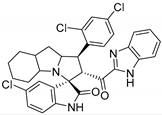
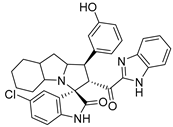
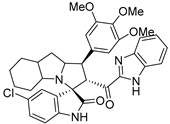
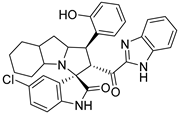
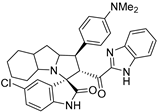
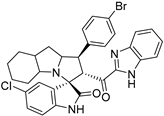
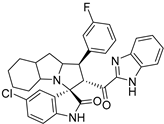
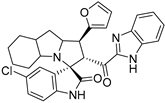
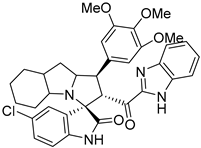
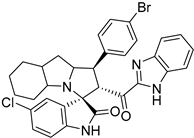
Abstract
Rational design for a new spiroxindoles, combined with a benzimidazole scaffold to identify a new murine double minute two (MDM2) inhibitor was synthesized and characterized. The desired spiroxindoles were achieved via a [3+2] cycloaddition reaction approach which afforded the cycloadducts with four asymmetric centers separated in an excellent regioselective and diastereoselective compound. The separated spiroxindoles were subjected to a set of biochemical assays including an NCI cell panel assay, MTT assay, and MDM2 binding analysis by a microscale thermophoresis assay. The anticancer reactivity for the tested compounds showed IC50 (µM) in the range between 3.797–6.879 µM, and compound 7d with IC50 = 3.797 ± 0.205 µM was the most active candidate between the series. The results showed promising results that identified that compound 7a could be inhibited the MDM2 with KD = 2.38 μm. Compound 7a developed a network of interactions with the MDM2 receptor studied in silico by molecular docking.
1. Introduction
The MDM2–p53 protein–protein interaction inhibitor is a hot research topic and has been gaining a lot of attention recently [1,2,3,4]. The inhibition of the interaction between the two proteins, p53, and MDM2, leads to reactivation of the p53 which has many functionalities, including DNA repairing, apoptosis, cell cycle arrest, senescence, metabolic alteration, and tumor suppresser [5,6]. The mutant p53 protein has been found in approximately 50% of human cancer cells [7,8]. The dislocation between the MDM2 protein and p53 protein is a challenge and is important to the development of a new chemotherapeutic agent.
Based on the literature survey, it has been reported so far that more than 20 chemotypes of molecules have been identified as MDM2–p53 inhibitors such as spirooxindoles [9], nutlins [10], isoquinoline-1-one [11], chalcone [12], pyrrolin-2-one [13], piperidine [14], morpholinone [15], imidazolyl indole [16], benzodiazpinedione [17], diketopiperazines [18], chromenotriazolopyrimidines [19], and other pharmacophores. For this, protein–protein interaction (PPI) inhibitors have progressed into clinical trials including spirooxindoles such as APG-115 [20], SAR405838 [21], and other pharmacophores such as RG7388 [22], HDM201 [23], RG7112 [24], and AMG-232 [25]. Inhibiting the p53–MDM2 interaction is a promising strategy for cancer treatment, as it can help to restore normal cell growth and death.
Protein–protein interaction inhibitors (PPIs) are typically small molecules that are designed to bind to the p53 and MDM2 proteins and prevent them from interacting. Several PPIs have been developed and are currently in clinical trials for a variety of cancers. Common side effects including fatigue, nausea, and anemia were observed.
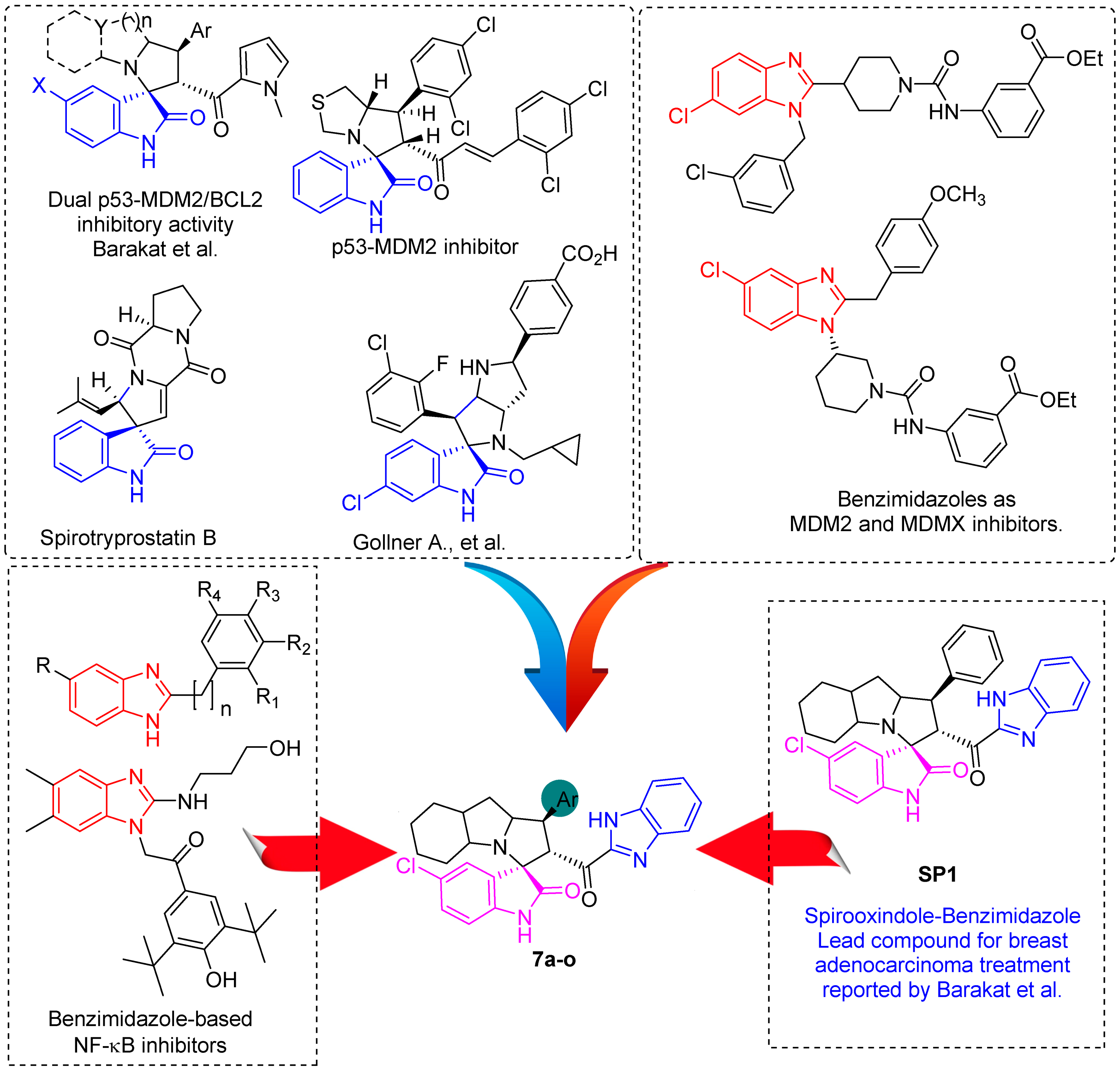
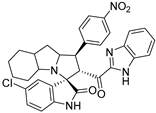
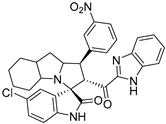
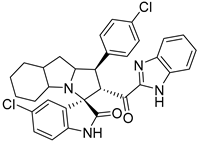
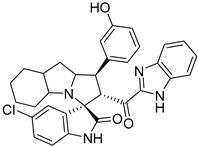
In between the small molecules reported as promising lead compounds for cancer research are the spirooxindoles. This scaffold is able to activate the p53 and bind with the MDM2 domain [26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47]. Spirotryprostatin B is an inspired natural product that exhibits anticancer reactivity [33] (Figure 1). Gollner A. et al. reported a novel chemically stable spiro [3H-indole-3, 2′-pyrrolidin]-2 (1H)-one lead compound and orally active inhibitors of the MDM2–p53 interaction [34]. Benzimidazole scaffold was introduced to many compounds which showed high efficacy against MDM2, MDMX, and NF-kB inhibitors [40,41,42,43,44,45,46]. Our research group has engaged in this research program for a couple of years and has been successful in designing and developing several molecules towards PPI [35,36,37,38,39]. Among the discovered molecules, a new spiroxindole [48], as a rigid structure with a combination of benzimidazole scaffold, has been discovered as a novel MDM2 protein inhibitor with dual effects of antimetastatic efficacy. Based on these findings, we have rationally designed and synthesized a new spirooxindoles-based benzimidazole unit as an MDM2 inhibitor.
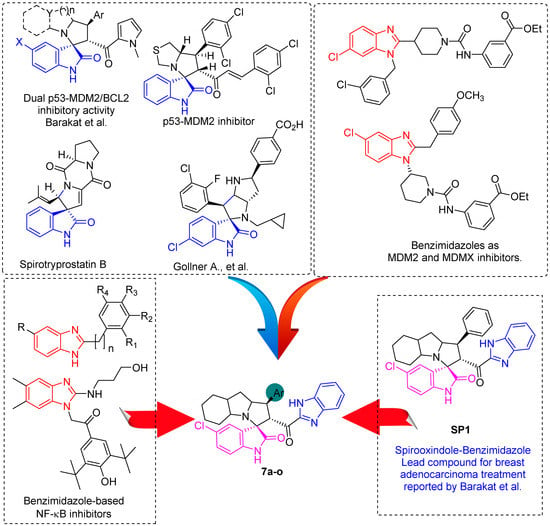
Figure 1.
Reported spirooxindoles and benzimidazoles with anticancer activity and our rationally designed compound 7a-o [34,36].
2. Materials and Methods
2.1. General
“All chemicals were purchased from Aldrich, Sigma-Aldrich and Fluka, which were used without further purification unless otherwise stated. All melting points were measured using a Gallenkamp melting point apparatus in open glass capillaries and were uncorrected. Crude products were purified by column chromatography on silica gel of 100–200 mesh. IR spectra were measured as KBr pellets using a Nicolet 6700 FT-IR spectrophotometer. The NMR spectra were recorded using a Varian Mercury Jeol-400 NMR spectrometer. 1H NMR (400 MHz) and 13C NMR (100 MHz) spectroscopy were performed in either deuterated dimethylsulfoxide (DMSO-d6) or deuterated chloroform (CDCl3). Chemical shifts (δ) are reported in terms of ppm and coupling constants J are given in Hz. Elemental analysis was carried out using an Elmer 2400 Elemental Analyzer in CHN mode”.
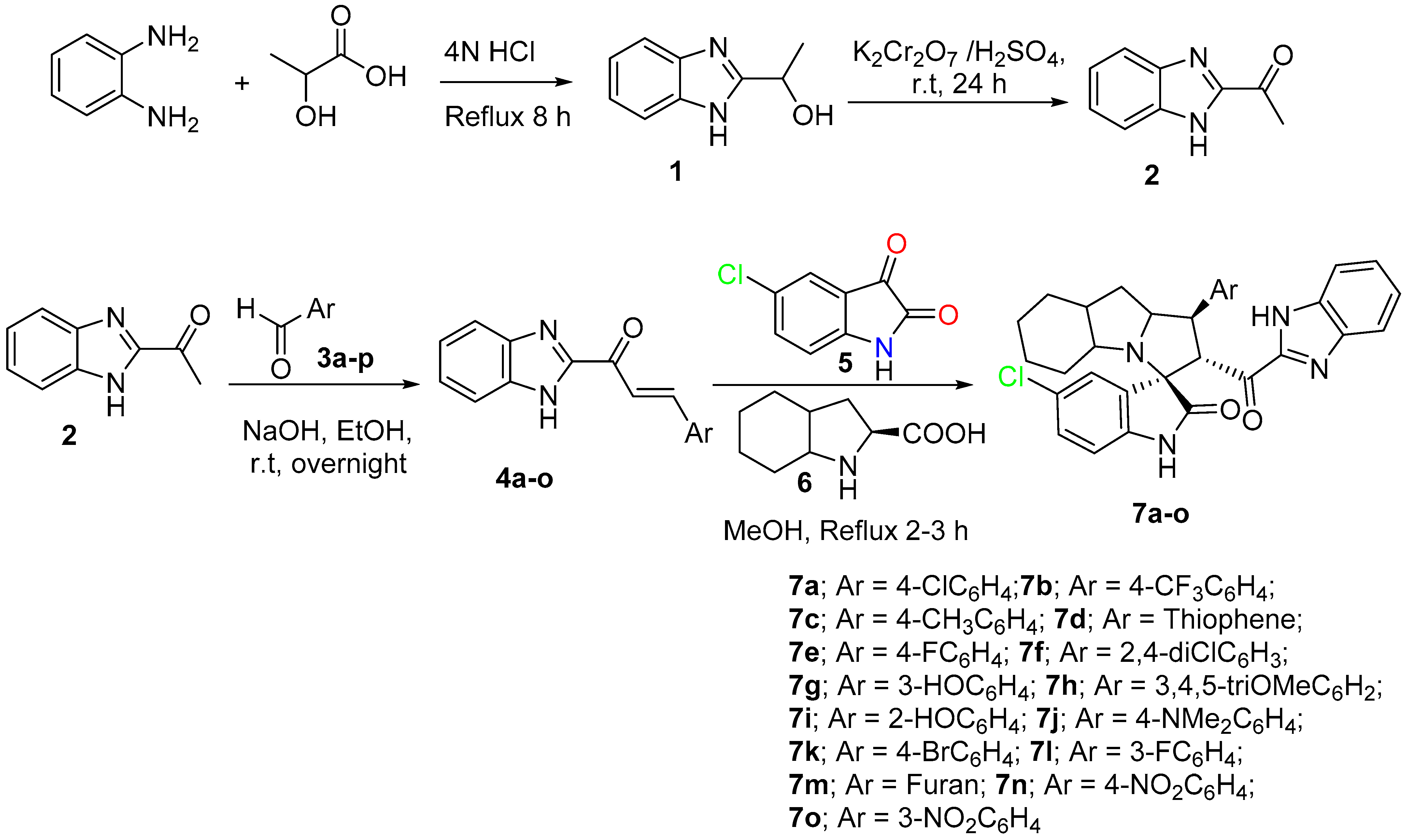
2.2. Synthesis of Spirooxindole Analogues (7a-o) General Procedure)
Chalcone derivative 4a-o (0.5 mmol), octahydroindole-2-carboxylic acid 6 (84.62 mg, 0.5 mmol), and 5-chlorisatin 5 (90.79 mg, 0.5 mmol) were mixed in 20 mL MeOH then, heated up at 60–65 °C for 2–3 h. After the reaction was completed, as monitored by TLC, the crude material was subjected to column chromatography using ethylacetate/n-hexane (2: 6), yielding spiro compounds in pure form.
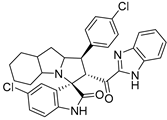
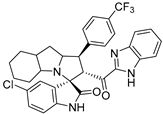
- 2’-(1H-Benzo[d]imidazole-2-carbonyl)-5-chloro-1’-(4-chlorophenyl)-1’,2’,4a’,5’,6’,7’,8’,8a’,9’,9a’-decahydrospiro[indoline-3,3’-pyrrolo [1,2-a]indol]-2-one (7a).
Pale yellow solid; yield (80%); m.p.:176–178 °C; IR (KBr, cm−1): 3434 (NH), 3277 (NH), 3093 (CH), 2929 (CH),1729 (CO), 1682 (CO); 1H-NMR (DMSO-d6, 400 MHz): δ 12.98 (1H, s, NH), 10.17 (1H, s, NH), 7.73 (1H, s, Ph-H) 7.45 (2H, d, J = 8.0 Hz, Ph-H), 7.42–7.26 (6H, m, Ph-H), 7.08 (1H, d, J = 8.0 Hz, Ph-H), 6.44 (1H, d, J = 8.0 Hz, Ph-H), 5.29 (1H, d, J = 11.9 Hz, CHCO), 4.00 (2H, m, CHN, CHPh), 3.23 (1H, d, J = 3.7 Hz), 2.16–2.08 (1H, m), 2.06–1.98 (1H, m), 1.55–0.70 (10H, m, aliphatic CH); Anal. for C32H28Cl2N4O2; calcd: C, 67.25; H, 4.94; N, 9.80 Exper.: C, 66.89; H, 5.03; N, 10.04.
- 2’-(1H-Benzo[d]imidazole-2-carbonyl)-5-chloro-1’-(4-(trifluoromethyl)phenyl)-1’,2’,4a’,5’,6’,7’,8’,8a’,9’,9a’-decahydrospiro[indoline-3,3’-pyrrolo[1,2-a]indol]-2-one (7b).
Pale yellow solid; yield (32%); m.p.:125–127 °C; IR (KBr, cm−1): 3429 (NH), 3282 (NH), 3099 (CH), 2927 (CH),1724 (CO), 1686 (CO); 1H-NMR (DMSO-d6, 400 MHz): δ 12.98 (1H, s, NH), 10.20 (1H, s, NH), 7.75 (1H, d, J = 8 Hz, Ph-H), 7.68 (4H, m, Ph-H), 7.48–7.20 (4H, m, Ph-H), 7.09 (1H, d, J = 8.8 Hz, Ph-H), 6.43 (1H, d, J = 8.3 Hz, Ph-H), 5.34 (1H, d, J = 11.7 Hz, CHCO), 4.11 (1H, m, CHN), 3.46 (1H, m, CHPh), 2.10 (2 H, d, J = 12.9Hz), 1.58–0.67 (10 H, m, aliphatic C-H); Anal. for C33H28ClF3N4O2; calcd: C, 65.51; H, 4.66; N, 9.26 Exper.: C, 66.09; H, 4.77; N, 9.14.
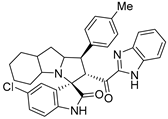
- 2’-(1H-Benzo[d]imidazole-2-carbonyl)-5-chloro-1’-(p-tolyl)-1’,2’,4a’,5’,6’,7’,8’,8a’,9’,9a’-decahydrospiro[indoline-3,3’-pyrrolo[1,2-a]indol]-2-one (7c).
Yellow solid; yield (41%); m.p.:163–165 °C; IR (KBr, cm−1): 3649 (NH), 3277 (NH), 3087(CH), 2926 (CH),1727 (CO), 1690 (CO); 1H-NMR (DMSO-d6, 400 MHz): δ 12.98 (1H, s, NH), 10.18 (1H, s, NH), 7.76 (1H, d, J = 8.0 Hz, Ph-H), 7.40 (1H, d, J = 2.2 Hz, Ph-H), 7.35–7.26 (5H, m, Ph-H), 7.10–7.06 (3H, d, J = 8.0 Hz, Ph-H), 6.44 (1H, d, J = 8 Hz, Ph-H), 5.34 (1H, d, J = 12.4 Hz, CHCO), 4.11–4.00 (1H, m, CHN), 3.96–3.86 (1H, m, CHPh), 2.20 (3H, s, CH3), 2.11 (2 H, d, J = 8.0Hz), 1.56–0.72 (10H, m, aliphatic C-H); 13C-NMR (DMSO-d6, 100 MHz): δ = 189.91, 180.05, 148.02, 143.07, 141.62, 136.60, 136.47, 135.18, 129.69, 127.85, 126.33, 125.17, 123.63, 111.02, 71.74, 63.94, 57.24, 52.79, 36.80, 28.19, 25.07; Anal. for C33H31ClN4O2; calcd: C, 71.92; H, 5.67; N, 10.17 Exper.: C, 71.59; H, 5.71; N, 10.44.
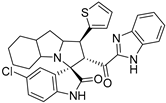
- 2’-(1H-Benzo[d]imidazole-2-carbonyl)-5-chloro-1’-(thiophen-2-yl)-1’,2’,4a’,5’,6’,7’,8’,8a’,9’,9a’-decahydrospiro[indoline-3,3’-pyrrolo[1,2-a]indol]-2-one (7d).
Pale yellow solid; yield (48%); m.p.:130–132 °C; IR (KBr, cm−1): 3624 (NH), 3258 (NH), 3091(CH), 2927 (CH),1728 (CO), 1689 (CO); 1H-NMR (CDCl3, 400 MHz): δ 10.18 (1H, s, NH), 8.64 (1H, s, NH), 7.84 (1H, d, J = 8.0 Hz, Ph-H), 7.34 (1H, d, J = 8.0 Hz, thio-H), 7.31–7.20 (4H, m, Ph-H), 6.99 (1H, d, J = 3.5 Hz, thio-H), 6.95 (1H, dd, J = 8.2, 1.7 Hz, thio-H), 6.90–6.85 (1H, m, Ph-H), 6.39 (1H, d, J = 8.6 Hz, Ph-H), 5.30 (1H, d, J = 12.0 Hz, CHCO), 4.53–4.34(1H, m, CHN), 4.12 (1H, t, J = 11.0 Hz, CHPh), 3.20 (1H, d, J = 4.0 Hz), 2.16 (1H, d, J = 5.0Hz), 1.83–0.82 (10 H, m, aliphatic C-H); 13C-NMR (CDCl3, 100 MHz): δ = 189.65, 181.65, 146.83, 143.16, 141.99, 139.94, 133.80, 129.28, 126.77, 125.88, 123.80, 111.02, 72.12, 71.33, 65.76, 57.93, 48.77, 37.52, 28.38, 27.86, 19.70; Anal. for C30H27ClN4O2S; calcd: C, 66.35; H, 5.01; N, 10.32 Exper.: C, 66.49; H, 5.20; N, 10.14.
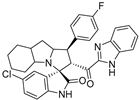
- 2’-(1H-Benzo[d]imidazole-2-carbonyl)-5-chloro-1’-(4-fluorophenyl)-1’,2’,4a’,5’,6’,7’,8’,8a’,9’,9a’-decahydrospiro[indoline-3,3’-pyrrolo[1,2-a]indol]-2-one (7e).
Pale yellow solid; yield (35%); m.p.:148–150 °C; IR (KBr, cm−1): 3431 (NH), 3268 (NH), 3096 (CH), 2960 (CH),1732 (CO), 1684 (CO); 1H-NMR (CDCl3, 400 MHz): δ 10.02 (1H, s, NH), 8.55 (1H, s, NH), 7.82 (1H, d, J = 8.3 Hz, Ph-H), 7.40 (2 H, dd, J = 8.5, 5.4 Hz), 7.35–7.12 (5H, m, Ph-H), 6.94 (3H, m, Ph-H), 6.36 (1H, d, J = 8 Hz, Ph-H), 5.32 (1H, d, J = 12.4 Hz, CHCO), 4.31 (1H, q, J = 7.4 Hz, CHN), 3.79 (1H, t, J = 12.4 Hz, CHPh), 3.19–0.87 (12 H, m, aliphatic C-H); 13C-NMR (CDCl3, 100 MHz): δ = 189.71, 181.92, 163.19, 160.76, 146.85, 143.08, 139.91, 134.65, 134.62, 133.69, 129.56, 129.49, 129.30, 127.35, 126.82, 126.64, 126.09, 123.68, 122.61, 115.63, 115.42, 112.08, 110.91, 72.01, 71.26, 65.28, 57.78, 53.18, 41.88, 37.45, 29.79, 28.43, 27.82, 24.79, 19.75; Anal. for C32H28ClFN4O2; calcd: C, 69.25; H, 5.08; N, 10.09 Exper.: C, 69.49; H, 5.18; N, 10.44.
- 2’-(1H-Benzo[d]imidazole-2-carbonyl)-5-chloro-1’-(2,4-dichlorophenyl)-1’,2’,4a’,5’,6’,7’,8’,8a’,9’,9a’-decahydrospiro[indoline-3,3’-pyrrolo[1,2-a]indol]-2-one (7f).
Pale yellow solid; yield (48%); m.p.:138–140 °C; IR (KBr, cm−1): 3436 (NH), 3281 (NH), 3090 (CH), 2925 (CH),1730 (CO), 1685 (CO); 1H-NMR (CDCl3, 400 MHz): δ 10.14 (1H, s, NH), 8.40 (1H, s, NH), 7.84 (1H, d, J = 8.0 Hz, Ph-H), 7.55 (1H, d, J = 8.7 Hz, Ph-H), 7.37 (1H, s, Ph-H), 7.35–7.14 (5H, m, Ph-H), 6.95 (1H, d, J = 8.0 Hz, Ph-H), 6.37 (1H, d, J = 8.0 Hz, Ph-H), 5.39 (1H, d, J = 12.3 Hz, CHCO), 4.47 (1H, t, J = 11.0 Hz, CHPh), 4.20 (1H, q, J = 8.5 Hz, CHN), 3.20 (1 H, d, J = 4.4 Hz), 2.14–0.89 (12 H, m, aliphatic C-H); 13C-NMR (CDCl3, 100 MHz): δ = 189.44, 181.66, 146.72, 143.03, 139.94, 135.42, 135.35, 133.76, 133.00, 129.56, 126.98, 125.98, 110.98, 71.87, 48.25, 27.66, 19.79; Anal. for C32H27Cl3N4O2; calcd: C, 63.43; H, 4.49; N, 9.25 Exper.: C, 63.59; H, 5.04; N, 9.04.
- 2’-(1H-Benzo[d]imidazole-2-carbonyl)-5-chloro-1’-(3-hydroxyphenyl)-1’,2’,4a’,5’,6’,7’,8’,8a’,9’,9a’-decahydrospiro[indoline-3,3’-pyrrolo[1,2-a]indol]-2-one (7g).
Pale yellow solid; yield (62%); m.p.:186–188 °C; IR (KBr, cm−1): 3626 (NH), 3257 (OH), 2931 (CH), 1726 (CO), 1688 (CO); 1H-NMR (CDCl3, 400 MHz): δ 9.00 (1H, s, NH), 8.25 (1H, s, NH), 7.54 (1H, d, J = 4.0 Hz, Ph-H), 7.46 (1H, s, OH), 7.22 (1H, d, J = 8.0 Hz, Ph-H), 7.08–7.04 (3H, m, Ph-H), 6.89–6.72 (5H, m, Ph-H), 6.23 (1H, d, J = 8.0 Hz, Ph-H), 5.31 (1H, d, J = 12.5 Hz, CHCO), 4.49–4.38 (1H, m, CHN), 3.75–3.65 (1H, m, CHPh), 3.14 (1H, d, J = 4 Hz), 2.16–0.81 (12 H, m, aliphatic C-H); Anal. for C32H29ClN4O3; calcd: C, 69.49; H, 5.29; N, 10.13 Exper.: C, 69.69; H, 5.14; N, 9.94.
- 2’-(1H-Benzo[d]imidazole-2-carbonyl)-5-chloro-1’-(3,4,5-trimethoxyphenyl)-1’,2’,4a’,5’,6’,7’,8’,8a’,9’,9a’-decahydrospiro[indoline-3,3’-pyrrolo[1,2-a]indol]-2-one (7h).
Pale yellow solid; yield (88%); m.p.:160–162 °C; IR (KBr, cm−1): 3430 (NH), 3269 (NH), 3093 (CH), 2996 (CH),1722 (CO), 1683 (CO); 1H-NMR (CDCl3, 400 MHz): δ 10.15 (1H, s, NH), 8.500 (1H, s, NH), 7.81 (1H, d, J = 8.0 Hz, Ph-H), 7.37–7.19 (4H, m, Ph-H), 6.97 (1H, d, J = 8.4 Hz, Ph-H), 6.66 (2H, s, Ph-H), 6.38 (1H, d, J = 8.5 Hz, Ph-H), 5.38 (1H, d, J = 12.3 Hz, CHCO), 4.33 (1H, m, CHPh), 3.78 (6H, s, OCH3), 3.73 (3H, s, OCH3), 3.19 (1H, m, CHN), 2.02–0.78 (12 H, m, aliphatic C-H); 13C-NMR (CDCl3, 100 MHz): δ = 189.75, 181.68, 153.30, 146.99, 143.14, 139.85, 136.95, 134.53, 133.80, 133.31, 126.78, 126.18, 123.39, 122.48, 110.86, 104.89, 72.25, 71.07, 60.86, 56.33, 56.18, 54.56, 41.84, 37.58, 28.44, 27.83, 24.76.; Anal. for C35H35ClN4O5; calcd: C, 67.03; H, 5.63; N, 8.93 Exper.: C, 67.59; H, 5.34; N, 9.05.
- 2’-(1H-Benzo[d]imidazole-2-carbonyl)-5-chloro-1’-(2-hydroxyphenyl)-1’,2’,4a’,5’,6’,7’,8’,8a’,9’,9a’-decahydrospiro[indoline-3,3’-pyrrolo[1,2-a]indol]-2-one (7i).
Yellow solid; yield (82%); m.p.:128–130 °C; IR (KBr, cm−1): 3311 (OH), 3063 (CH), 2925 (CH),1716 (CO), 1667 (CO); 1H-NMR (CDCl3, 400 MHz): δ 10.57 (1H, s, NH), 8.93 (1H, s, NH), 7.80 (1H, d, J = 8.0 Hz, Ph-H), 7.49 (1H, d, J = 8.0 Hz, Ph-H), 7.29 (2H, d, J = 7.3 Hz, Ph-H), 7.23 (1H, s, OH), 7.08 (2H, t, J = 7.7 Hz, Ph-H), 6.97 (2H, d, J = 8 Hz, Ph-H), 6.87 (2H, d, J = 8 Hz, Ph-H), 6.45 (1H, d, J = 8 Hz, Ph-H), 5.07 (1H, dd, J = 11.7, 6.6 Hz, CHCO), 4.54 (1H, q, J = 7.3 Hz, CHN), 4.43 (1H, t, J = 11.0 Hz, CHPh), 3.22 (1H, d, J = 4.4 Hz), 2.16–0.83 (12 H, m, aliphatic C-H); 13C-NMR (CDCl3, 100 MHz): δ = 191.67, 181.97, 154.84, 146.69, 142.54, 139.78, 133.34, 129.39, 128.16, 127.44, 126.93, 125.55, 125.42, 124.12, 122.07, 121.41, 118.44, 112.22, 111.31, 85.63, 85.25, 83.06, 73.14, 57.83, 46.34, 41.88, 40.99, 37.30, 28.55, 28.44, 27.76, 24.73, 23.97; Anal. for C32H29ClN4O3; calcd: C, 69.49; H, 5.29; N, 10.13 Exper.: C, 69.55; H, 5.16; N, 10.34.
- 2’-(1H-Benzo[d]imidazole-2-carbonyl)-5-chloro-1’-(4-(dimethylamino)phenyl)-1’,2’,4a’,5’,6’,7’,8’,8a’,9’,9a’-decahydrospiro[indoline-3,3’-pyrrolo[1,2-a]indol]-2-one (7j).
Orange solid; yield (45%); m.p.:140–142 °C; IR (KBr, cm−1): 3434 (NH), 3275 (NH), 3094 (CH), 2929 (CH),1729 (CO), 1682 (CO); 1H-NMR (CDCl3, 400 MHz): δ 9.81 (1H, s, NH), 8.01 (1H, s, NH), 7.78 (1H, d, J = 8.0 Hz, Ph-H), 7.31 (5H, m, Ph-H), 7.17 (1H, d, J = 2.2 Hz, Ph-H), 6.94 (1H, dd, J = 8.3, 2.3 Hz, Ph-H), 6.65 (2H, d, J = 8.8 Hz, Ph-H), 6.31 (1H, d, J = 8.6 Hz, Ph-H), 5.32 (1H, d, J = 12.4 Hz, CHCO), 4.38–4.27 (1H, m, CHN), 4.10 (1H, t, J = 7.3 Hz, CHPh), 3.19 (1H, d, J = 3.8 Hz), 2.87 (6H, s, NCH3), 2.07–0.83 (12 H, m, aliphatic C-H); 13C-NMR (CDCl3, 100 MHz): δ = 189.78, 181.66, 149.77, 147.05, 139.62, 133.54, 129.10, 128.85, 128.72, 127.56, 126.77, 126.36, 112.88, 110.59, 72.04, 71.13, 65.09, 57.80, 53.18, 40.70, 31.67, 28.50, 27.81, 24.84, 22.74, 19.86; Anal. for C34H34ClN5O2; calcd: C, 70.39; H, 5.91; N, 12.07 Exper.: C, 70.65; H, 6.06; N, 11.94.
- 2’-(1H-Benzo[d]imidazole-2-carbonyl)-1’-(4-bromophenyl)-5-chloro-1’,2’,4a’,5’,6’,7’,8’,8a’,9’,9a’-decahydrospiro[indoline-3,3’-pyrrolo[1,2-a]indol]-2-one (7k).
Yellow solid; yield (72%); m.p.:159–161 °C; IR (KBr, cm−1): 3625 (NH), 3422 (NH), 3088 (CH), 2913 (CH),1725 (CO), 1682 (CO); 1H-NMR (CDCl3, 400 MHz): δ 10.08 (1H, s, NH), 8.46 (1H, s, NH), 7.80 (1H, d, J = 8.2 Hz, Ph-H), 7.36 (2H, d, J = 8.6 Hz, Ph-H), 7.31–7.16 (7H, m, Ph-H), 6.99–6.93 (1H, m, Ph-H), 6.34 (1H, d, J = 8.2 Hz, Ph-H), 5.30 (1H, d, J = 12.3 Hz, CHCO), 4.36–4.24 (1H, m, CHN), 3.75 (1H, t, J = 12.4 Hz, CHPh), 3.19 (1H, d, J = 3.9 Hz), 2.22–0.82 (12 H, m, aliphatic C-H); 13C-NMR (CDCl3, 100 MHz): δ = 189.58, 181.78, 146.77, 143.06, 139.92, 138.08, 133.72, 131.76, 129.87, 129.35, 127.32, 126.82, 126.65, 126.03, 122.61, 120.92, 112.11, 110.94, 71.97, 71.19, 65.20, 57.78, 53.43, 41.86, 37.41, 28.43, 27.81, 24.78, 19.75; Anal. for C32H28BrClN4O2; calcd: C, 62.40; H, 4.58; N, 9.10 Exper.: C, 62.60; H, 4.34; N, 9.03.
- 2’-(1H-Benzo[d]imidazole-2-carbonyl)-5-chloro-1’-(3-fluorophenyl)-1’,2’,4a’,5’,6’,7’,8’,8a’,9’,9a’-decahydrospiro[indoline-3,3’-pyrrolo[1,2- a]indol]-2-one (7l).
Pale yellow solid; yield (42%); m.p.:143–145 °C; IR (KBr, cm−1): 3625 (NH), 3422 (NH), 3088 (CH), 2913 (CH),1725 (CO), 1682 (CO); 1H-NMR (CDCl3, 400 MHz): δ 9.51 (1H, s, NH), 7.86 (1H, d, J = 8.0 Hz, Ph-H), 7.42 (1H, s, NH), 7.32 (2H, d, J = 4 Hz, Ph-H), 7.29–7.27 (2H, m, Ph-H), 7.23–7.13 (3H, m, Ph-H), 6.96 (1H, dd, J = 8.5, 2.2 Hz, Ph-H), 6.91–6.85 (1H, m, Ph-H), 6.32 (1H, d, J = 8.4 Hz, Ph-H), 5.31 (1H, d, J = 12.0 Hz, CHCO), 4.41–4.30 (1H, m, CHN), 3.79 (1H, t, J = 12.4 Hz, CHPh), 3.22 (1H, d, J = 4.3 Hz, CHPh), 2.17–0.88 (12 H, m, aliphatic C-H); Anal. for C32H28ClFN4O2; calcd: C, 69.25; H, 5.08; N, 10.09 Exper.: C, 69.40; H, 4.94; N, 10.13.
- 2’-(1H-Benzo[d]imidazole-2-carbonyl)-5-chloro-1’-(furan-2-yl)-1’,2’,4a’,5’,6’,7’,8’,8a’,9’,9a’-decahydrospiro[indoline-3,3’-pyrrolo[1,2- a]indol]-2-one (7m).
Pale yellow solid; yield (62%); m.p.:165–167 °C; IR (KBr, cm−1): 3435 (NH), 3256 (NH), 3090 (CH), 2928 (CH),1729 (CO), 1687 (CO); 1H-NMR (CDCl3, 400 MHz): δ 10.31 (1H, s, NH), 8.77 (1H, s, NH), 7.83 (1H, d, J = 8 Hz, Ph-H), 7.36 (1H, d, J = 8 Hz, Ph-H), 7.28 (1H, t, J = 7.5 Ph-H), 7.22 (2H, d, J = 7.9 Hz, Ar), 7.11 (1H, s, Ph-H), 6.93 (1H, d, J = 8.7 Hz, Ph-H), 6.39 (1H, d, J = 8.7 Hz, Ph-H), 6.20 (1H, t, J = 1.6 Hz, fur-H), 6.12 (1H, d, J = 3.6 Hz, fur-H), 5.37 (1H, d, J = 12.3 Hz, CHCO), 4.39 (1H, q, J = 8.3 Hz, CHN), 3.97 (1H, t, J = 11.4 Hz, CHPh), 3.17 (1H, d, J = 4.4 Hz), 2.16–0.85 (12 H, m, aliphatic C-H); 13C-NMR (CDCl3, 100 MHz): δ = 189.68, 181.80, 153.04, 146.83, 143.19, 141.75, 141.71, 141.67, 139.98, 133.89, 129.32, 127.40, 126.74, 126.63, 125.97, 123.69, 122.69, 112.20, 111.05, 110.27, 106.01, 71.94, 68.30, 62.85, 57.68, 46.89, 41.94, 37.74, 28.39, 27.79, 24.80, 19.69; Anal. for C30H27ClN4O3; calcd: C, 68.37; H, 5.16; N, 10.63 Exper.: C, 68.60; H, 4.94; N, 10.33.
- 2’-(1H-Benzo[d]imidazole-2-carbonyl)-5-chloro-1’-(4-nitrophenyl)-1’,2’,4a’,5’,6’,7’,8’,8a’,9’,9a’-decahydrospiro[indoline-3,3’-pyrrolo[1,2- a]indol]-2-one (7n).
Pale yellow solid; yield (46%); m.p.:165–167 °C; IR (KBr, cm−1): 3437 (NH), 3255 (NH), 3094 (CH), 2929 (CH),1728 (CO), 1686 (CO); 1H-NMR (CDCl3, 400 MHz): δ 9.81 (1H, s, NH), 8.12 (1H, s, NH), 8.12 (2H, d, J = 8.6 Hz, Ph-H), 7.80 (1H, d, J = 8.0 Hz, Ph-H), 7.63 (2H, d, J = 8.6 Hz, Ph-H), 7.33–7.23 (3H, m, Ph-H), 7.14 (1H, s, Ph-H), 6.97 (1H, dd, J = 8.3, 1.8 Hz, Ph-H), 6.36 (1H, d, J = 8.6 Hz, Ph-H), 5.35 (1H, d, J = 11.9 Hz, CHCO), 4.41–4.35 (1H, m, CHN), 3.90 (1H, t, J = 12 Hz, CHPh), 3.23 (1 H, d, J = 3.9 Hz), 2.03- 0.84 (12 H, m, aliphatic C-H); 13C-NMR (CDCl3, 100 MHz): δ = 189.08, 181.27, 147.16, 146.89, 146.55, 143.00, 139.84, 133.60, 129.53, 129.10, 129.04, 126.98, 126.86, 125.72, 123.92, 122.54, 112.06, 110.85, 71.90, 71.25, 65.46, 57.83, 53.73, 41.86, 37.38, 31.67, 28.38, 27.80, 24.73, 23.95; Anal. for C32H28ClN5O4; calcd: C, 66.03; H, 4.85; N, 12.03 Exper.: C, 66.10; H, 4.74; N, 12.11.
- 2’-(1H-benzo[d]imidazole-2-carbonyl)-5-chloro-1’-(3-nitrophenyl)-1’,2’,4a’,5’,6’,7’,8’,8a’,9’,9a’-decahydrospiro[indoline-3,3’-pyrrolo[1,2- a]indol]-2-one (7o).
Pale yellow solid; yield (52%); m.p.:135–137 °C; IR (KBr, cm−1): 3438 (NH), 3257 (NH), 3092 (CH), 2930 (CH),1729 (CO), 1688 (CO); 1H-NMR (CDCl3, 400 MHz): δ 9.55 (1H, s, NH), 8.36 (1H, s, NH), 8.06 (1H, dd, J = 8.1, 2.3 Hz, Ph-H), 7.86 (2H, d, J = 8.2 Hz, Ph-H), 7.57 (1H, s, Ph-H), 7.47 (1H, t, J = 8 Hz, Ph-H), 7.31 (3H, d, J = 4.0 Hz, Ph-H), 7.14 (1H, d, J = 2.1 Hz, Ph-H), 6.98 (1H, dd, J = 8.3, 2.4 Hz, Ph-H), 6.33 (1H, d, J = 8.0 Hz, Ph-H), 5.31 (1H, d, J = 12.0 Hz, CHCO), 4.46–4.36 (1H, m, CHN), 3.92 (1H, t, J = 12.0 Hz, CHPh), 3.24 (1H, d, J = 4.3 Hz), 2.19–0.81 (12 H, m, aliphatic C-H); Anal. for C32H28ClN5O4; calcd: C, 66.03; H, 4.85; N, 12.03 Exper.: C, 66.09; H, 4.73; N, 12.10.
2.3. NCI Screening
The compounds have been processed according to the standard method NCI-60 Human Tumor Cell Lines Screen for the organic compound at the development therapeutic program (DTP) (see Supplementary Materials, Table S2; Figures S1 and S2).
2.4. Anticancer Activity Protocol
The anticancer activity protocol was carried out according to the method reported in [48]. “The cytotoxicity of tested compounds was investigated on a human normal lung fibroblast (Wi-38) cell line, triple-negative breast (MDA-MB 231) cells, and prostate cancer (PC3) cells. These cells were cultured in DMEM containing 10% fetal bovine serum. After cell seeding (10,000, 4000, and 5000 cells, respectively, per well) in 96-well cell culture plates and incubating for 24 h in a 5% CO2 incubator, serial dilutions (2,4,6,8, and 10 μM) of the tested compounds were added. Following 48 h in a 5% CO2 incubator, 20 µL of MTT (5 mg/mL) was added and incubated for 4 h, then this solution was removed and 100 µL of DMSO was added. The absorbance was measured at 590 nm (BMG LabTech, Ortenberg, Germany). The half-inhibitory growth concentration (IC50) was calculated by GraphPad Prism software” [48].
2.5. MDM2 Binding Analysis by Microscale Thermophoresis (MST) Assay
The full protocol has been provided in SI and the binding curve is shown in the Supplementary Material (Figures S3–S6).
2.6. Methodology for Molecular Docking
Two-dimensional structures of compounds 7a, 7g, 7h, and 7k were drawn via the builder and subjected to preliminary structure preparation, namely energy minimization with the force field MMFF94x and the subsequent application of partial charges. The X-Ray crystal structure of MDM2 with PDB ID: 5LAZ, having a cocrystallized ligand with structural similarity to the studied compounds, was retrieved from the RCSB Protein Data Bank for the docking studies. Structure preparation of the protein was brought about by energy minimization with force field Amber10:EHT and partial charges were then applied to the protein. The cocrystallized ligand (6ST), after the necessary structure preparations, was used as a reference compound for validation of the results in docking studies of the aforementioned compounds [34]. Benchmarking of the docking protocols was performed well before the docking studies. Redocking of the 6ST ligand was brought about to observe the deviation of the ligand conformation from the original one (SI Figure S7). All the operations were performed in the molecular operating environment (MOE 2019.01) [49], which was chosen based on the RMSD value (0.14 Å) between the coordinates of the cognate ligand and the simulated pose. Induced fit docking was directed to the ligand atoms, brought about by placement of the ligand into the binding site of MDM2 utilizing the triangle matcher algorithm followed by determining the scores of the generated fifteen conformations through the London dG scoring function. Finally, five top-scored conformations were retained and evaluated by the GBVI/WSA dG scoring function. Thereafter, the same protocols were followed for the docking simulation of the studied compounds. The interaction patterns of the ligands with binding site residues were analyzed by the Protein-Ligand Interaction Profiler [50].
2.7. Statistical Analysis
The data are expressed as mean ± standard error of the mean (SEM) and values were considered significantly different at p < 0.05, using one-way analysis of variance (ANOVA) and Tukey’s test (SPSS software version 16).
3. Results and Discussion
3.1. Chemistry
Based on the recently published idea of spiroxindole having the benzimidazole nucleus and showing promising results against cancer cell lines and an antimetastatic effect. To study the cytotoxicity and structure reactivity relationship, a new library 7a-o has been synthesized and characterized (Scheme 1). Different electronic effects on the aromatic ring, including electron-donating and electron-withdrawing effects, also achieved a heterocycle aromatic ring and were explored. The synthetic methodology was carried out based on a multicomponent one-pot reaction via the [3+2] cycloaddition reaction approach [48]. The desired dipolarphiles were synthesized from orthophenylene diamine in a consequential step. Mixing the chalcones 4a-o with the 5-chloroisatin, 5 and the key amino acid 6 in methanol under reflux for 2–3 h, afforded the final compounds in a high chemical yield and a regio- and diasetero-selective manner. The stereochemistry for the final cycloadduct is matched with the previous lead compound published by our research group [48].
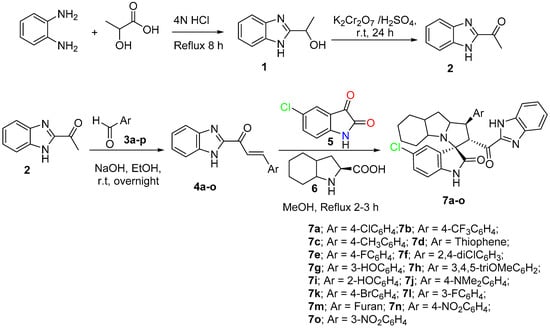
Scheme 1.
Synthetic methodology for the desired spirooxindole derivative 7a-o.
3.2. In Vitro Anti-Cancer Activity Assays
3.2.1. NCI Screening (Development Therapeutic Program, DTP)
The successfully synthesized spirooxindoles (7a-o) were submitted for NCI for screening against 60 various cancer cell lines, classified into nine subpanels: breast, kidney, melanoma colon, prostate, CNS, ovary, melanoma, leukemia, and lung cancers. The initial single dose assay for the assessed spirooxindoles were tested at a 10 µM and the results were then expressed as a percentage of growth inhibition (GI%) (Table S2). As observed, the synthesized compounds inhibited the growth of the NCI cell-line panel according to the following order: breast > renal > leukemia cancer cell lines > other tested cancer cell lines (Table S2). The initial results afforded the most active compound, 7g, which entered the five dose assays, and the results are shown in Supporting Materials (Figures S1 and S2).
3.2.2. MTT Assay
In order to determine the IC50 (µM) of the synthesized spirooxindoles, 7a-o were subjected to an MTT assay in vitro against the two-cancer cell line MDA-MB 231 and PC3 cells, and the data reported in Table 1. For the breast cancer line (MDA-MB 231) the compounds are shown in IC50 (µM) in the range between 3.797–6.879 µM; the most active candidate between the series was compound 7d with IC50 = 3.797 ± 0.205 µM, the chemical structure compromises a thiophene ring. On the other hand, the least reactivity was compound 7n with IC50 = 6.879 ± 0.308 µM. All other compounds are shown in the range of 4 µM reactivity. In the case of prostate cancer (PC3), the reactivity of the synthesized compounds exhibited the range of IC50 = 4.252 to 7.567 µM. In the case of compound 7a with the Cl-atom in the fourth position showed IC50 = 4.763 ± 0.069 and 4.574 ± 0.011 µM, for both tested two-cancer cells MDA-MB 231, and PC3, respectively. The reactivity slightly improved compared with compound 7a when the CF3-group was introduced into the aromatic ring, as indicated in compound 7b which showed IC50 = 4.284 ± 0.007 and 4.404 ± 0.008 µM; 7e contain the p-fluoro atom on the aromatic ring compared with 7l having the m-fluoro atom on the aromatic, where no differences were observed in the cytotoxicity. In the isosteric analog of the compound 7d, which replaced the thiophene with furan heterocycle, as shown in compound 7m, the cytotoxicity dropped to IC50 = 6.039 ± 0.111 and 5.098 ± 0.119 µM with less than 1.6 and 1.18 times, compared to the compound 7d. Introducing the electron-withdrawing effect of the NO2 group either in the para- or meta-position, we observed that NO2 in the meta (compound 7o) was more active than the para-position (compound 7n) with 1.66 and 1.77 folds. The existence of electron donating groups such as methyl group (compound 7c); hydroxyl group (compound 7g or compound 7c); trimethoxy groups (compound 7h); and dimethyl amine (compound 7j) did not alter the reactivity.

Table 1.
The estimated IC50 (µM) of 7a-o on Wi-38 viability, the growth of MDA-MB 231, and PC3 cells.
3.2.3. Microscale Thermophoresis Assay (MST) for MDM2 Binding Detection
Compounds 7a, 7g, 7h, and 7k shown very good anti-cancer potential as well as high safety profiles as obtained by the MTT assay. Accordingly, the compound 7a, 7g, 7h, and 7k were then tested for MDM2 binding analysis. The tested compound was incubated with a fluorescently labeled MDM2 at increasing concentrations (0.763 nM to 25 μM; Solubility play a crucial role to reach maximum concentration). MST binding curves showed that spirooxindole-based benzimidazole 7a, 7g, and 7h showed moderate binding affinity in the range of (KD; 2.38–38 μm). Compound 7a showed better binding reactivity compared to our previous spirooxindole-based benzimidazole, which has been shown (KD; 7.94 μM) [48]. The 7k did not show any binding detection (Table 2).

Table 2.
MST binding assay results of MDM2.
3.3. Molecular Docking of the Studied Compound
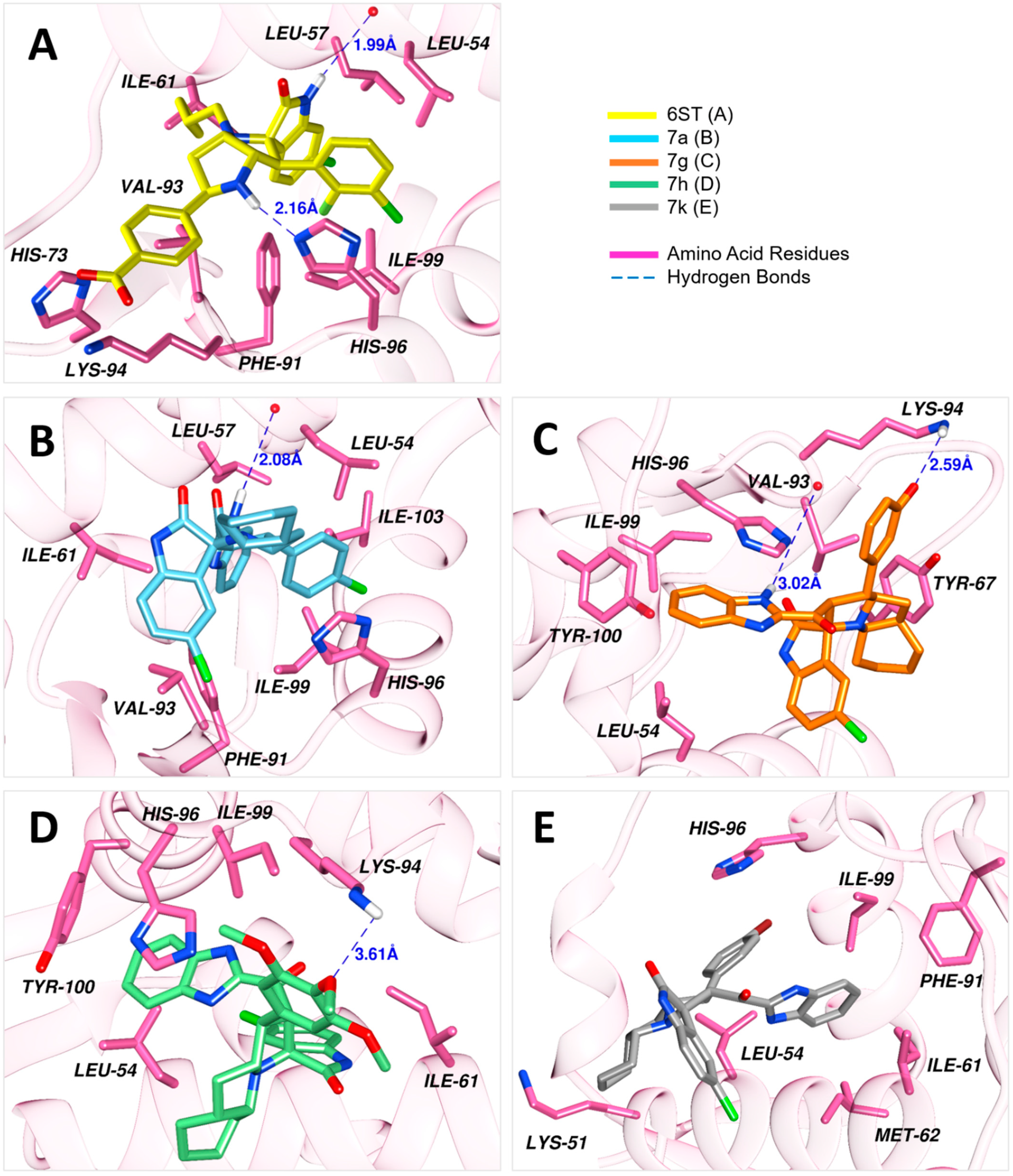
Results of the docking simulations of the spirooxindoles molecules were validated relative to the reference compound (6ST), which was firmly held into the binding site by a diverse network of interactions. The nitrogen of the imidazole ring of His96 and oxygen in the carboxylic acid moiety of Leu54 accepted hydrogens from the N-H of indolinone and the N-H of pyrrolidine rings, resulting in hydrogen bonds with 1.99 Å and 2.16 Å lengths, respectively. Additionally, His96 had π-stacking and halogen bonding with the chloroindolinone moiety. There were formed salt bridges by the carboxylate group of this compound with His73 and Lys94. This binding was further enhanced by hydrophobic interactions with seven amino acid residues.
Compound 7a developed a network of interactions similar to that of the reference compound. A hydrogen bond was established with the ligand at a distance of 2.08 Å, where hydrogen was donated by the N-H of benzimidazole to Leu54. Furthermore, His96 formed π-stacking and halogen bonding with the chlorobenzene ring the same way it was created with the reference compound. Similarly, hydrophobic interactions formed by seven residues well accommodated this compound into the binding site. In contrast to 7a, compound 7g formed two hydrogen bonds. One of the hydrogen bonds between benzimidazole moiety and Val93 was of 3.02 Å length, and the other bond resulted between the amino group of Lys96, being the donor, and phenolic group of the ligand was accepting at 2.59 Å distance. His96 provided π-stacking with the aromatic rings of benzimidazole moiety. However, fewer hydrophobic interactions were observed for this compound as compared to 7a. In the case of 7h, only one hydrogen bond was formed, which was between the amino group of Lys94 and the central methoxy group of the trimethoxybenzene substituent in the compound. The benzimidazole of this compound was anchored by π-stacking, twice with His96 and once with Tyr100. Three hydrophobic interactions were also developed. For compound 7k, the observed interactions included a halogen bond between its bromobenzene substituent and His96, and hydrophobic interactions with six residues of the MDM2 binding site.
The docked poses of the studied compounds are presented in Figure 2, and Table 3 enlists all the observed interactions, the interacting groups, and docking scores.
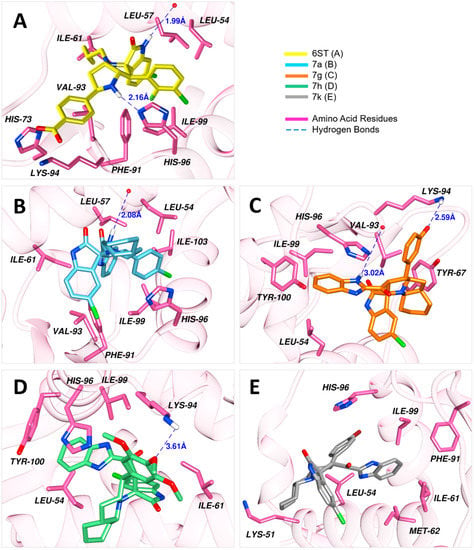
Figure 2.
Docked conformations of the ligands into the binding site of MDM2 (PDB ID: 5LAZ).

Table 3.
Docking scores and network of interactions formed between the ligands and residues of MDM2.
4. Conclusions
New spiroxindoles, combined with a benzimidazole scaffold, were synthesized, characterized, and identified as an MDM2 inhibitor. The requisite spiroxindoles were successfully achieved via the [3+2] cycloaddition reaction approach, which separated in an excellent regioselective and diastereoselective manner. The separated spirooxindoles showed promising results against cancer cells including MDA-MB 231 and PC3 in microscale reactivity. The anticancer reactivity for compound 7d showed potential activity with IC50 = 3.797±0.205 µM and was recognized as the most active candidate in the series. MDM2 binding analysis showed that compound 7a could be inhibited by the MDM2 with KD = 2.38 μm. This finding could be of possible use for cancer research development in the future.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/separations10040225/s1, Figure S1: one dose for compound 7g; Figure S2: Five doses for the compound 7g; Figure S3–S6: Binding curve for MST assay; Figure S7: validation of the docking protocol. Figure S8–S38: Selected NMR and IR spectrum; Table S1: Characterization of the chalcones 4a-o; Table S2: GI % at 10 μM concentration for compounds 7a-o; Table S3: The percentage of Wi-38 viability and the toxicity percentage of MDA-MB231, and PC3 cells after incubation with 5 µM of different tested compounds (7a-o).
Author Contributions
Conceptualization, A.B. and A.D.; methodology, S.A., M.A. and A.S.A.; software, M.S. and Z.U.-H.; validation, S.A., M.A. and A.S.A.; formal analysis, S.A., M.A. and A.S.A.; investigation, S.A., M.A. and A.S.A.; resources, A.B.; data curation, S.A. and Z.U.-H.; Biological activity assays: A.D. and M.M.A.-S.; writing—original draft preparation, A.B.; writing—review and editing, A.B.; visualization, A.M.A.-M.; supervision, A.M.A.-M.; project administration, M.A.; funding acquisition, A.B. All authors have read and agreed to the published version of the manuscript.
Funding
The authors extend their appreciation to the Deputyship for Research & Innovation, Ministry of Education in Saudi Arabia for funding this research work through the project number (IFKSURG–2-361).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The authors extend their appreciation to the Deputyship for Research & Innovation, Ministry of Education in Saudi Arabia for funding this research work through the project number (IFKSURG–2-361).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Xue, P.; Zhang, Z.; Sun, S. Targeting p53-MDM2 Interaction by Small Molecule Inhibitors: A Promising Strategy for Cancer Treatment. Int. J. Mol. Sci. 2019, 20, 64. [Google Scholar]
- Masuda, H.; Takahashi, M.; Takahashi, R. Small-molecule inhibitors targeting the p53-MDM2 interaction for cancer therapy. Drug Discov. Today 2018, 23, 774–784. [Google Scholar]
- Zhou, X.; Xu, X.; Huang, P. Small-molecule inhibitors targeting the p53-MDM2 interaction for cancer therapy: A review. Bioorg. Med. Chem. 2016, 24, 4710–4717. [Google Scholar]
- Zhu, H.; Gao, H.; Ji, Y.; Zhou, Q.; Du, Z.; Tian, L.; Jiang, Y.; Yao, K.; Zhou, Z. Targeting p53–MDM2 interaction by small-molecule inhibitors: Learning from MDM2 inhibitors in clinical trials. J. Hematol. Oncol. 2022, 15, 91. [Google Scholar] [CrossRef]
- Zhuang, C.; Miao, Z.; Zhu, L.; Dong, G.; Guo, Z.; Wang, S.; Zhang, Y.; Wu, Y.; Yao, J.; Sheng, C.; et al. Discovery, synthesis, and biological evaluation of orally active pyrrolidone derivatives as novel inhibitors of p53-MDM2 protein-protein interaction. J. Med. Chem. 2012, 55, 9630–9642. [Google Scholar] [CrossRef]
- Ding, K.; Lu, Y.; Nikolovska-Coleska, Z.; Wang, G.; Qiu, S.; Shangary, S.; Gao, W.; Qin, D.; Stuckey, J.; Krajewski, K.; et al. Structure-based design of spiro-oxindoles as potent, specific small-molecule inhibitors of the MDM2-p53 interaction. J. Med. Chem. 2006, 49, 3432–3435. [Google Scholar] [CrossRef]
- Sanz, G.; Singh, M.; Peuget, S.; Selivanova, G. Inhibition of p53 inhibitors: Progress, challenges and perspectives. J. Mol. Cell Biol. 2019, 11, 586–599. [Google Scholar] [CrossRef]
- Zhang, Q.; Zeng, S.X.; Lu, H. Targeting p53-MDM2-MDMX loop for cancer therapy. In Mutant p53 and MDM2 in Cancer; Springer: Berlin/Heidelberg, Germany, 2014; pp. 281–319. [Google Scholar]
- Ding, K.; Lu, Y.; Nikolovska-Coleska, Z.; Qiu, S.; Ding, Y.; Gao, W.; Stuckey, J.; Krajewski, K.; Roller, P.P.; Tomita, Y.; et al. Structure-based design of potent non-peptide MDM2 inhibitors. J. Am. Chem. Soc. 2005, 127, 10130–10131. [Google Scholar] [CrossRef]
- Fry, D.C.; Emerson, S.D.; Palme, S.; Vu, B.T.; Liu, C.M.; Podlaski, F. NMR structure of a complex between MDM2 and a small molecule inhibitor. J. Biomol. NMR 2004, 30, 163–173. [Google Scholar] [CrossRef]
- Rothweiler, U.; Czarna, A.; Krajewski, M.; Ciombor, J.; Kalinski, C.; Khazak, V.; Ross, G.; Skobeleva, N.; Weber, L.; Holak, T.A. Isoquinolin-1-one inhibitors of the MDM2-p53 interaction. Chem. Med. Chem. 2008, 3, 1118–1128. [Google Scholar] [CrossRef]
- Kumar, S.K.; Hager, E.; Pettit, C.; Gurulingappa, H.; Davidson, N.E.; Khan, S.R. Design, Synthesis, and Evaluation of Novel Boronic-Chalcone Derivatives as Antitumor Agents. J. Med. Chem. 2003, 46, 2813–2815. [Google Scholar] [CrossRef]
- Surmiak, E.; Twarda-Clapa, A.; Zak, K.M.; Musielak, B.; Tomala, M.D.; Kubica, K.; Grudnik, P.; Madej, M.; Jablonski, M.; Potempa, J.; et al. A Unique Mdm2-Binding Mode of the 3-Pyrrolin-2-one- and 2-Furanone-Based Antagonists of the p53-Mdm2 Interaction. ACS Chem. Biol. 2016, 11, 3310–3318. [Google Scholar] [CrossRef]
- Yang, M.C.; Peng, C.; Huang, H.; Yang, L.; He, X.H.; Huang, W.; Cui, H.L.; He, G.; Han, B. Organocatalytic Asymmetric Synthesis of Spiro-oxindole Piperidine Derivatives That Reduce Cancer Cell Proliferation by Inhibiting MDM2-p53 Interaction. Org. Lett. 2017, 19, 6752–6755. [Google Scholar] [CrossRef]
- Gonzalez, A.Z.; Eksterowicz, J.; Bartberger, M.D.; Beck, H.P.; Canon, J.; Chen, A.; Chow, D.; Duquette, J.; Fox, B.M.; Fu, J.; et al. Selective and potent morpholinone inhibitors of the MDM2-p53 protein-protein interaction. J. Med. Chem. 2014, 57, 2472–2488. [Google Scholar] [CrossRef]
- Furet, P.; Chene, P.; De Pover, A.; Valat, T.S.; Lisztwan, J.H.; Kallen, J.; Masuya, K. The central valine concept provides an entry in a new class of non peptide inhibitors of the p53-MDM2 interaction. Bioorg. Med. Chem. Lett. 2012, 22, 3498–3502. [Google Scholar] [CrossRef]
- Koblish, H.K.; Zhao, S.; Franks, C.F.; Donatelli, R.R.; Tominovich, R.M.; LaFrance, L.V.; Leonard, K.A.; Gushue, J.M.; Parks, D.J.; Calvo, R.R.; et al. Benzodiazepinedione inhibitors of the Hdm2:p53 complex suppress human tumor cell proliferation in vitro and sensitize tumors to doxorubicin in vivo. Mol. Cancer Ther. 2006, 5, 160–169. [Google Scholar] [CrossRef]
- Pettersson, M.; Quant, M.; Min, J.; Iconaru, L.; Kriwacki, R.W.; Waddell, M.B.; Guy, R.K.; Luthman, K.; Grotli, M. Design, Synthesis and Evaluation of 2,5-Diketopiperazines as Inhibitors of the MDM2-p53 Interaction. PLoS ONE 2015, 10, e0137867. [Google Scholar] [CrossRef]
- Allen, J.G.; Bourbeau, M.P.; Wohlhieter, G.E.; Bartberger, M.D.; Michelsen, K.; Hungate, R.; Gadwood, R.C.; Gaston, R.D.; Evans, B.; Mann, L.W.; et al. Discovery and optimization of chromenotriazolopyrimidines as potent inhibitors of the mouse double minute 2-tumor protein 53 protein-protein interaction. J. Med. Chem. 2009, 52, 7044–7053. [Google Scholar] [CrossRef]
- Aguilar, A.; Lu, J.; Liu, L.; Du, D.; Bernard, D.; McEachern, D.; Przybranowski, S.; Li, X.; Luo, R.; Wen, B.; et al. Discovery of 4-((30R,40S,50R)-6′-Chloro-40-(3-chloro-2-fluorophenyl)-10-ethyl-2′-oxodispiro[cyclohexane-1,20pyrrolidine-30,3′-indoline]-50—carboxamido)bicyclo[2.2.2]octane-1-carboxylic Acid (AA-115/APG-115): A Potent and Orally Active Murine Double Minute 2 (MDM2) Inhibitor in Clinical Development. J. Med. Chem. 2017, 60, 2819–2839. [Google Scholar]
- De Jonge, M.; de Weger, V.A.; Dickson, M.A.; Langenberg, M.; Le Cesne, A.; Wagner, A.J.; Hsu, K.; Zheng, W.; Mace, S.; Tuffal, G.; et al. A phase I study of SAR405838, a novel human double minute 2 (HDM2) antagonist, in patients with solid tumours. Eur. J. Cancer 2017, 76, 144–151. [Google Scholar] [CrossRef]
- Zanjirband, M.; Edmondson, R.J.; Lunec, J. Pre-clinical efficacy and synergistic potential of the MDM2-p53 antagonists, Nutlin-3 and RG7388, as single agents and in combined treatment with cisplatin in ovarian cancer. Oncotarget 2016, 7, 40115–40134. [Google Scholar] [CrossRef] [PubMed]
- Novartis Pharmaceuticals. Study of Safety and Efficacy of HDM201 in Combination with LEE011 in Patients with Liposarcoma. ClinicalTrials. Gov Identifier: NCT02343172. 2015. Available online: https://clinicaltrials.gov/ct2/show/NCT02343172 (accessed on 13 February 2023).
- Andreeff, M.; Kelly, K.R.; Yee, K.; Assouline, S.; Strair, R.; Popplewell, L.; Bowen, D.; Martinelli, G.; Drummond, M.W.; Vyas, P.; et al. Results of the Phase I Trial of RG7112, a Small-Molecule MDM2 Antagonist in Leukemia. Clin. Cancer Res. 2016, 22, 868–876. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Li, Z.; Rew, Y.; Gribble, M.; Bartberger, M.D.; Beck, H.P.; Canon, J.; Chen, A.; Chen, X.; Chow, D.; et al. Discovery of AMG 232, a potent, selective, and orally bioavailable MDM2-p53 inhibitor in clinical development. J. Med. Chem. 2014, 57, 1454–1472. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Liu, L.; Sun, W.; Lu, J.; McEachern, D.; Li, X.; Yu, S.; Bernard, D.; Ochsenbein, P.; Ferey, V.; et al. Diastereomeric spirooxindoles as highly potent and efficacious MDM2 inhibitors. J. Am. Chem. Soc. 2013, 135, 7223–7234. [Google Scholar] [CrossRef]
- Tovar, C.; Graves, B.; Packman, K.; Filipovic, Z.; Xia, B.H.M.; Tardell, C.; Garrido, R.; Lee, E.; Kolinsky, K.; To, K.H.; et al. MDM2 small-molecule antagonist RG7112 activates p53 signaling and regresses human tumors in preclinical cancer models. Cancer Res. 2013, 73, 2587–2597. [Google Scholar] [CrossRef]
- Beloglazkina, A.; Zyk, N.; Majouga, A.; Beloglazkina, E. Recent small-molecule inhibitors of the p53–MDM2 protein–protein interaction. Molecules 2020, 25, 1211. [Google Scholar] [CrossRef]
- Riedinger, C.; McDonnell, J.M. Inhibitors of MDM2 and MDMX: A structural perspective. Future Med. Chem. 2009, 1, 1075–1094. [Google Scholar] [CrossRef]
- Millard, M.; Pathania, D.; Grande, F.; Xu, S.; Neamati, N. Small-molecule inhibitors of p53-MDM2 interaction: The 2006–2010 update. Curr. Pharm. Des. 2011, 17, 536–559. [Google Scholar] [CrossRef]
- Vassilev, L.T. MDM2 inhibitors for cancer therapy. Trends Mol. Med. 2007, 13, 23–31. [Google Scholar] [CrossRef]
- Zhang, Z.; Chu, X.J.; Liu, J.J.; Ding, Q.; Zhang, J.; Bartkovitz, D.; Jiang, N.; Karnachi, P.; So, S.S.; Tovar, C.; et al. Discovery of potent and orally active p53-MDM2 inhibitors RO5353 and RO2468 for potential clinical development. ACS Med. Chem. Lett. 2014, 5, 124–127. [Google Scholar] [CrossRef]
- Cui, C.B.; Kakeya, H.; Osada, H. Spirotryprostatin B, a novel mammalian cell cycle inhibitor produced by Aspergillus fumigatus. J. Antibiot. 1996, 49, 832–835. [Google Scholar] [CrossRef]
- Gollner, A.; Rudolph, D.; Arnhof, H.; Bauer, M.; Blake, S.M.; Boehmelt, G.; Cockroft, X.L.; Dahmann, G.; Ettmayer, P.; Gerstberger, T.; et al. Discovery of novel spiro [3 H-indole-3, 2′-pyrrolidin]-2 (1 H)-one compounds as chemically stable and orally active inhibitors of the MDM2–p53 interaction. J. Med. Chem. 2016, 59, 10147–10162. [Google Scholar] [CrossRef]
- Islam, M.S.; Al-Majid, A.M.; El-Senduny, F.F.; Badria, F.A.; Rahman, A.F.M.; Barakat, A.; Elshaier, Y.A. Synthesis, Anticancer Activity, and Molecular Modeling of New Halogenated Spiro [pyrrolidine-thiazolo-oxindoles] Derivatives. Appl. Sci. 2020, 10, 2170. [Google Scholar] [CrossRef]
- Barakat, A.; Islam, M.S.; Ghawas, H.M.; Al-Majid, A.M.; El-Senduny, F.F.; Badria, F.A.; Elshaier, Y.A.; Ghabbour, H.A. Design and synthesis of new substituted spirooxindoles as potential inhibitors of the MDM2–p53 interaction. Bioorg. Chem. 2019, 86, 598–608. [Google Scholar] [CrossRef]
- Islam, M.S.; Ghawas, H.M.; El-Senduny, F.F.; Al-Majid, A.M.; Elshaier, Y.A.; Badria, F.A.; Barakat, A. Synthesis of new thiazolo-pyrrolidine–(spirooxindole) tethered to 3-acylindole as anticancer agents. Bioorg. Chem. 2019, 82, 423–430. [Google Scholar] [CrossRef]
- Lotfy, G.; Aziz, Y.M.A.; Said, M.M.; El Sayed, H.; El Sayed, H.; Abu-Serie, M.M.; Teleb, M.; Dömling, A.; Barakat, A. Molecular hybridization design and synthesis of novel spirooxindole-based MDM2 inhibitors endowed with BCL2 signaling attenuation; a step towards the next generation p53 activators. Bioorg. Chem. 2021, 117, 105427. [Google Scholar] [CrossRef]
- Aziz, Y.M.A.; Lotfy, G.; Said, M.M.; El Ashry, E.S.H.; El Tamany, E.S.H.; Soliman, S.; Abu-Serie, M.M.; Teleb, M.; Yousuf, S.; Dömling, A. Design, synthesis, chemical and biochemical insights on to novel hybrid spirooxindoles-based p53-MDM2 inhibitors with potential Bcl2 signaling attenuation. Front. Chem. 2021, 9, 915. [Google Scholar] [CrossRef]
- Popowicz, G.M.; Dömling, A.; Holak, T.A. The structure-based design of MDM2/MDMX–p53 inhibitors gets serious. Angew. Chem. Int. Ed. 2011, 50, 2680–2688. [Google Scholar] [CrossRef]
- Mrkvová, Z.; Uldrijan, S.; Pombinho, A.; Bartůněk, P.; Slaninová, I. Benzimidazoles downregulate Mdm2 and MdmX and activate p53 in MdmX overexpressing tumor cells. Molecules 2019, 24, 2152. [Google Scholar] [CrossRef]
- Wu, L.T.; Jiang, Z.; Shen, J.J.; Yi, H.; Zhan, Y.C.; Sha, M.Q.; Wang, Z.; Xue, S.T.; Li, Z.R. Design, synthesis and biological evaluation of novel benzimidazole-2-substituted phenyl or pyridine propyl ketene derivatives as antitumour agents. Eur. J. Med. Chem. 2016, 114, 328–336. [Google Scholar] [CrossRef]
- Zaytsev, A.; Dodd, B.; Magnani, M.; Ghiron, C.; Golding, B.T.; Griffin, R.J.; Liu, J.; Lu, X.; Micco, I.; Newell, D.R.; et al. Searching for Dual Inhibitors of the MDM 2- p53 and MDMX- p53 Protein–Protein Interaction by a Scaffold- Hopping Approach. Chem. Biol. Drug. Des. 2015, 86, 180–189. [Google Scholar] [CrossRef] [PubMed]
- Boggu, P.; Venkateswararao, E.; Manickam, M.; Kim, Y.; Jung, S.H. Exploration of SAR for novel 2-benzylbenzimidazole analogs as inhibitor of transcription factor NF-κB. Arch. Pharm. Res. 2017, 40, 469–479. [Google Scholar] [CrossRef] [PubMed]
- Błaszczak-Świątkiewicz, K. Antiproliferative aspect of benzimidazole derivatives’ activity and their impact on NF-κB expression. Molecules 2019, 24, 3902. [Google Scholar] [CrossRef] [PubMed]
- Boggu, P.; Venkateswararao, E.; Manickam, M.; Kwak, D.; Kim, Y.; Jung, S.H. Exploration of 2-benzylbenzimidazole scaffold as novel inhibitor of NF-κB. Bioorg. Med. Chem. 2016, 24, 1872–1878. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Viallet, J.; Haura, E.B. A small molecule pan-Bcl-2 family inhibitor, GX15-070, induces apoptosis and enhances cisplatin-induced apoptosis in non-small cell lung cancer cells. Cancer Chemother. Pharmacol. 2008, 61, 525–534. [Google Scholar] [CrossRef]
- Barakat, A.; Alshahrani, S.; Al-Majid, A.M.; Alamary, A.S.; Haukka, M.; Abu-Serie, M.M.; Dömling, A.; Mazyed, E.A.; Badria, F.A.; El-Senduny, F.F. Novel spirooxindole based benzimidazole scaffold: In vitro, nanoformulation and in vivo studies on anticancer and antimetastatic activity of breast adenocarcinoma. Bioorg. Chem. 2022, 129, 106124. [Google Scholar] [CrossRef]
- Vilar, S.; Cozza, G.; Moro, S. Medicinal chemistry and the molecular operating environment (MOE): Application of QSAR and molecular docking to drug discovery. Curr. Top. Med. Chem. 2008, 8, 1555–1572. [Google Scholar] [CrossRef]
- Salentin, S.; Schreiber, S.; Haupt, V.J.; Adasme, M.F.; Schroeder, M. PLIP: Fully automated protein–ligand interaction profiler. Nucleic Acids Res. 2015, 43, W443–W447. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).