R.; Ulrich, E.M.; Williams, A.J.; Newton, S.R. Demonstrating the Use of Non-targeted Analysis for Identification of Unknown Chemicals in Rapid Response Scenarios. Environ. Sci. Technol. 2023, 57, 3075–3084. [Google Scholar] [CrossRef]
- Zhang, C.X.; Wang, X.Y.; Lin, Z.Z.; Wang, H.D.; Qian, Y.X.; Li, W.W.; Yang, W.Z.; Guo, D.A. Highly selective monitoring of in-source fragmentation sapogenin product ions in positive mode enabling group-target ginsenosides profiling and simultaneous identification of seven Panax herbal medicines. J. Chromatogr. A 2020, 1618, 460850. [Google Scholar] [CrossRef]
- Zhang, L.; Saber, F.R.; Rocchetti, G.; Zengin, G.; Hashem, M.M.; Lucini, L. UHPLC-QTOF-MS based metabolomics and biological activities of different parts of Eriobotrya japonica. Food Res. Int. 2021, 143, 110242. [Google Scholar] [CrossRef]
- Li, Y.; Li, H.; Luo, T.; Lin, G.; Li, L. Intensity-dependent mass search for improving metabolite database matches in chemical isotope labeling LC-QTOF-MS-based metabolomics. Anal. Chim. Acta 2023, 1272, 341467. [Google Scholar] [CrossRef]
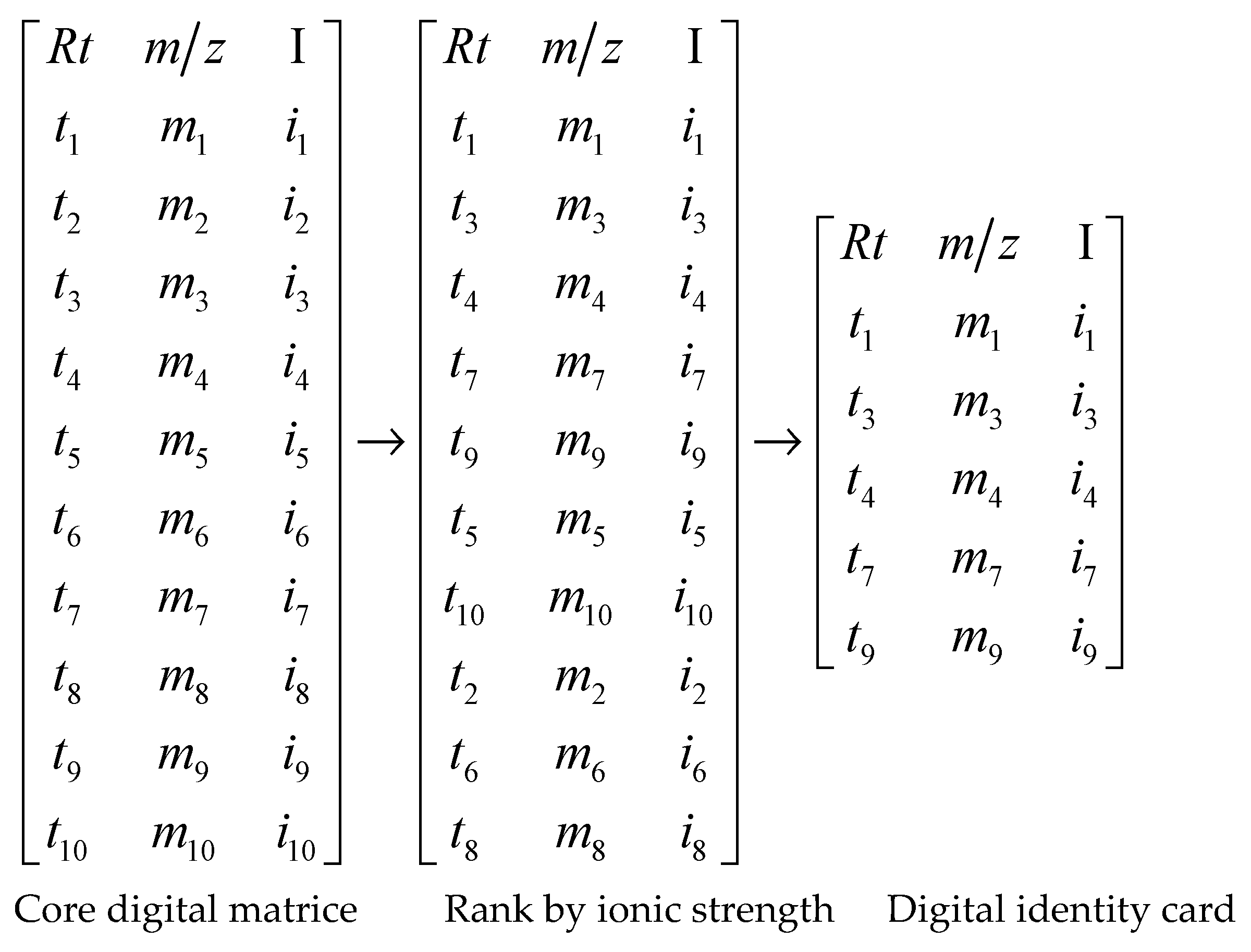
| TCM | Batch | Number | RSD | TCM | Batch | Number | RSD |
|---|---|---|---|---|---|---|---|
| PR | 20141101 | 5287 | 14.5% | GRR | 20111001 | 7683 | 3.6% |
| 20141102 | 5496 | 20111002 | 7909 | ||||
| 20161201 | 5669 | 20121101 | 8519 | ||||
| 20161202 | 5736 | 20121102 | 8025 | ||||
| 20211301 | 7320 | 20171201 | 8203 | ||||
| 20211302 | 7146 | 20171202 | 8257 | ||||
| NRR | 20110801 | 8145 | 4.6% | PQR | 20060801 | 7127 | 8.4% |
| 20110802 | 7898 | 20060802 | 7005 | ||||
| 20140901 | 7787 | 20130901 | 6413 | ||||
| 20140902 | 7507 | 20130902 | 6590 | ||||
| 20181001 | 7121 | 20181001 | 7892 | ||||
| 20181002 | 7577 | 20181002 | 7769 |
| TCM | Match Units | Number of Units in “Matrix Characteristics” | TCM | M(%) |
|---|---|---|---|---|
| PR | 194 | 200 | PR | 97.00 |
| 29 | 200 | GRR | 14.50 | |
| 8 | 200 | PQR | 4.00 | |
| 7 | 200 | NRR | 3.50 | |
| GRR | 200 | 200 | GRR | 100.00 |
| 139 | 200 | PQR | 69.50 | |
| 131 | 200 | NRR | 65.50 | |
| 55 | 200 | PR | 27.50 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J.; He, F.; Wang, X.; Jing, W.; Li, M.; Guo, X.; Cheng, X.; An, F.; Wei, F. Identification of Ginseng Radix et Rhizoma, Panacis Quinquefolii Radix, Notoginseng Radix et Rhizoma, and Platycodonis Radix Based on UHPLC-QTOF-MS and “Matrix Characteristics”. Separations 2024, 11, 304. https://doi.org/10.3390/separations11110304
Zhang J, He F, Wang X, Jing W, Li M, Guo X, Cheng X, An F, Wei F. Identification of Ginseng Radix et Rhizoma, Panacis Quinquefolii Radix, Notoginseng Radix et Rhizoma, and Platycodonis Radix Based on UHPLC-QTOF-MS and “Matrix Characteristics”. Separations. 2024; 11(11):304. https://doi.org/10.3390/separations11110304
Chicago/Turabian StyleZhang, Jiating, Fangliang He, Xianrui Wang, Wenguang Jing, Minghua Li, Xiaohan Guo, Xianlong Cheng, Fudong An, and Feng Wei. 2024. "Identification of Ginseng Radix et Rhizoma, Panacis Quinquefolii Radix, Notoginseng Radix et Rhizoma, and Platycodonis Radix Based on UHPLC-QTOF-MS and “Matrix Characteristics”" Separations 11, no. 11: 304. https://doi.org/10.3390/separations11110304
APA StyleZhang, J., He, F., Wang, X., Jing, W., Li, M., Guo, X., Cheng, X., An, F., & Wei, F. (2024). Identification of Ginseng Radix et Rhizoma, Panacis Quinquefolii Radix, Notoginseng Radix et Rhizoma, and Platycodonis Radix Based on UHPLC-QTOF-MS and “Matrix Characteristics”. Separations, 11(11), 304. https://doi.org/10.3390/separations11110304






