Optimization of the Preparation of Hydrophilic Poly(DHPMA-co-MBA) Monolithic Capillary Columns: A New Support for Affinity Chromatography
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents and Buffers
2.2. Monolithic Capillary Column Synthesis
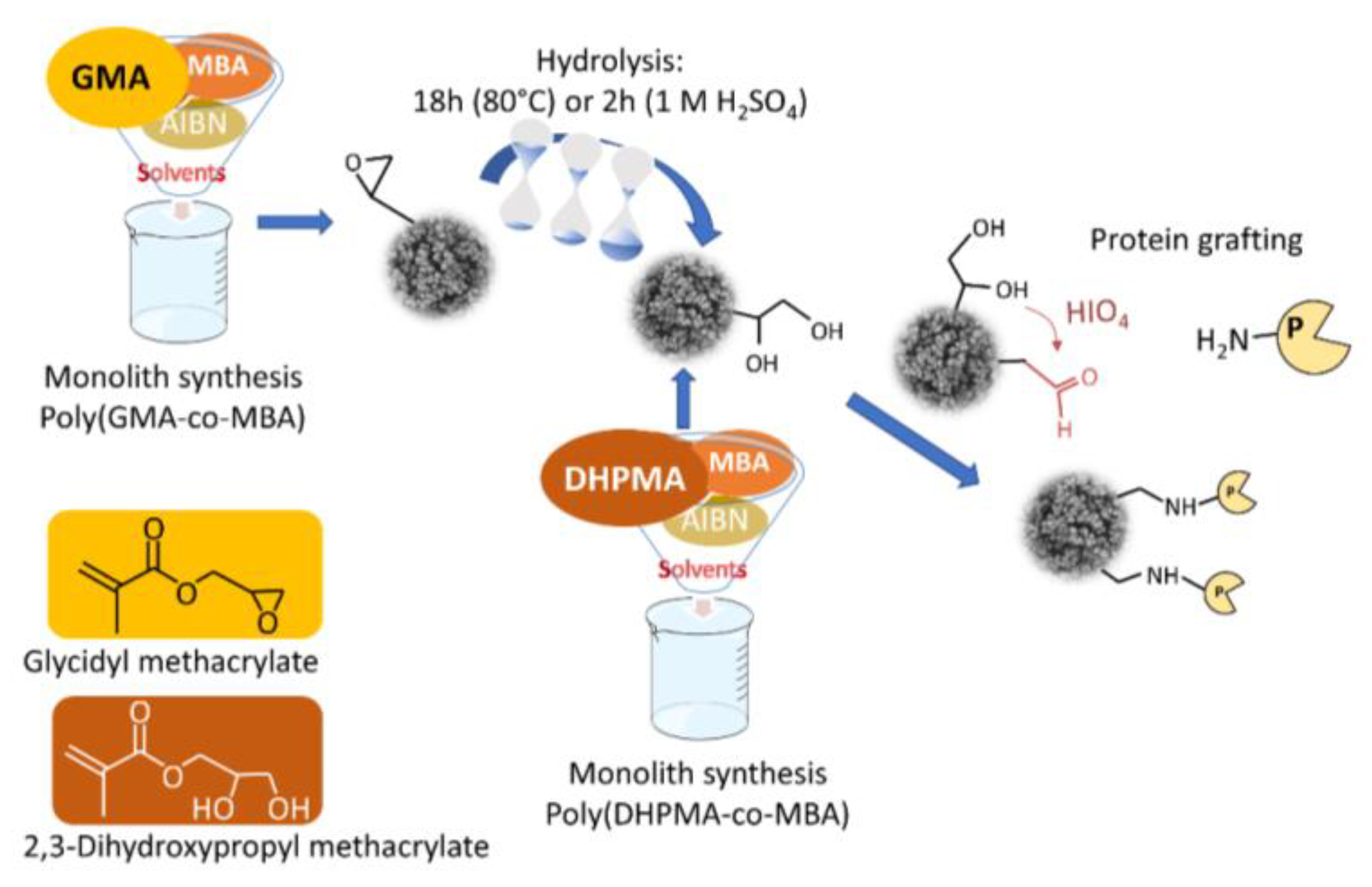
2.2.1. In-Capillary Poly(GMA-co-MBA) Monolith Synthesis
2.2.2. In-Capillary Poly(DHPMA-co-MBA) Monolith Synthesis
2.3. Column Biofunctionalization
2.3.1. Preparation of Streptavidin-Functionalized Monolithic Capillary Columns
2.3.2. Preparation of Concanavalin a Monolithic Capillary Columns
2.4. Nano-LC Experiments
2.4.1. Nano-FAC Experiments: Evaluation of Non-Specific Interactions
2.4.2. Quantification of the Number of Protein Active Binding Sites (Streptavidin or Concanavalin Columns)
3. Results and Discussion
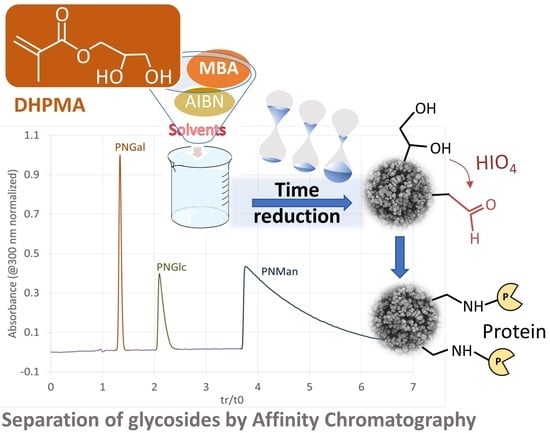
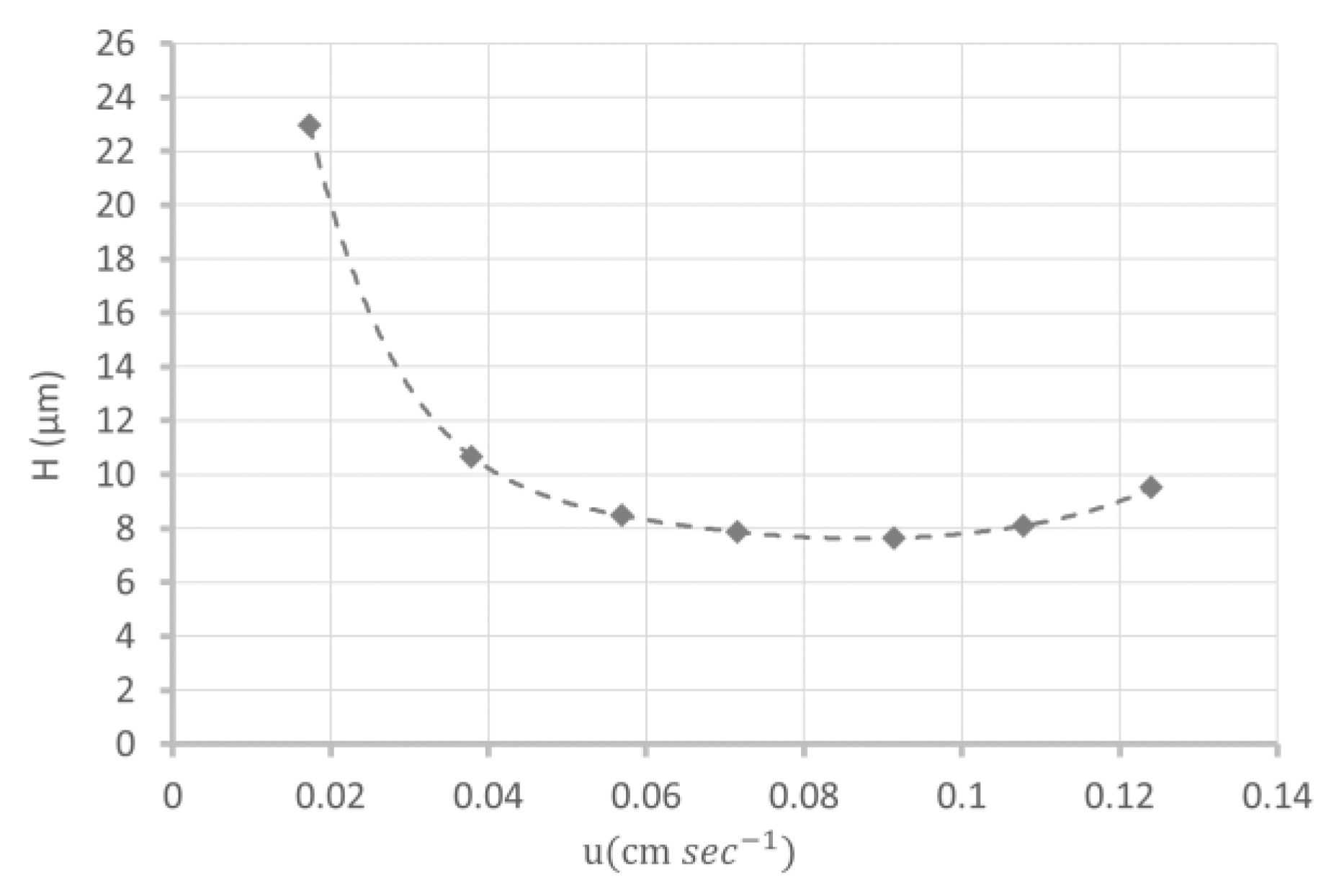
3.1. Synthesis and Characterization of DHPMA-co-MBA Monoliths
3.2. Characterization of the Number of Active Binding Sites after Protein Immobilization
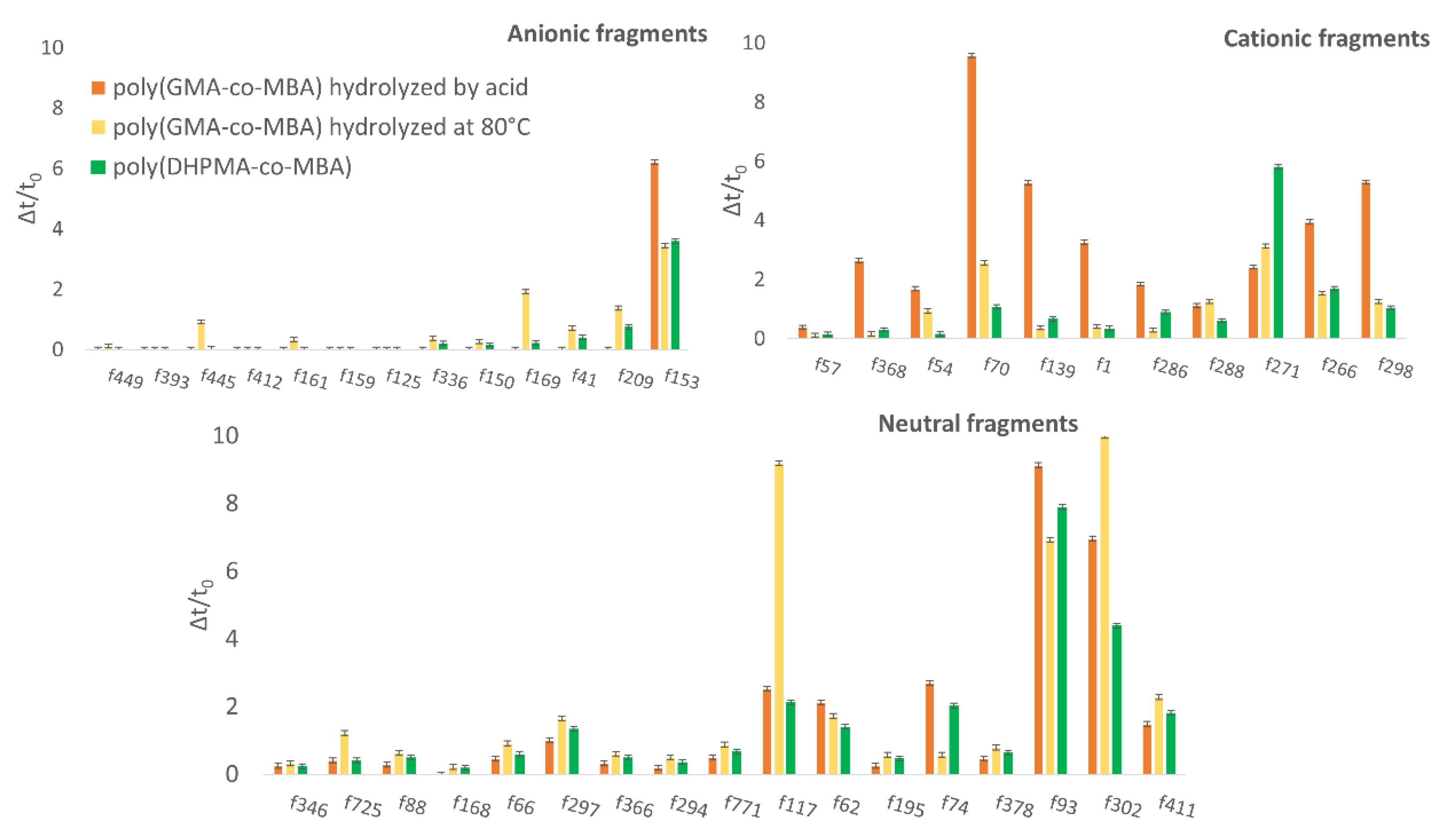
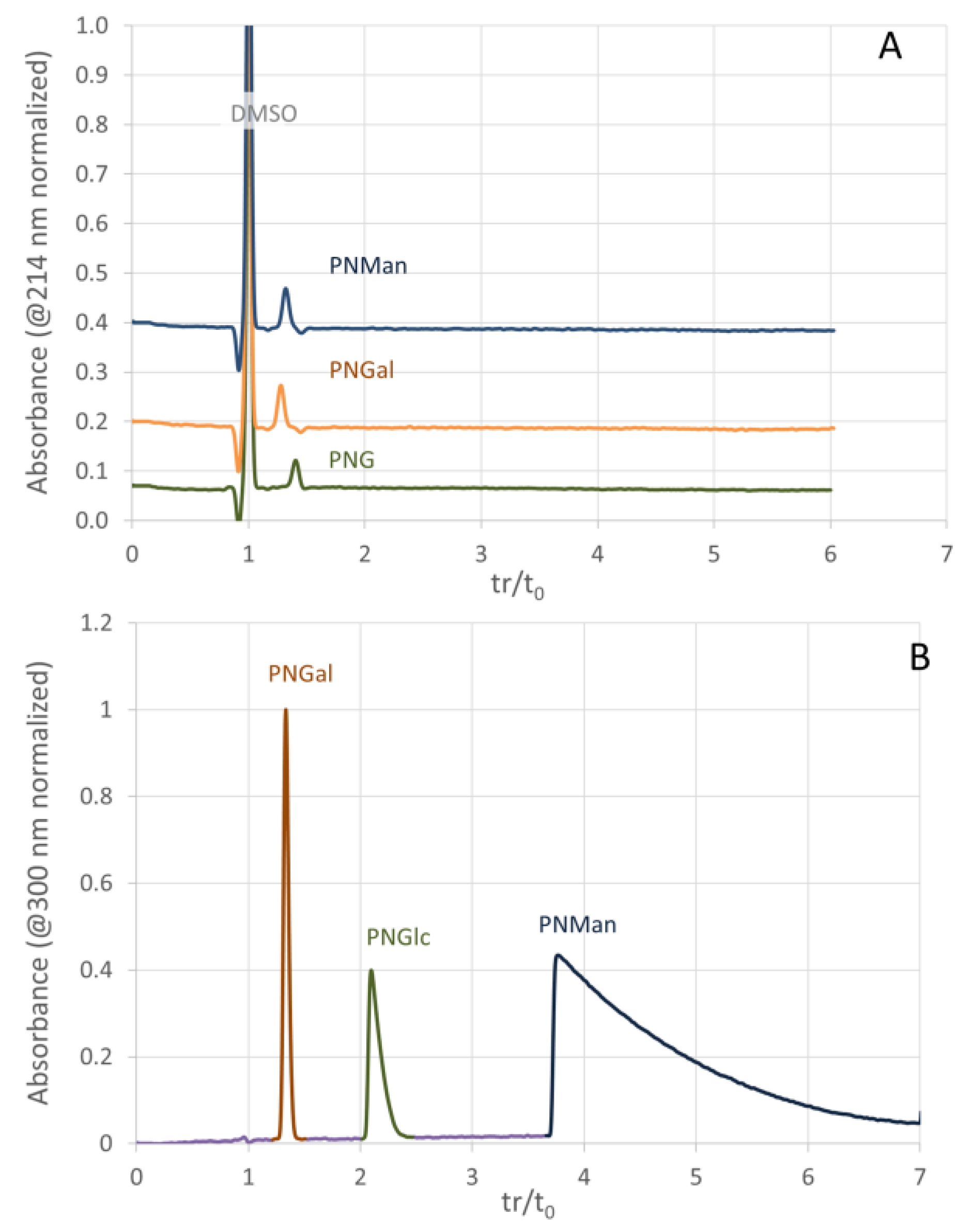
3.3. Evaluation of the Non-Specific Interactions on Poly(DHPMA-co-MBA) Monoliths and Comparison with Poly(GMA-co-MBA) Monoliths
3.4. Separation of Concanavalin Ligands by Weak Affinity Chromatography
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Pfaunmiller, E.L.; Paulemond, M.L.; Dupper, C.M.; Hage, D.S. Affinity Monolith Chromatography: A Review of Principles and Recent Analytical Applications. Anal. Bioanal. Chem. 2013, 405, 2133–2145. [Google Scholar] [CrossRef] [PubMed]
- Calleri, E.; Temporini, C.; Massolini, G. Frontal Affinity Chromatography in Characterizing Immobilized Receptors. J. Pharm. Biomed. Anal. 2011, 54, 911–925. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, E.L.; Poddar, S.; Iftekhar, S.; Suh, K.; Woolfork, A.G.; Ovbude, S.; Pekarek, A.; Walters, M.; Lott, S.; Hage, D.S. Affinity Chromatography: A Review of Trends and Developments over the Past 50 Years. J. Chromatogr. B 2020, 1157, 122332. [Google Scholar] [CrossRef] [PubMed]
- Hage, D.S. Affinity Chromatography: A Review of Clinical Applications. Clin. Chem. 1999, 45, 593–615. [Google Scholar] [CrossRef]
- Alla, A.J.; Stine, K.J. Recent Strategies for Using Monolithic Materials in Glycoprotein and Glycopeptide Analysis. Separations 2022, 9, 44. [Google Scholar] [CrossRef]
- Salim, H.; Pero-Gascon, R.; Giménez, E.; Benavente, F. On-Line Coupling of Aptamer Affinity Solid-Phase Extraction and Immobilized Enzyme Microreactor Capillary Electrophoresis-Mass Spectrometry for the Sensitive Targeted Bottom-Up Analysis of Protein Biomarkers. Anal. Chem. 2022, 94, 6948–6956. [Google Scholar] [CrossRef]
- Espina-Benitez, M.B.; Randon, J.; Demesmay, C.; Dugas, V. Development and Application of a New In-Line Coupling of a Miniaturized Boronate Affinity Monolithic Column with Capillary Zone Electrophoresis for the Selective Enrichment and Analysis of Cis-Diol-Containing Compounds. J. Chromatogr. A 2017, 1494, 65–76. [Google Scholar] [CrossRef]
- Pero-Gascon, R.; Benavente, F.; Minic, Z.; Berezovski, M.V.; Sanz-Nebot, V. On-Line Aptamer Affinity Solid-Phase Extraction Capillary Electrophoresis-Mass Spectrometry for the Analysis of Blood α-Synuclein. Anal. Chem. 2020, 92, 1525–1533. [Google Scholar] [CrossRef]
- Pfaunmiller, E.L.; Hartmann, M.; Dupper, C.M.; Soman, S.; Hage, D.S. Optimization of Human Serum Albumin Monoliths for Chiral Separations and High-Performance Affinity Chromatography. J. Chromatogr. A 2012, 1269, 198–207. [Google Scholar] [CrossRef]
- Temporini, C.; Massolini, G.; Marucci, G.; Lambertucci, C.; Buccioni, M.; Volpini, R.; Calleri, E. Development of New Chromatographic Tools Based on A2A Adenosine Receptor Subtype for Ligand Characterization and Screening by FAC-MS. Anal. Bioanal. Chem. 2013, 405, 837–845. [Google Scholar] [CrossRef]
- Lecas, L.; Hartmann, L.; Caro, L.; Mohamed-Bouteben, S.; Raingeval, C.; Krimm, I.; Wagner, R.; Dugas, V.; Demesmay, C. Miniaturized Weak Affinity Chromatography for Ligand Identification of Nanodiscs-Embedded G-Protein Coupled Receptors. Anal. Chim. Acta 2020, 1113, 26–35. [Google Scholar] [CrossRef]
- Lecas, L.; Randon, J.; Berthod, A.; Dugas, V.; Demesmay, C. Monolith Weak Affinity Chromatography for Μg-Protein-Ligand Interaction Study. J. Pharm. Biomed. Anal. 2019, 166, 164–173. [Google Scholar] [CrossRef] [PubMed]
- Hage, D.S.; Chen, J. Quantitative Affinity Chromatography: Practical Aspects. In Handbook of Affinity Chromatography; CRC Press: Boca Raton, FL, USA, 2006; pp. 596–628. [Google Scholar]
- Kasai, K. Frontal Affinity Chromatography: An Excellent Method of Analyzing Weak Biomolecular Interactions Based on a Unique Principle. Biochim. Biophys. Biochim. Biophys. Acta-Gen. Subj. 2021, 1865, 129761. [Google Scholar] [CrossRef] [PubMed]
- Gunasena, D.N.; El Rassi, Z. Hydrophilic Diol Monolith for the Preparation of Immuno-Sorbents at Reduced Nonspecific Interactions. J. Sep. Sci. 2011, 34, 2097–2105. [Google Scholar] [CrossRef] [PubMed]
- Khadka, S.; El Rassi, Z. Postpolymerization Modification of a Hydroxy Monolith Precursor. Part III. Activation of Poly(Hydroxyethyl Methacrylate-Co-Pentaerythritol Triacrylate) Monolith with Epoxy Functionalities Followed by Bonding of Glycerol, Polyamines, and Hydroxypropyl-β-Cyclodextrin for Hydrophilic Interaction and Chiral Capillary Electrochromatography. Electrophoresis 2016, 37, 3178–3185. [Google Scholar] [CrossRef] [PubMed]
- Poddar, S.; Sharmeen, S.; Hage, D.S. Affinity Monolith Chromatography: A Review of General Principles and Recent Developments. Electrophoresis 2021, 42, 2577–2598. [Google Scholar] [CrossRef] [PubMed]
- Svec, F. Porous Polymer Monoliths: Amazingly Wide Variety of Techniques Enabling Their Preparation. J. Chromatogr. A 2010, 1217, 902–924. [Google Scholar] [CrossRef]
- Li, Z.; Rodriguez, E.; Azaria, S.; Pekarek, A.; Hage, D.S. Affinity Monolith Chromatography: A Review of General Principles and Applications. Electrophoresis 2017, 38, 2837–2850. [Google Scholar] [CrossRef]
- GIL, J.; Krimm, I.; Dugas, V.; Demesmay, C. Preparation of Miniaturized Hydrophilic Affinity Monoliths: Towards a Reduction of Non-Specific Interactions and an Increased Target Protein Density. J. Chromatogr. A 2023, 1687, 463670. [Google Scholar] [CrossRef]
- Jancǒ, M.; Xie, S.; Peterson, D.S.; Allington, R.W.; Svec, F.; Fréchet, J.M.J. Effect of Porosity and Surface Chemistry on the Characterization of Synthetic Polymers by HPLC Using Porous Polymer Monolithic Columns. J. Sep. Sci. 2002, 25, 909–916. [Google Scholar] [CrossRef]
- Selvaraju, S.; El Rassi, Z. Tandem Lectin Affinity Chromatography Monolithic Columns with Surface Immobilised Concanavalin A, Wheat Germ Agglutinin and Ricinus Communis Agglutinin-I for Capturing Sub-Glycoproteomics from Breast Cancer and Disease-Free Human Sera. J. Sep. Sci. 2012, 35, 1785–1795. [Google Scholar] [CrossRef] [PubMed]
- Gottardini, A.; Netter, C.; Dugas, V.; Demesmay, C. Two Original Experimental Setups for Staircase Frontal Affinity Chromatography at the Miniaturized Scale. Anal. Chem. 2021, 93, 16981–16986. [Google Scholar] [CrossRef] [PubMed]
- Gray, R.D.; Glew, R.H. The Kinetics of Carbohydrate Binding to Concanavalin A. J. Biol. Chem. 1973, 248, 7547–7551. [Google Scholar] [CrossRef] [PubMed]
- Oda, Y.; Kasai, K.; Ishii, S. Studies on the Specific Interaction of Concanavalin A and Saccharides by Affinity Chromatography. Application of Quantitative Affinity Chromatography to a Multivalent System. J. Biochem. 1981, 89, 285–296. [Google Scholar] [CrossRef]
- Ohyama, Y.; Kasai, K.; Nomoto, H.; Inoue, Y. Frontal Affinity Chromatography of Ovalbumin Glycoasparagines on a Concanavalin A-Sepharose Column. A Quantitative Study of the Binding Specificity of the Lectin. J. Biol. Chem. 1985, 260, 6882–6887. [Google Scholar] [CrossRef]
- Skudas, R.; Grimes, B.A.; Thommes, M.; Unger, K.K. Flow-through Pore Characteristics of Monolithic Silicas and Their Impact on Column Performance in High-Performance Liquid Chromatography. J. Chromatogr. A 2009, 1216, 2625–2636. [Google Scholar] [CrossRef]
- Svec, F. Quest for Organic Polymer-Based Monolithic Columns Affording Enhanced Efficiency in High Performance Liquid Chromatography Separations of Small Molecules in Isocratic Mode. J. Chromatogr. A 2012, 1228, 250–262. [Google Scholar] [CrossRef]
- Aydoğan, C.; Beltekin, B.; Demir, N.; Yurt, B.; El Rassi, Z. Nano-Liquid Chromatography with a New Monolithic Column for the Analysis of Coenzyme Q10 in Pistachio Samples. Molecules 2023, 28, 1423. [Google Scholar] [CrossRef]

| Weight (mg) | % | |
|---|---|---|
| DHPMA | 120 | 6.38 |
| MBA | 80 | 4.26 |
| AIBN | 2 | 1 (of monomers) |
| DMSO | 888 | 89.4 |
| 1,4-butanediol | 395 | |
| Dodecanol | 397 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gil, J.; Passalacqua, G.; Deloche, A.; Vidal, F.-X.; Dugas, V.; Demesmay, C. Optimization of the Preparation of Hydrophilic Poly(DHPMA-co-MBA) Monolithic Capillary Columns: A New Support for Affinity Chromatography. Separations 2023, 10, 437. https://doi.org/10.3390/separations10080437
Gil J, Passalacqua G, Deloche A, Vidal F-X, Dugas V, Demesmay C. Optimization of the Preparation of Hydrophilic Poly(DHPMA-co-MBA) Monolithic Capillary Columns: A New Support for Affinity Chromatography. Separations. 2023; 10(8):437. https://doi.org/10.3390/separations10080437
Chicago/Turabian StyleGil, Julie, Gaëtan Passalacqua, Adrien Deloche, François-Xavier Vidal, Vincent Dugas, and Claire Demesmay. 2023. "Optimization of the Preparation of Hydrophilic Poly(DHPMA-co-MBA) Monolithic Capillary Columns: A New Support for Affinity Chromatography" Separations 10, no. 8: 437. https://doi.org/10.3390/separations10080437
APA StyleGil, J., Passalacqua, G., Deloche, A., Vidal, F.-X., Dugas, V., & Demesmay, C. (2023). Optimization of the Preparation of Hydrophilic Poly(DHPMA-co-MBA) Monolithic Capillary Columns: A New Support for Affinity Chromatography. Separations, 10(8), 437. https://doi.org/10.3390/separations10080437