Steric Exclusion Chromatography for Purification of Biomolecules—A Review
Abstract
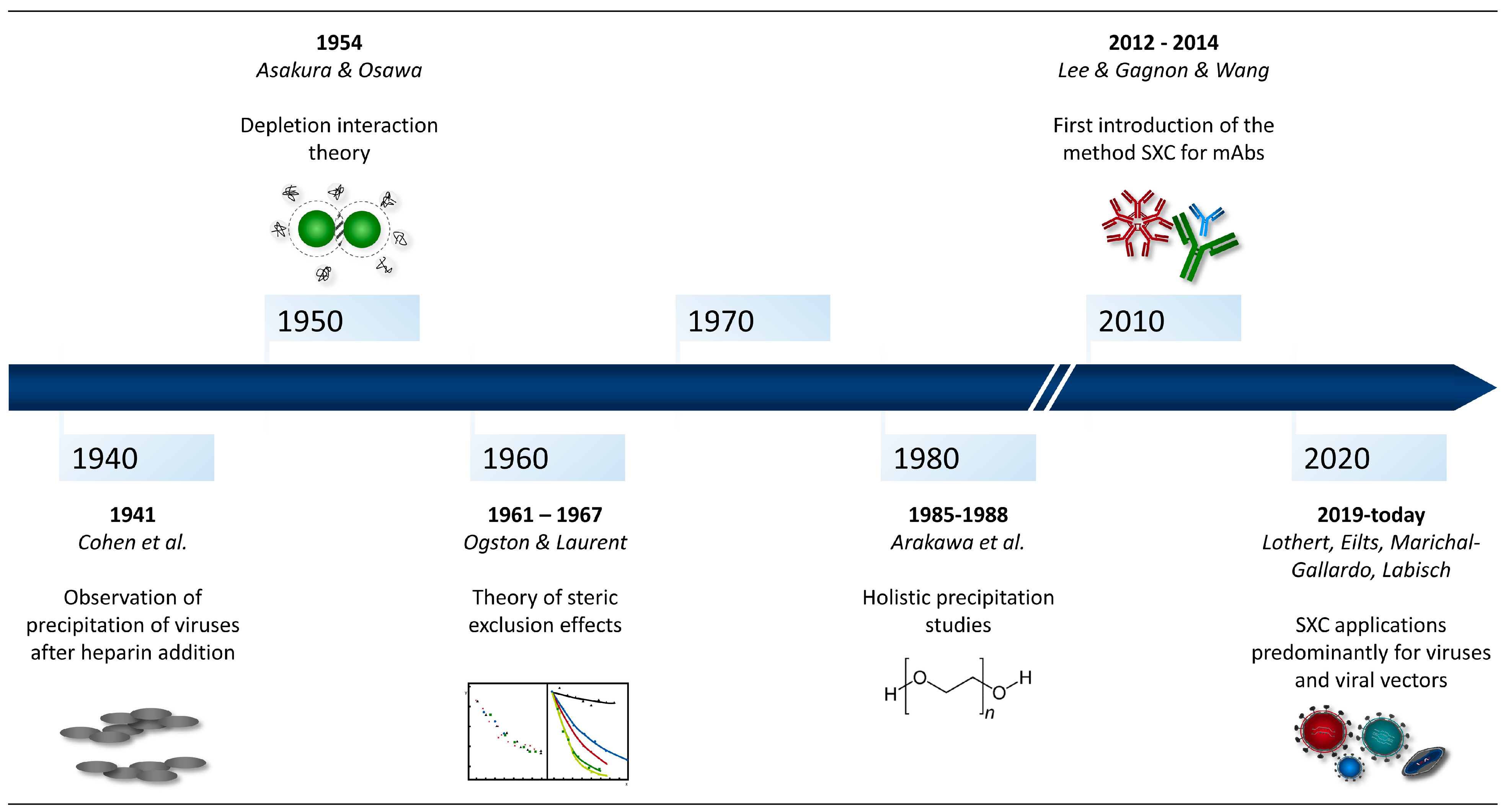
1. Introduction
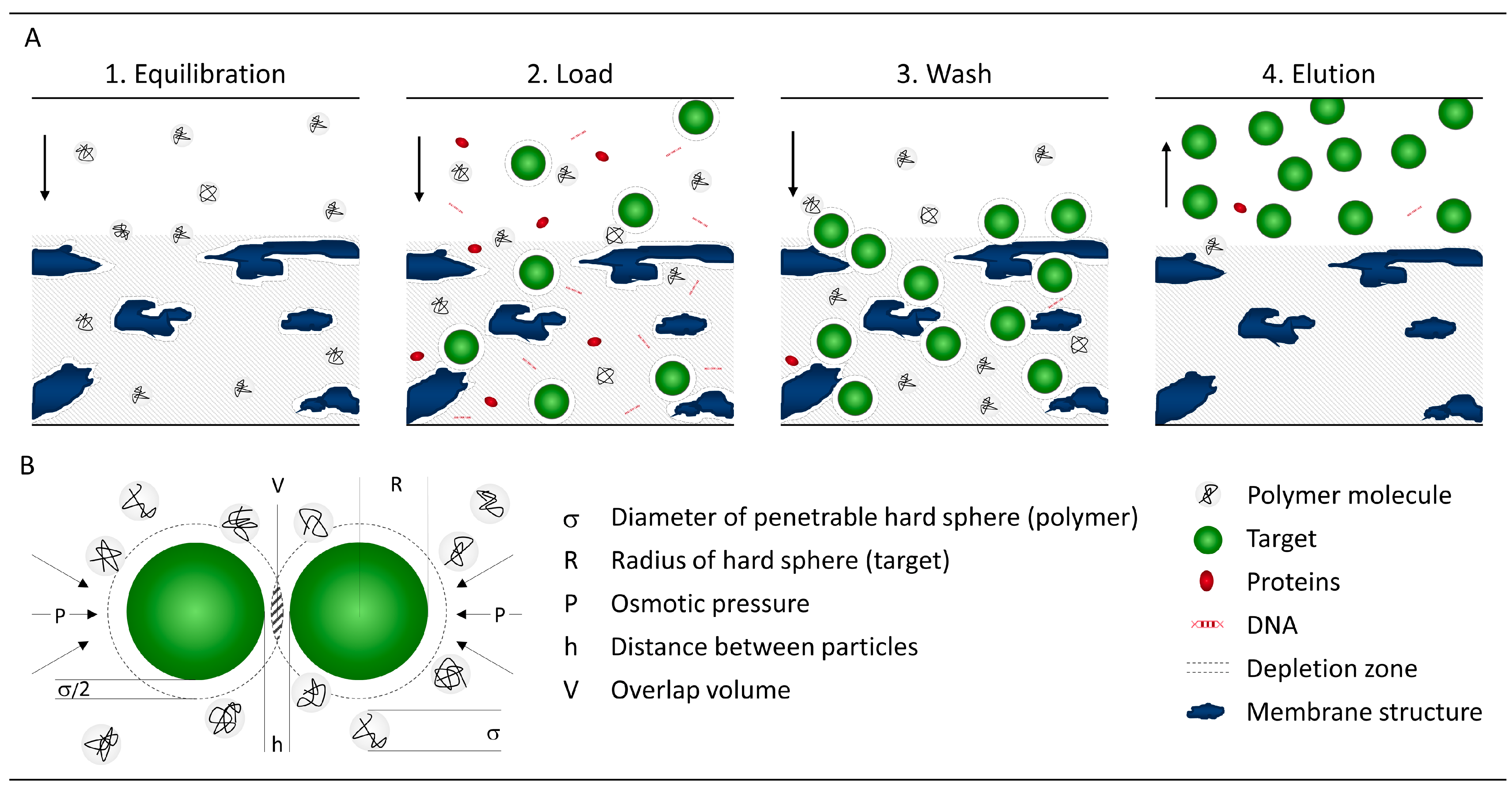
2. Method Fundamentals
3. Process Parameters and Optimization of SXC
3.1. Technical Requirements
3.1.1. Equipment
3.1.2. Stationary Phases
| Stationary Phase | Size | Pore Size | Supplier | Reference |
|---|---|---|---|---|
| CIMmultus® OH monolith | 0.34 mL, 1 mL 1 | 1.3 µm 2, 2 µm | Sartorius BIA Separations | [1,22,31] |
| Polyacrylamide monolith | 0.83 mL 3 | 10–100 µm | Self-fabricated | [21] |
| Whatman® regenerated cellulose membrane filters | 13 mm or 25 mm diameter, 10; 15, or 20 layers | 1 µm | Cytiva | [18,22,23,24,25,26,27,28] |
| Hydrosart® stabilized cellulose membrane | 13 mm diameter with 10 or 15 layers; 25 mm diameter with 5 layers | 3–5 µm | Sartorius | [16,25,29] |
| Glass fiber filters | 13 mm diameter, 15 layers | 1.6 µm | VWR | [29] |
| Ultipor® Nylon polyamide membrane | 13 mm diameter, 15 layers | 5 µm | Pall | [29] |
| Starch-coated magnetic nanoparticles | - | - | Chemicell | [2] |
3.2. Buffers
3.2.1. PEG Size and Concentration
3.2.2. Mixing Approaches of PEG with the Feed Solution
3.2.3. Buffer Systems and Influence of Salt
3.3. Scalability
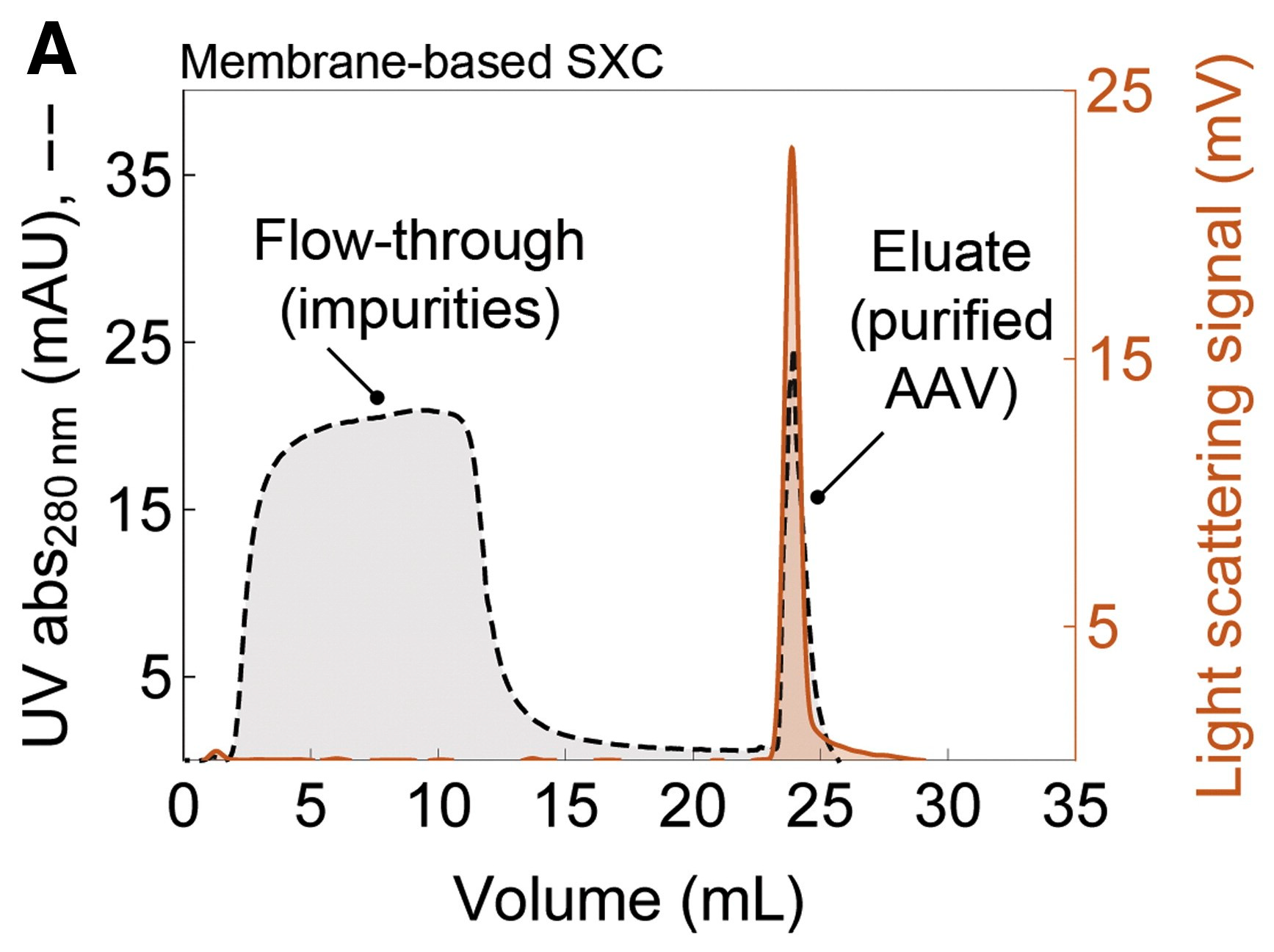
4. Applications of SXC
5. Conclusions and Future Perspectives
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Lee, J.; Gan, H.T.; Latiff, S.M.A.; Chuah, C.; Lee, W.Y.; Yang, Y.-S.; Loo, B.; Ng, S.K.; Gagnon, P. Principles and applications of steric exclusion chromatography. J. Chromatogr. A 2012, 1270, 162–170. [Google Scholar] [CrossRef] [PubMed]
- Gagnon, P.; Toh, P.; Lee, J. High productivity purification of immunoglobulin G monoclonal antibodies on starch-coated magnetic nanoparticles by steric exclusion of polyethylene glycol. J. Chromatogr. A 2013, 1324, 171–180. [Google Scholar] [CrossRef]
- Cohen, S.S. The isolation and crystallization of plant viruses and other protein macro molecules by means of hydrophilic colloids. J. Biol. Chem. 1942, 144, 353–362. [Google Scholar] [CrossRef]
- Polson, A.; Potgieter, G.M.; Largier, J.F.; Mears, G.E.; Joubert, F.J. The fractionation of protein mixtures by linear polymers of high molecular weight. Biochim. Biophys. Acta 1964, 82, 463–475. [Google Scholar] [CrossRef] [PubMed]
- Ogston, A.G.; Preston, B.N. The exclusion of protein by hyaluronic acid. J. Biol. Chem. 1966, 241, 17–19. [Google Scholar] [CrossRef]
- Iverius, P.H.; Laurent, T.C. Precipitation of some plasma proteins by the addition of dextran or polyethylene glycol. Biochim. Biophys. Acta 1967, 133, 371–373. [Google Scholar] [CrossRef] [PubMed]
- Laurent, T.C. The interaction between polysaccharides and other macromolecules. 5. The solubility of proteins in the prescence of dextran. Biochem. J. 1963, 89, 253–257. [Google Scholar] [CrossRef]
- Ogston, A.G. Excluded-Volume Interactions of Neutral Polymers in Solution. In Chemistry and Technology of Water-Soluble Polymers; Finch, C.A., Ed.; Springer: Boston, MA, USA, 1983; pp. 203–213. ISBN 978-1-4757-9663-6. [Google Scholar]
- Ogston, A.G.; Phelps, C.F. The partition of solutes between buffer solutions and solutions containing hyaluronic acid. Biochem. J. 1961, 78, 827–833. [Google Scholar] [CrossRef]
- Polson, A. A theory for the displacement of proteins and viruses with polyethylene glycol. Prep. Biochem. 1977, 7, 129–154. [Google Scholar] [CrossRef]
- Asakura, S.; Oosawa, F. On interaction between two bodies immersed in a solution of macromolecules. J. Chem. Phys. 1954, 22, 1255–1256. [Google Scholar] [CrossRef]
- Vrij, A. Polymers at interfaces and the interactions in colloidal dispersions. Pure Appl. Chem. 1976, 4, 471–483. [Google Scholar] [CrossRef]
- Arakawa, T.; Timasheff, S.N. Mechanism of poly(ethylene glycol) interaction with proteins. Biochemistry 1985, 24, 6756–6762. [Google Scholar] [CrossRef]
- Timasheff, S.N.; Arakawa, T. Mechanism of protein precipitation and stabilization by co-solvents. J. Cryst. Growth 1988, 90, 39–46. [Google Scholar] [CrossRef]
- Sim, S.-L.; He, T.; Tscheliessnig, A.; Mueller, M.; Tan, R.B.H.; Jungbauer, A. Protein precipitation by polyethylene glycol: A generalized model based on hydrodynamic radius. J. Biotechnol. 2012, 157, 315–319. [Google Scholar] [CrossRef]
- Labisch, J.J.; Kassar, M.; Bollmann, F.; Valentic, A.; Hubbuch, J.; Pflanz, K. Steric exclusion chromatography of lentiviral vectors using hydrophilic cellulose membranes. J. Chromatogr. A 2022, 17, 463148. [Google Scholar] [CrossRef]
- Lekkerkerker, H.N.W.; Tuinier, R. Colloids and the Depletion Interaction; Springer: Dordrecht, The Netherlands, 2011. [Google Scholar]
- Marichal-Gallardo, P.; Börner, K.; Pieler, M.M.; Sonntag-Buck, V.; Obr, M.; Bejarano, D.; Wolff, M.W.; Kräusslich, H.-G.; Reichl, U.; Grimm, D. Single-use capture purification of adeno-associated viral gene transfer vectors by membrane-based steric exclusion chromatography. Hum. Gene Ther. 2021, 32, 959–974. [Google Scholar] [CrossRef]
- Tuinier, R.; Rieger, J.; de Kruif, C.G. Depletion-induced phase separation in colloid–polymer mixtures. Adv. Colloid Interface Sci. 2003, 103, 1–31. [Google Scholar] [CrossRef]
- Atha, D.H.; Ingham, K.C. Mechanism of precipitation of proteins by polyethylene glycols. Analysis in terms of excluded volume. J. Biol. Chem. 1981, 256, 12108–12117. [Google Scholar] [PubMed]
- Wang, C.; Bai, S.; Tao, S.-P.; Sun, Y. Evaluation of steric exclusion chromatography on cryogel column for the separation of serum proteins. J. Chromatogr. A 2014, 1333, 54–59. [Google Scholar] [CrossRef] [PubMed]
- Lothert, K.; Sprick, G.; Beyer, F.; Lauria, G.; Czermak, P.; Wolff, M.W. Membrane-based steric exclusion chromatography for the purification of a recombinant baculovirus and its application for cell therapy. J. Virol. Methods 2020, 275, 113756. [Google Scholar] [CrossRef] [PubMed]
- Eilts, F.; Steger, M.; Lothert, K.; Wolff, M.W. The suitability of latex particles to evaluate critical process parameters in steric exclusion chromatography. Membranes 2022, 12, 488. [Google Scholar] [CrossRef] [PubMed]
- Hein, M.D.; Kollmus, H.; Marichal-Gallardo, P.; Püttker, S.; Benndorf, D.; Genzel, Y.; Schughart, K.; Kupke, S.Y.; Reichl, U. OP7, a novel influenza A virus defective interfering particle: Production, purification, and animal experiments demonstrating antiviral potential. Appl. Microbiol. Biotechnol. 2021, 105, 129–146. [Google Scholar] [CrossRef] [PubMed]
- Lothert, K.; Offersgaard, A.F.; Pihl, A.F.; Mathiesen, C.K.; Jensen, T.B.; Alzua, G.P.; Fahnøe, U.; Bukh, J.; Gottwein, J.M.; Wolff, M.W. Development of a downstream process for the production of an inactivated whole hepatitis C virus vaccine. Sci. Rep. 2020, 10, 16261. [Google Scholar] [CrossRef] [PubMed]
- Eilts, F.; Lothert, K.; Orbay, S.; Pagallies, F.; Amann, R.; Wolff, M.W. A summary of practical considerations for the application of the steric exclusion chromatography for the purification of the Orf viral vector. Membranes 2022, 12, 1070. [Google Scholar] [CrossRef] [PubMed]
- Lothert, K.; Pagallies, F.; Eilts, F.; Sivanesapillai, A.; Hardt, M.; Moebus, A.; Feger, T.; Amann, R.; Wolff, M.W. A scalable downstream process for the purification of the cell culture-derived Orf virus for human or veterinary applications. J. Biotechnol. 2020, 323, 221–230. [Google Scholar] [CrossRef] [PubMed]
- Lothert, K.; Pagallies, F.; Feger, T.; Amann, R.; Wolff, M.W. Selection of chromatographic methods for the purification of cell culture-derived Orf virus for its application as a vaccine or viral vector. J. Biotechnol. 2020, 323, 62–72. [Google Scholar] [CrossRef]
- Marichal-Gallardo, P.; Pieler, M.M.; Wolff, M.W.; Reichl, U. Steric exclusion chromatography for purification of cell culture-derived influenza A virus using regenerated cellulose membranes and polyethylene glycol. J. Chromatogr. A 2017, 1483, 110–119. [Google Scholar] [CrossRef]
- ContiVir. Membrane-Based Steric Exclusion Chromatography (SXC): Rapid Cycling & Single-Use Universal Purification Method for Virus Particles. Available online: https://www.contivir.com/dsp-sxc (accessed on 1 December 2022).
- Levanova, A.; Poranen, M.M. Application of steric exclusion chromatography on monoliths for separation and purification of RNA molecules. J. Chromatogr. A 2018, 1574, 50–59. [Google Scholar] [CrossRef]
- Sartorius BIA Separations. OH-HIC. Available online: https://www.biaseparations.com/en/products/monolithic-columns/products-for-preparative-applications/69/oh-hic (accessed on 6 December 2022).
- Jungbauer, A. Chromatographic media for bioseparation. J. Chromatogr. A 2005, 1065, 3–12. [Google Scholar] [CrossRef]
- Barut, M.; Podgornik, A.; Brne, P.; Strancar, A. Convective interaction media short monolithic columns: Enabling chromatographic supports for the separation and purification of large biomolecules. J. Sep. Sci. 2005, 28, 1876–1892. [Google Scholar] [CrossRef]
- Zydney, A.L. New developments in membranes for bioprocessing—A review. J. Membr. Sci. 2021, 620, 118804. [Google Scholar] [CrossRef]
- Hoffmann, D.; Leber, J.; Loewe, D.; Lothert, K.; Oppermann, T.; Zitzmann, J.; Weidner, T.; Salzig, D.; Wolff, M.; Czermak, P. Purification of new biologicals using membrane-based processes. In Current Trends and Future Developments on (Bio-) Membranes; Elsevier: Amsterdam, The Netherlands, 2019; pp. 123–150. [Google Scholar]
- Perry, C.; Rayat, A.C.M.E. Lentiviral vector bioprocessing. Viruses 2021, 13, 268. [Google Scholar] [CrossRef] [PubMed]
- Minh, A.D.; Kamen, A.A. Critical assessment of purification and analytical technologies for enveloped viral vector and vaccine processing and their current limitations in resolving co-expressed extracellular vesicles. Vaccines 2021, 9, 823. [Google Scholar] [CrossRef] [PubMed]
- Nestola, P.; Peixoto, C.; Silva, R.R.J.S.; Alves, P.M.; Mota, J.P.B.; Carrondo, M.J.T. Improved virus purification processes for vaccines and gene therapy. Biotechnol. Bioeng. 2015, 112, 843–857. [Google Scholar] [CrossRef] [PubMed]
- Gaudin, T.; Ma, H. The macroscopic viscosity approximation: A first-principle relationship between molecular diffusion and viscosity. AIP Adv. 2020, 10, 35321. [Google Scholar] [CrossRef]
- D’souza, A.A.; Shegokar, R. Polyethylene glycol (PEG): A versatile polymer for pharmaceutical applications. Expert Opin. Drug Deliv. 2016, 13, 1257–1275. [Google Scholar] [CrossRef]
- Miekka, S.I.; Ingham, K.C. Influence of self-association of proteins on their precipitation by poly(ethylene glycol). Arch. Biochem. Biophys. 1978, 191, 525–536. [Google Scholar] [CrossRef]
- Labisch, J.J.; Paul, R.; Wiese, G.; Pflanz, K. Scaling Up of Steric Exclusion Membrane Chromatography for Lentiviral Vector Purification. Membranes 2023, 13, 149. [Google Scholar] [CrossRef]
- Sim, S.-L.; He, T.; Tscheliessnig, A.; Mueller, M.; Tan, R.B.H.; Jungbauer, A. Branched polyethylene glycol for protein precipitation. Biotechnol. Bioeng. 2012, 109, 736–746. [Google Scholar] [CrossRef]
- Matter, F.; Luna, A.L.; Niederberger, M. From colloidal dispersions to aerogels: How to master nanoparticle gelation. Nano Today 2020, 30, 100827. [Google Scholar] [CrossRef]
- Thomas, D.K.; Charlesby, A. Viscosity relationship in solutions of polyethylene glycols. J. Polym. Sci. 1960, 42, 195–202. [Google Scholar] [CrossRef]
- Eilts, F.; Steger, M.; Pagallies, F.; Rziha, H.-J.; Hardt, M.; Amann, R.; Wolff, M.W. Comparison of sample preparation techniques for the physicochemical characterization of Orf virus particles. J. Virol. Methods 2022, 310, 114614. [Google Scholar] [CrossRef]
- Hofmeister, F. Zur Lehre von der Wirkung der Salze. Arch. Für Exp. Pathol. Und Pharmakol. 1888, 24, 247–260. [Google Scholar] [CrossRef]
- Tan, C.; Albright, J.G.; Annunziata, O. Determination of preferential interaction parameters by multicomponent diffusion. applications to poly(ethylene glycol)-salt-water ternary mixtures. J. Phys. Chem. B 2008, 112, 4967–4974. [Google Scholar] [CrossRef]
- Ingham, K.C. Precipitation of proteins with polyethylene glycol: Characterization of albumin. Arch. Biochem. Biophys. 1978, 186, 106–113. [Google Scholar] [CrossRef] [PubMed]
- Lothert, K. Steric exclusion chromatography: Advancement of a laboratory-based platform technology into a key component of viral vector and vaccine production processes. Ph.D. Thesis, Forschungscampus Mittelhessen, Gießen, Germany, 2022. [Google Scholar]
- Saber, R.; Sarkar, S.; Gill, P.; Nazari, B.; Faridani, F. High resolution imaging of IgG and IgM molecules by scanning tunneling microscopy in air condition. Sci. Iran. 2011, 18, 1643–1646. [Google Scholar] [CrossRef]
- Choi, H.S.; Liu, W.; Misra, P.; Tanaka, E.; Zimmer, J.P.; Itty Ipe, B.; Bawendi, M.G.; Frangioni, J.V. Renal clearance of quantum dots. Nat. Biotechnol. 2007, 25, 1165–1170. [Google Scholar] [CrossRef]
- Rodrigues, T.; Carrondo, M.J.T.; Alves, P.M.; Cruz, P.E. Purification of retroviral vectors for clinical application: Biological implications and technological challenges. J. Biotechnol. 2007, 127, 520–541. [Google Scholar] [CrossRef]
- Segura, M.M.; Kamen, A.A.; Garnier, A. Overview of current scalable methods for purification of viral vectors. Methods Mol. Biol. 2011, 737, 89–116. [Google Scholar] [CrossRef]
- Zimmermann, K.; Scheibe, O.; Kocourek, A.; Muelich, J.; Jurkiewicz, E.; Pfeifer, A. Highly efficient concentration of lenti- and retroviral vector preparations by membrane adsorbers and ultrafiltration. BMC Biotechnol. 2011, 11, 55. [Google Scholar] [CrossRef]
- Labisch, J.J.; Wiese, G.P.; Barnes, K.; Bollmann, F.; Pflanz, K. Infectious titer determination of lentiviral vectors using a temporal immunological real-time imaging approach. PLoS ONE 2021, 16, e0254739. [Google Scholar] [CrossRef]
- Weiss, K.; Salzig, D.; Röder, Y.; Gerstenberger, J.; Mühlebach, M.D.; Cichutek, K.; Pörtner, R.; Czermak, P. Influence of process conditions on measles virus stability. Am. J. Biochem. Biotechnol. 2013, 9, 243–254. [Google Scholar] [CrossRef]
- Loewe, D.; Häussler, J.; Grein, T.A.; Dieken, H.; Weidner, T.; Salzig, D.; Czermak, P. Forced degradation studies to identify critical process parameters for the purification of infectious measles virus. Viruses 2019, 11, 725. [Google Scholar] [CrossRef]
- Tinch, S.; Szczur, K.; Swaney, W.; Reeves, L.; Witting, S.R. A scalable lentiviral vector production and purification method using Mustang Q chromatography and tangential flow filtration. Methods Mol. Biol. 2019, 1937, 135–153. [Google Scholar] [CrossRef]
- Bandeira, V.; Peixoto, C.; Rodrigues, A.F.; Cruz, P.E.; Alves, P.M.; Coroadinha, A.S.; Carrondo, M.J.T. Downstream processing of lentiviral vectors: Releasing bottlenecks. Hum. Gene Ther. Methods 2012, 23, 255–263. [Google Scholar] [CrossRef]
- Morenweiser, R. Downstream processing of viral vectors and vaccines. Gene Ther. 2005, 12 (Suppl 1), S103–S110. [Google Scholar] [CrossRef]
- Moleirinho, M.G.; Silva, R.J.S.; Alves, P.M.; Carrondo, M.J.T.; Peixoto, C. Current challenges in biotherapeutic particles manufacturing. Expert Opin. Biol. Ther. 2020, 20, 451–465. [Google Scholar] [CrossRef]
- Liu, H.F.; Ma, J.; Winter, C.; Bayer, R. Recovery and purification process development for monoclonal antibody production. MAbs 2010, 2, 480–499. [Google Scholar] [CrossRef]
- Ramos-de-la-Peña, A.M.; González-Valdez, J.; Aguilar, O. Protein A chromatography: Challenges and progress in the purification of monoclonal antibodies. J. Sep. Sci. 2019, 42, 1816–1827. [Google Scholar] [CrossRef]
- McNally, D.J.; Darling, D.; Farzaneh, F.; Levison, P.R.; Slater, N.K.H. Optimised concentration and purification of retroviruses using membrane chromatography. J. Chromatogr. A 2014, 1340, 24–32. [Google Scholar] [CrossRef]
- Valkama, A.J.; Oruetxebarria, I.; Lipponen, E.M.; Leinonen, H.M.; Käyhty, P.; Hynynen, H.; Turkki, V.; Malinen, J.; Miinalainen, T.; Heikura, T.; et al. Development of large-scale downstream processing for lentiviral vectors. Mol. Ther. Methods Clin. Dev. 2020, 17, 717–730. [Google Scholar] [CrossRef]
- Swierczewska, M.; Lee, K.C.; Lee, S. What is the future of PEGylated therapies? Expert Opin. Emerg. Drugs 2015, 20, 531–536. [Google Scholar] [CrossRef]
- d’Avanzo, N.; Celia, C.; Barone, A.; Carafa, M.; Di Marzio, L.; Santos, H.A.; Fresta, M. Immunogenicity of polyethylene glycol based nanomedicines: Mechanisms, clinical implications and systematic approach. Adv. Therap. 2020, 3, 1900170. [Google Scholar] [CrossRef]
- Hohmann-Jeddi, C. PEG als ein Anaphylaxie-Auslöser Nach Covid-Impfung Bestätigt. 2021. Available online: https://www.pharmazeutische-zeitung.de/peg-als-ein-anaphylaxie-ausloeser-nach-covid-impfung-bestaetigt-124884/ (accessed on 13 February 2023).

| Target | Size | Shape | Maximum Recovery | Impurity Removal | References |
|---|---|---|---|---|---|
| Bacteriophage M13KO7 | 916 × 7.2 nm | rod | >90% | >99% HCP, 93% DNA | [1] |
| IgM | 25–35 nm diameter 4 | radial pentamer | ≤90% | 95% HCP | [1] |
| IgG | 12 nm diameter 5 | Y-shaped | 87% | 99% HCP | [2] |
| γ-globulin | 12–35 nm diameter | various | n.a. | n.a. | [21] |
| Influenza A virus | 80–120 nm diameter | spherical, sometimes filamentous | >95% | >99% HCD, 92% HCP | [22] |
| ssRNA dsRNA | 700–6374 nt and 500–6374 bp length | various | >90% retained on the column | n.a. | [31] |
| Baculovirus 1 | 200–300 nm × 30–60 nm | rod | 91% | >99% total protein, 85% total DNA | [29] |
| Parapoxvirus ovis (Orf) | 220–300 nm × 140–200 nm | rod | 67% to >90% | >98% total protein, >60% total DNA | [23,24,25,47] |
| Hepatitis C virus | 30–80 nm diameter | spherical | >97% | >99% total protein, 84% total DNA | [26] |
| OP7 2 | 80–90 nm diameter | spherical | n.a. | 89% total protein | [27] |
| Adeno-associated virus (AAV) | 20 nm diameter | near spherical | >95% | ≥80% total protein, ≥94% total DNA | [18] |
| Lentiviral vector | 120 nm diameter | spherical | ≥86% | 80% total protein and total DNA | [16] |
| Latex particles 3 | 190 nm diameter | spherical | ≤21% for CPS, ≤7% for CPH | n.a. | [28] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Labisch, J.J.; Wiese, G.P.; Pflanz, K. Steric Exclusion Chromatography for Purification of Biomolecules—A Review. Separations 2023, 10, 183. https://doi.org/10.3390/separations10030183
Labisch JJ, Wiese GP, Pflanz K. Steric Exclusion Chromatography for Purification of Biomolecules—A Review. Separations. 2023; 10(3):183. https://doi.org/10.3390/separations10030183
Chicago/Turabian StyleLabisch, Jennifer J., G. Philip Wiese, and Karl Pflanz. 2023. "Steric Exclusion Chromatography for Purification of Biomolecules—A Review" Separations 10, no. 3: 183. https://doi.org/10.3390/separations10030183
APA StyleLabisch, J. J., Wiese, G. P., & Pflanz, K. (2023). Steric Exclusion Chromatography for Purification of Biomolecules—A Review. Separations, 10(3), 183. https://doi.org/10.3390/separations10030183






