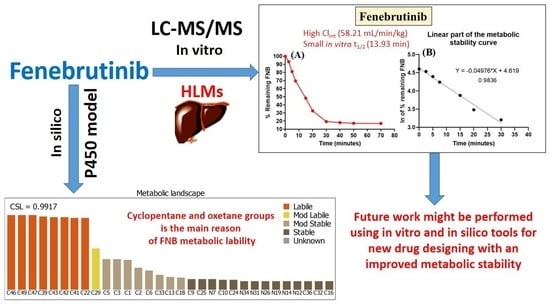
Development of a Fast and Sensitive UPLC–MS/MS Analytical Methodology for Fenebrutinib Estimation in Human Liver Microsomes: In Vitro and In Silico Metabolic Stability Evaluation
Abstract
1. Introduction
2. Materials, Instruments and Methods
2.1. Materials and Instruments
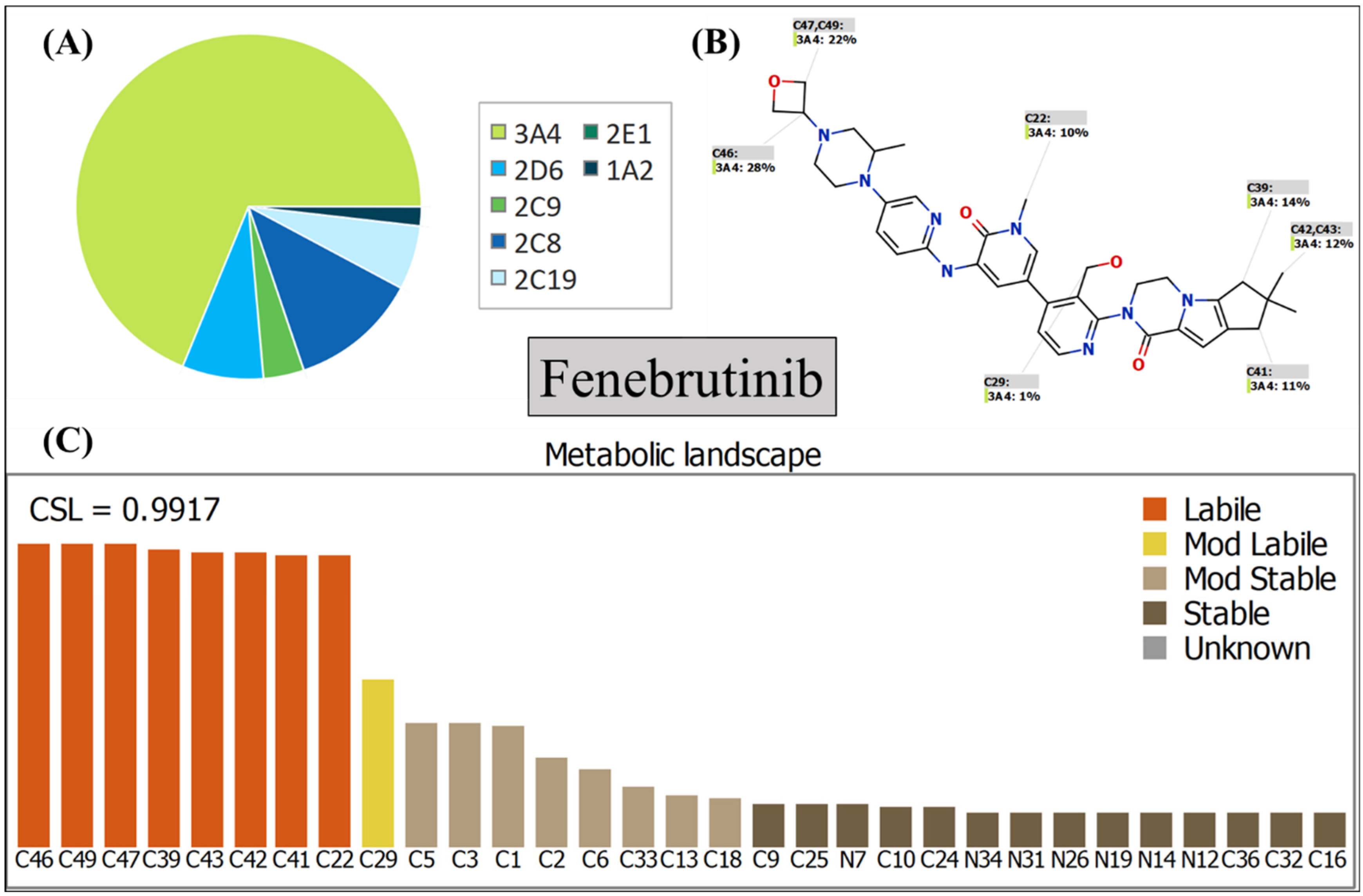
2.2. In Silico Assessment of FNB Metabolic Lability
2.3. UPLC-MS/MS Adjusted Characteristics
2.4. FNB Working Solutions
2.5. FNB Plotted Calibration Standards
2.6. Protein Precipitation Methodology for Extracting FNB and SVB from the HLMs Incubation Matrix
2.7. Validation Parameters of the Developed UPLC-MS/MS Methodology
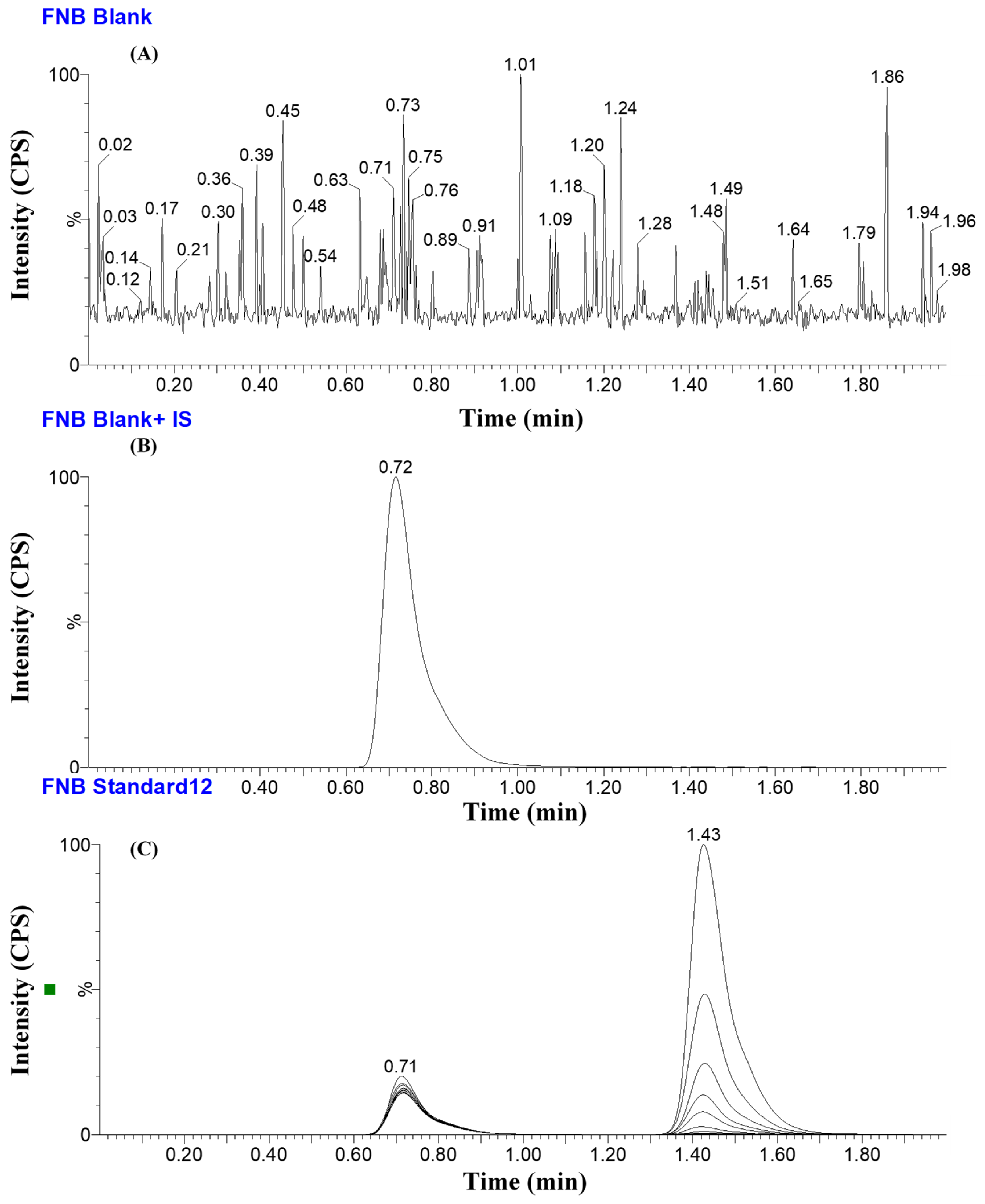
2.7.1. Specificity of the Developed UPLC-MS/MS Method
2.7.2. Sensitivity and Linearity of the Developed UPLC-MS/MS Method
2.7.3. Precision and Accuracy of the Established UPLC-MS/MS Analytical Method
2.7.4. Extraction Recovery and Matrix Effect of the Developed UPLC-MS/MS Method
2.8. Stability of the FNB Stack Samples
2.9. In Vitro Determination of FNB Metabolic Stability
3. Results and Discussion
3.1. In Silico Metabolic Stability Assessment of FNB Using P450 Metabolic Model
3.2. Establishing Steps of UPLC–MS/MS Methodology
3.3. Validation Parameters of the Current UPLC-MS/MS Analytical Methodology
3.3.1. Specificity of the Developed UPLC-MS/MS Analytical Methodology
3.3.2. Linearity and Sensitivity of the Developed UPLC-MS/MS Analytical Methodology
3.3.3. Precision and Accuracy of the UPLC-MS/MS Analytical Method
3.3.4. Matrix Effects of HLMs and FNB Extraction Recovery
3.3.5. Stability of FNB in DMSO and HLMs Matrix
3.4. In Vitro Metabolic Incubations of FNB
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Ethical Approval
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Isenberg, D.; Furie, R.; Jones, N.S.; Guibord, P.; Galanter, J.; Lee, C.; McGregor, A.; Toth, B.; Rae, J.; Hwang, O.; et al. Efficacy, Safety, and Pharmacodynamic Effects of the Bruton’s Tyrosine Kinase Inhibitor Fenebrutinib (GDC-0853) in Systemic Lupus Erythematosus: Results of a Phase II, Randomized, Double-Blind, Placebo-Controlled Trial. Arthritis Rheumatol. 2021, 73, 1835–1846. [Google Scholar] [CrossRef] [PubMed]
- Weber, M.; Harp, C.; Bremer, M.; Goodyear, A.; Crawford, J.; Johnson, A.; Bar-Or, A. Fenebrutinib demonstrates the highest potency of bruton tyrosine kinase inhibitors (BTKis) in phase 3 clinical development for multiple sclerosis (MS)(4437). Neurology 2021, 96, 4473. [Google Scholar]
- Bag-Ozbek, A.; Hui-Yuen, J.S. Emerging B-cell therapies in systemic lupus erythematosus. Ther. Clin. Risk Manag. 2021, 17, 39–54. [Google Scholar] [CrossRef] [PubMed]
- Crawford, J.J.; Zhang, H. Discovery and Development of Non-Covalent, Reversible Bruton’s Tyrosine Kinase Inhibitor Fenebrutinib (GDC-0853). In Complete Accounts of Integrated Drug Discovery and Development: Recent Examples from the Pharmaceutical Industry Volume 2; ACS Publications: Washington, DC, USA, 2019; pp. 239–266. [Google Scholar]
- Houston, J.B. Utility of in vitro drug metabolism data in predicting in vivo metabolic clearance. Biochem. Pharm. 1994, 47, 1469–1479. [Google Scholar] [CrossRef] [PubMed]
- Obach, R.S.; Baxter, J.G.; Liston, T.E.; Silber, B.M.; Jones, B.C.; MacIntyre, F.; Rance, D.J.; Wastall, P. The prediction of human pharmacokinetic parameters from preclinical and in vitro metabolism data. J. Pharm. Exp. 1997, 283, 46–58. [Google Scholar]
- Attwa, M.W.; Kadi, A.A.; Darwish, H.W.; Amer, S.M.; Alrabiah, H. A reliable and stable method for the determination of foretinib in human plasma by LC-MS/MS: Application to metabolic stability investigation and excretion rate. Eur. J. Mass Spectrom. 2018, 24, 344–351. [Google Scholar] [CrossRef]
- Darwish, H.W.; Kadi, A.A.; Attwa, M.W.; Almutairi, H.S. Investigation of metabolic stability of the novel ALK inhibitor brigatinib by liquid chromatography tandem mass spectrometry. Clin. Chim. Acta 2018, 480, 180–185. [Google Scholar] [CrossRef]
- Attwa, M.W.; Darwish, H.W.; Alhazmi, H.A.; Kadi, A.A. Investigation of metabolic degradation of new ALK inhibitor: Entrectinib by LC-MS/MS. Clin. Chim. Acta 2018, 485, 298–304. [Google Scholar] [CrossRef]
- Amer, S.M.; Kadi, A.A.; Darwish, H.W.; Attwa, M.W. LC–MS/MS method for the quantification of masitinib in RLMs matrix and rat urine: Application to metabolic stability and excretion rate. Chem. Cent. J. 2017, 11, 136. [Google Scholar] [CrossRef]
- Jones, N.S.; Yoshida, K.; Salphati, L.; Kenny, J.R.; Durk, M.R.; Chinn, L.W. Complex DDI by fenebrutinib and the use of transporter endogenous biomarkers to elucidate the mechanism of DDI. Clin. Pharmacol. Ther. 2020, 107, 269–277. [Google Scholar] [CrossRef]
- Durk, M.R.; Jones, N.S.; Liu, J.; Nagapudi, K.; Mao, C.; Plise, E.G.; Wong, S.; Chen, J.Z.; Chen, Y.; Chinn, L.W.; et al. Understanding the Effect of Hydroxypropyl-β-Cyclodextrin on Fenebrutinib Absorption in an Itraconazole–Fenebrutinib Drug–Drug Interaction Study. Clin. Pharmacol. Ther. 2020, 108, 1224–1232. [Google Scholar] [CrossRef]
- Estupiñán, H.Y.; Berglöf, A.; Zain, R.; Smith, C.E. Comparative analysis of BTK inhibitors and mechanisms underlying adverse effects. Front. Cell Dev. Biol. 2021, 9, 630942. [Google Scholar] [CrossRef] [PubMed]
- Krishna, M.V.; Padmalatha, K.; Madhavi, G. In Vitro Metabolic Stability of Drugs and Applications of LC-MS in Metabolite Profiling. In Drug Metabolism; Dunnington, K., Ed.; IntechOpen: Rijeka, Croatia, 2021. [Google Scholar]
- Wadhwa, G.; Krishna, K.V.; Taliyan, R.; Tandon, N.; Yadav, S.S.; Katiyar, C.; Dubey, S.K. Pre-clinical pharmacokinetic and pharmacodynamic modelling study of 4-hydroxyisoleucine using validated ultra-performance liquid chromatography-tandem mass spectrometry. RSC Adv. 2020, 10, 5525–5532. [Google Scholar] [CrossRef] [PubMed]
- Baranczewski, P.; Stańczak, A.; Sundberg, K.; Svensson, R.; Wallin, A.; Jansson, J.; Garberg, P.; Postlind, H. Introduction to in vitro estimation of metabolic stability and drug interactions of new chemical entities in drug discovery and development. Pharm. Rep. 2006, 58, 453–472. [Google Scholar]
- Tyzack, J.D.; Kirchmair, J. Computational methods and tools to predict cytochrome P450 metabolism for drug discovery. Chem. Biol. Drug Des. 2019, 93, 377–386. [Google Scholar] [CrossRef]
- Attwa, M.W.; AlRabiah, H.; Alsibaee, A.M.; Abdelhameed, A.S.; Kadi, A.A. An UPLC–ESI–MS/MS Bioanalytical Methodology for the Quantification of Gilteritinib in Human Liver Microsomes: Application to In Vitro and In Silico Metabolic Stability Estimation. Separations 2023, 10, 278. [Google Scholar] [CrossRef]
- Alrabiah, H.; Kadi, A.A.; Attwa, M.W.; Abdelhameed, A.S. A simple liquid chromatography-tandem mass spectrometry method to accurately determine the novel third-generation EGFR-TKI naquotinib with its applicability to metabolic stability assessment. Rsc Adv. 2019, 9, 4862–4869. [Google Scholar] [CrossRef]
- Kadi, A.A.; Darwish, H.W.; Abuelizz, H.A.; Alsubi, T.A.; Attwa, M.W. Identification of reactive intermediate formation and bioactivation pathways in Abemaciclib metabolism by LC-MS/MS: In vitro metabolic investigation. R. Soc. Open Sci. 2019, 6, 181714. [Google Scholar] [CrossRef] [PubMed]
- Attwa, M.W.; Kadi, A.A.; Abdelhameed, A.S.; Alhazmi, H.A. Metabolic stability assessment of new parp inhibitor talazoparib using validated lc–ms/ms methodology: In silico metabolic vulnerability and toxicity studies. Drug Des. Dev. Ther. 2020, 14, 783–793. [Google Scholar] [CrossRef]
- Busby, W.F., Jr.; Ackermann, J.M.; Crespi, C.L. Effect of methanol, ethanol, dimethyl sulfoxide, and acetonitrile on in vitro activities of cDNA-expressed human cytochromes P-450. Drug Metab. Dispos. 1999, 27, 246–249. [Google Scholar]
- Störmer, E.; Roots, I.; Brockmöller, J. Benzydamine N-oxidation as an index reaction reflecting FMO activity in human liver microsomes and impact of FMO3 polymorphisms on enzyme activity. Br. J. Clin. Pharm. 2000, 50, 553–561. [Google Scholar] [CrossRef] [PubMed]
- Fouin-Fortunet, H.; Tinel, M.; Descatoire, V.; Letteron, P.; Larrey, D.; Geneve, J.; Pessayre, D. Inactivation of cytochrome P-450 by the drug methoxsalen. J. Pharm. Exp. 1986, 236, 237–247. [Google Scholar]
- United State of America–Food and Drug Administration. Bioanalytical Method Validation Guidance for Industry. 2018. Available online: https://www.fda.gov/ucm/groups/fdagov-public/@fdagov-drugsgen/documents/document/ucm070107.pdf (accessed on 17 February 2022).
- Abdelhameed, A.S.; Attwa, M.W.; Kadi, A.A. Identification of Iminium Intermediates Generation in the Metabolism of Tepotinib Using LC-MS/MS: In Silico and Practical Approaches to Bioactivation Pathway Elucidation. Molecules 2020, 25, 5004. [Google Scholar] [CrossRef] [PubMed]
- McNaney, C.A.; Drexler, D.M.; Hnatyshyn, S.Y.; Zvyaga, T.A.; Knipe, J.O.; Belcastro, J.V.; Sanders, M. An automated liquid chromatography-mass spectrometry process to determine metabolic stability half-life and intrinsic clearance of drug candidates by substrate depletion. Assay Drug Dev. Technol. 2008, 6, 121–129. [Google Scholar] [CrossRef]
- Słoczyńska, K.; Gunia-Krzyżak, A.; Koczurkiewicz, P.; Wójcik-Pszczoła, K.; Żelaszczyk, D.; Popiół, J.; Pękala, E. Metabolic stability and its role in the discovery of new chemical entities. Acta Pharm. 2019, 69, 345–361. [Google Scholar] [CrossRef]
- Tan, L.; Kirchmair, J. Software for Metabolism Prediction. In Drug Metabolism Prediction; Kirchmair, J., Ed.; Wiley-VCH Verlag GmbH & Co. KGaA: Weinheim, Germany, 2014; pp. 27–52. [Google Scholar]
- Hunt, P.A.; Segall, M.D.; Tyzack, J.D. WhichP450: A multi-class categorical model to predict the major metabolising CYP450 isoform for a compound. J. Comput. Mol. Des. 2018, 32, 537–546. [Google Scholar] [CrossRef]
- Shin, Y.G.; Le, H.; Khojasteh, C.; Hop, C.E.C.A. Comparison of metabolic soft spot predictions of CYP3A4, CYP2C9 and CYP2D6 substrates using MetaSite and StarDrop. Comb. Chem. High Throughput Screen. 2011, 14, 811–823. [Google Scholar] [CrossRef]
- Leahy, D.E. Integrating invitro ADMET data through generic physiologically based pharmacokinetic models. Expert Opin. Drug Metab. Toxicol. 2006, 2, 619–628. [Google Scholar] [CrossRef]
- Attwa, M.W.; Al-Shakliah, N.S.; AlRabiah, H.; Kadi, A.A.; Abdelhameed, A.S. Estimation of zorifertinib metabolic stability in human liver microsomes using LC–MS/MS. J. Pharm. Biomed. Anal. 2022, 211, 114626. [Google Scholar] [CrossRef]
- Attwa, M.W.; Abdelhameed, A.S.; Alsaif, N.A.; Kadi, A.A.; AlRabiah, H. A validated LC-MS/MS analytical method for the quantification of pemigatinib: Metabolic stability evaluation in human liver microsomes. RSC Adv. 2022, 12, 20387–20394. [Google Scholar] [CrossRef]
- Mostafa, G.A.E.; Kadi, A.A.; AlMasoud, N.; Attwa, M.W.; Al-Shakliah, N.S.; AlRabiah, H. LC-MS/MS method for the quantification of the anti-cancer agent infigratinib: Application for estimation of metabolic stability in human liver microsomes. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2021, 1179, 122806. [Google Scholar] [CrossRef] [PubMed]
- Attwa, M.W.; Abdelhameed, A.S.; Al-Shakliah, N.S.; Kadi, A.A. Lc-ms/ms estimation of the anti-cancer agent tandutinib levels in human liver microsomes: Metabolic stability evaluation assay. Drug Des. Dev. Ther. 2020, 14, 4439–4449. [Google Scholar] [CrossRef] [PubMed]

| Acquity UPLC Chromatographic System (H10UPH) | Acquity TQD MS Mass Analyzer System (QBB1203) | |||
|---|---|---|---|---|
| Isocratic mobile phase | 50% acetonitrile | ESI | Cone gas: 100 L/H | |
| 0.1% HCOOH | 50% | Positive ESI | ||
| pH: 3.2 | ||||
| Injection volume: 5.0 μL | Drying gas: nitrogen of high purity at 650 L/H flow rate at 350 °C | |||
| Flow rate: 0.2 mL/min. | The extractor voltage: 3.0 (V) | |||
| ZORBAX Eclipse plus-C18 column | 2.1 mm i.d. | The RF lens voltage: 0.1 (V) | ||
| 1.8 μm particle size | Capillary voltage: 4 KV | |||
| T: 22.0 ± 2.0 °C | Mode | MRM | ||
| 50.0 mm long | Collision cell | Argon gas (high purity) at 0.15 mL/min. | ||
| Time | Retention Time | Analyte | MRM Transitions | ||
|---|---|---|---|---|---|
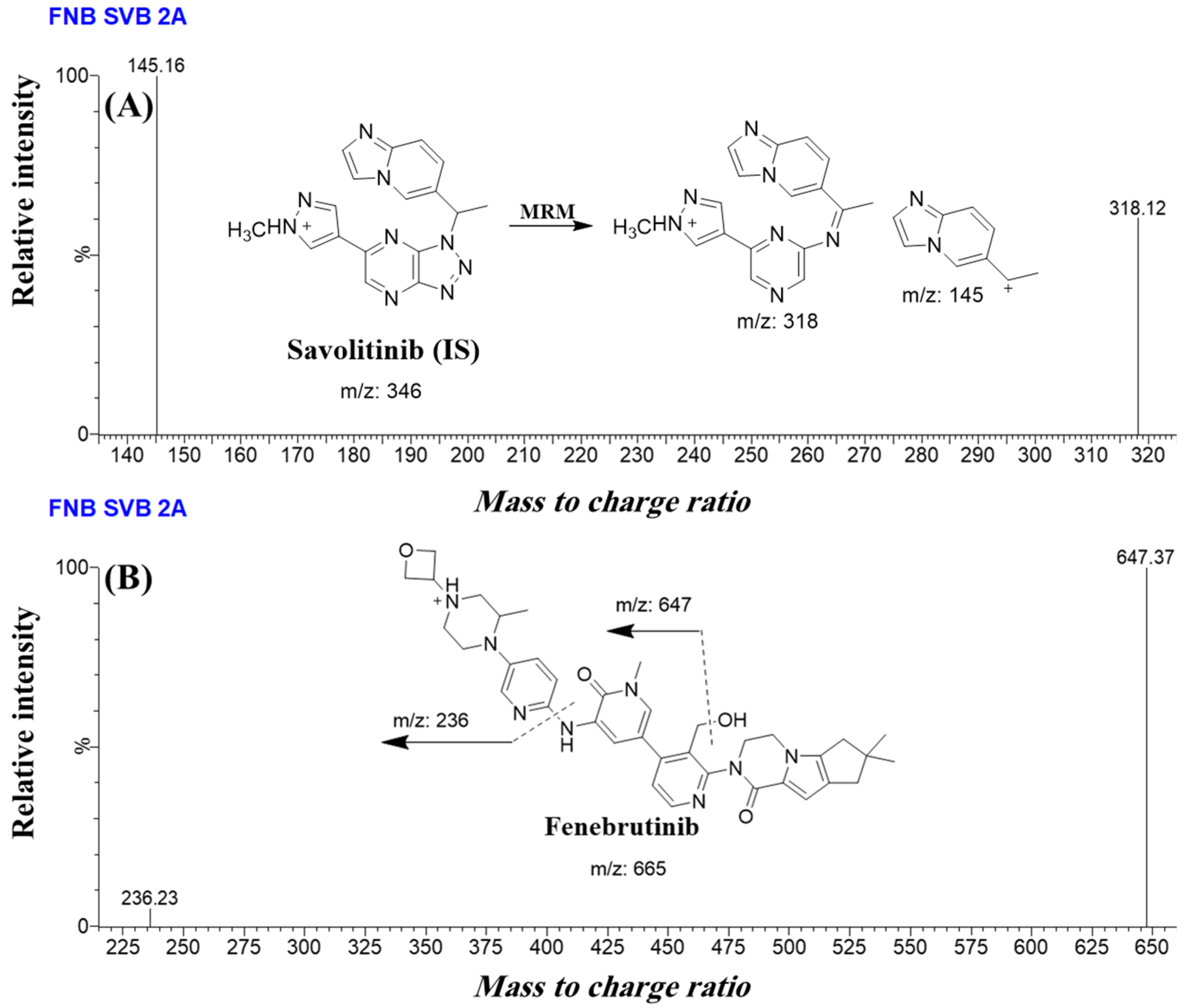
| Mass spectra segment | 0.0 to 1.2 min | SVB (0.71 min) | Savolitinib (SVB, IS) | Qualification traces (m/z) | 346.1 → 318.1 CV a: 30 and CE b: 10 |
| Quantification traces (m/z) | 346.1 → 145.2 CV: 30 and CE: 16 | ||||
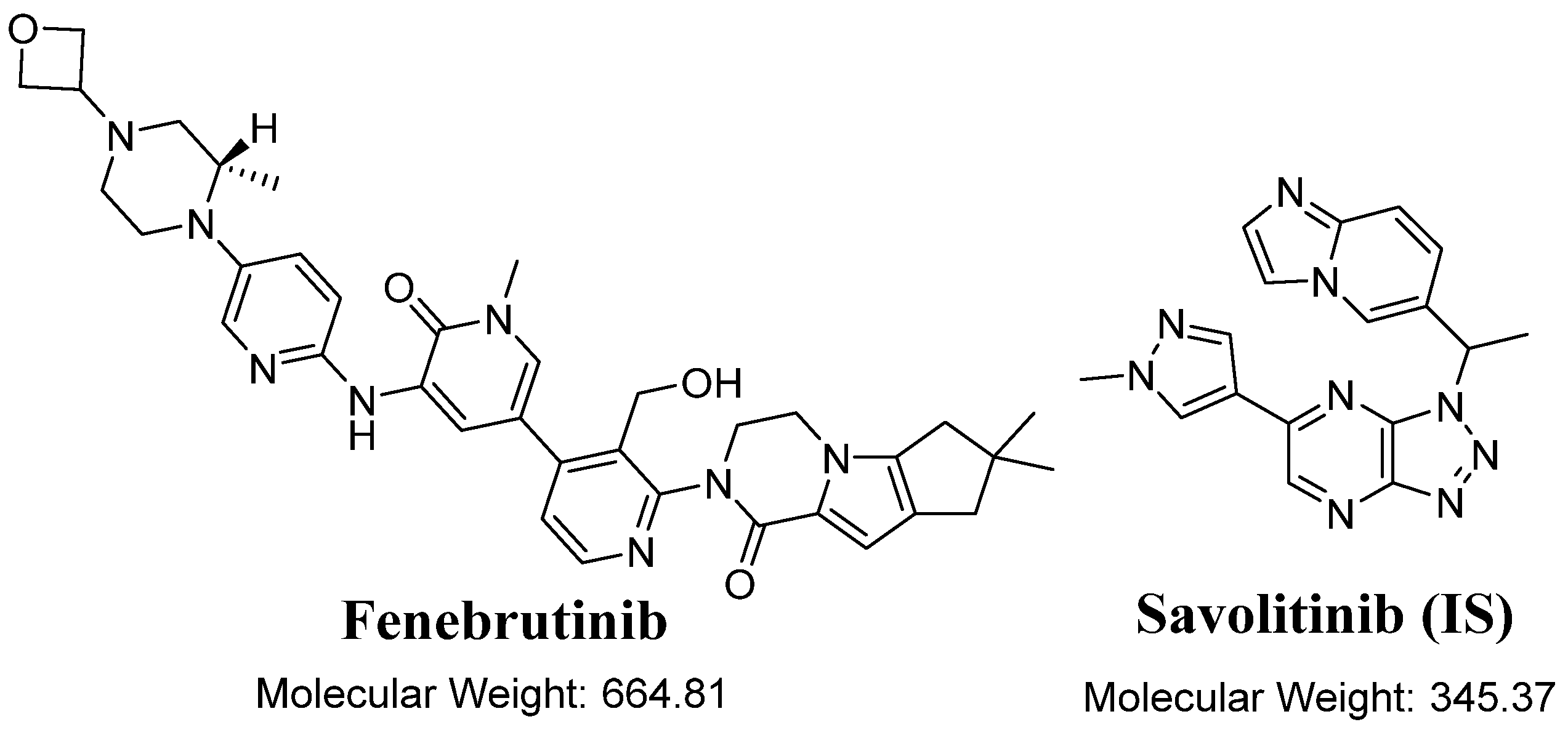
| 1.2 to 2.0 min | FNB (1.43 min) | Analyte: Fenebrutinib (FNB, IS) | Qualification traces (m/z) | 665.3 → 647.4 CV: 58 and CE: 26 | |
| Quantification traces (m/z) | 665.3 → 236.2 CV: 58 and CE: 58 | ||||
| Analytes | Methanol | ACN | Solid Phase Extraction | Protein Precipitation | C8 Column | C18 Column |
|---|---|---|---|---|---|---|
| SVB | 0.85 min | 0.71 min | Low recovery | High recovery | 1.23 min | 0.71 min |
| Tailed peaks | Good peak shape | Not precise | Precise results | Tailed peaks | Perfect peak shape | |
| FNB | 1.15 min | 1.43 min | Low recovery | High recovery | 1.38 min | 1.43 min |
| Overlapped | Good peak shape | Not precise | Precise results | Perfect peak shape | Perfect peak shape |
| FNB (ng/mL) | Average | SD | RSD % | RE % | Recovery |
|---|---|---|---|---|---|
| 1(LLQC) | 1.14 | 0.01 | 0.88 | 14.00 | 114.00 |
| 3(LQC) | 2.88 | 0.06 | 1.92 | −3.99 | 96.01 |
| 15 | 14.75 | 0.49 | 3.33 | −1.65 | 98.35 |
| 30 | 30.60 | 1.34 | 4.37 | 2.00 | 102.00 |
| 50 | 49.01 | 0.80 | 1.64 | −1.97 | 98.03 |
| 150 | 150.52 | 2.01 | 1.34 | 0.35 | 100.35 |
| 300 | 299.62 | 2.92 | 0.98 | −0.13 | 99.87 |
| 500 | 495.66 | 3.24 | 0.65 | −0.87 | 99.13 |
| 900 (MQC) | 890.94 | 4.76 | 0.53 | −1.01 | 98.99 |
| 1500 | 1512.40 | 5.13 | 0.34 | 0.83 | 100.83 |
| 2400 (HQC) | 2411.73 | 12.64 | 0.52 | 0.49 | 100.49 |
| 3000 | 3021.05 | 4.88 | 0.16 | 0.70 | 100.70 |
| % Recovery | 100.73 ± 4.47 |
| FNB (ng/mL) | Intra-Day Assay | Inter-Day Assay | ||||||
|---|---|---|---|---|---|---|---|---|
| 1 | 3 | 900 | 2400 | 1 | 3 | 900 | 2400 | |
| Average | 1.14 | 2.88 | 890.94 | 2411.73 | 1.13 | 2.90 | 892.63 | 2412.10 |
| SD | 0.01 | 0.06 | 4.76 | 12.64 | 0.01 | 0.11 | 7.16 | 37.93 |
| Precision (%RSD) | 0.88 | 1.92 | 0.53 | 0.52 | 1.25 | 3.83 | 0.80 | 1.57 |
| Accuracy (% RE) | 14.00 | −3.99 | −1.01 | 0.49 | 13.00 | −3.27 | −0.82 | 0.50 |
| Recovery (%) | 114.00 | 96.01 | 98.99 | 100.49 | 113.00 | 96.73 | 99.18 | 100.50 |
| Stability Parameter | LQC (3.0) | HQC (2400.0) | LQC (3.0) | HQC (2400.0) | LQC (3.0) | HQC (2400.0) | LQC (3.0) | HQC (2400.0) |
|---|---|---|---|---|---|---|---|---|
| Mean | SD | RSD (%) | Accuracy (%) | |||||
| Short-Term Stability (4 h at room temperature) | 2.95 | 2420.97 | 0.09 | 18.63 | 2.92 | 0.77 | −1.67 | 0.87 |
| Freeze–Thaw Stability (three cycles at −80 °C) | 2.94 | 2416.77 | 0.04 | 28.02 | 1.47 | 1.16 | −2.11 | 0.70 |
| Auto-sampler Stability (24 h at 15 °C) | 2.96 | 2408.35 | 0.03 | 27.54 | 0.90 | 1.14 | −1.22 | 0.35 |
| Long-Term Stability (−80 °C for 28 d) | 2.94 | 2415.30 | 0.04 | 22.73 | 1.27 | 0.94 | −2.17 | 0.64 |
| Time (min.) | Mean a (ng/mL) | X b | ln X | Linearity Parameters |
|---|---|---|---|---|
| 0 | 623.45 | 100.00 | 4.61 | Regression equation: y = −0.04976x + 4.619 |
| 2.5 | 578.23 | 93.26 | 4.54 | |
| 5 | 517.38 | 81.09 | 4.40 | R2 = 0.9836 |
| 7.5 | 443.97 | 69.59 | 4.24 | |
| 15 | 308.74 | 48.39 | 3.88 | Slope: −0.04976 |
| 20 | 207.98 | 32.60 | 3.48 | |
| 30 | 123.85 | 19.41 | 2.97 | t1/2: 13.93 min and |
| 40 | 115.75 | 18.14 | 2.90 | Clint: 58.21 mL/min/kg |
| 50 | 110.38 | 17.30 | 2.85 | |
| 70 | 108.87 | 17.06 | 2.84 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Attwa, M.W.; Alsibaee, A.M.; Aljohar, H.I.; Abdelhameed, A.S.; Kadi, A.A. Development of a Fast and Sensitive UPLC–MS/MS Analytical Methodology for Fenebrutinib Estimation in Human Liver Microsomes: In Vitro and In Silico Metabolic Stability Evaluation. Separations 2023, 10, 302. https://doi.org/10.3390/separations10050302
Attwa MW, Alsibaee AM, Aljohar HI, Abdelhameed AS, Kadi AA. Development of a Fast and Sensitive UPLC–MS/MS Analytical Methodology for Fenebrutinib Estimation in Human Liver Microsomes: In Vitro and In Silico Metabolic Stability Evaluation. Separations. 2023; 10(5):302. https://doi.org/10.3390/separations10050302
Chicago/Turabian StyleAttwa, Mohamed W., Aishah M. Alsibaee, Haya I. Aljohar, Ali S. Abdelhameed, and Adnan A. Kadi. 2023. "Development of a Fast and Sensitive UPLC–MS/MS Analytical Methodology for Fenebrutinib Estimation in Human Liver Microsomes: In Vitro and In Silico Metabolic Stability Evaluation" Separations 10, no. 5: 302. https://doi.org/10.3390/separations10050302
APA StyleAttwa, M. W., Alsibaee, A. M., Aljohar, H. I., Abdelhameed, A. S., & Kadi, A. A. (2023). Development of a Fast and Sensitive UPLC–MS/MS Analytical Methodology for Fenebrutinib Estimation in Human Liver Microsomes: In Vitro and In Silico Metabolic Stability Evaluation. Separations, 10(5), 302. https://doi.org/10.3390/separations10050302