Antibiotic Resistance Gene Transformation and Ultrastructural Alterations of Lettuce (Lactuca sativa L.) Resulting from Sulfadiazine Accumulation in Culture Solution
Abstract
1. Introduction
2. Materials and Methods
3. Results
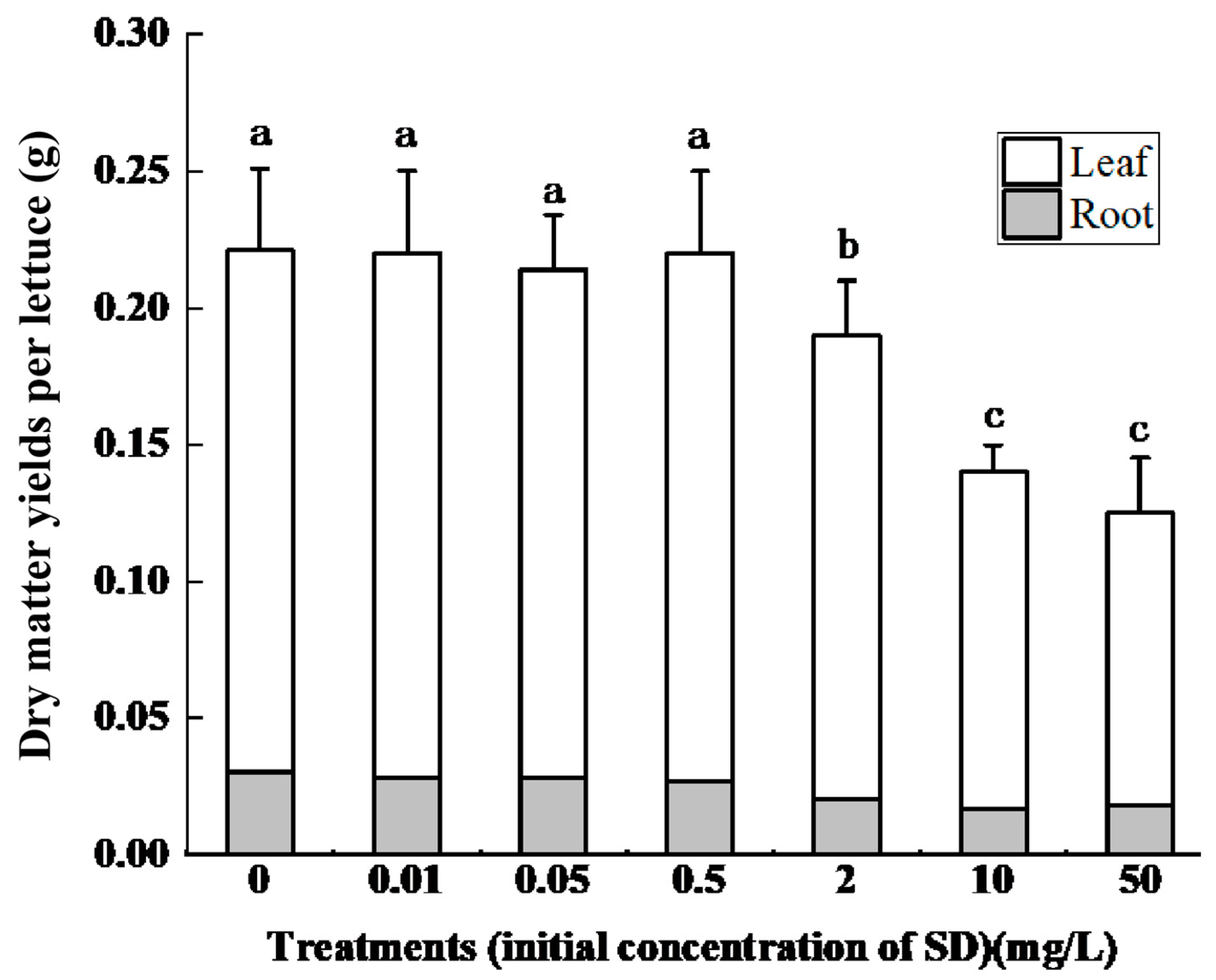
3.1. Effect of SD Accumulation on Lettuce Growth
3.2. Extent of SD Accumulation in Lettuce Organs
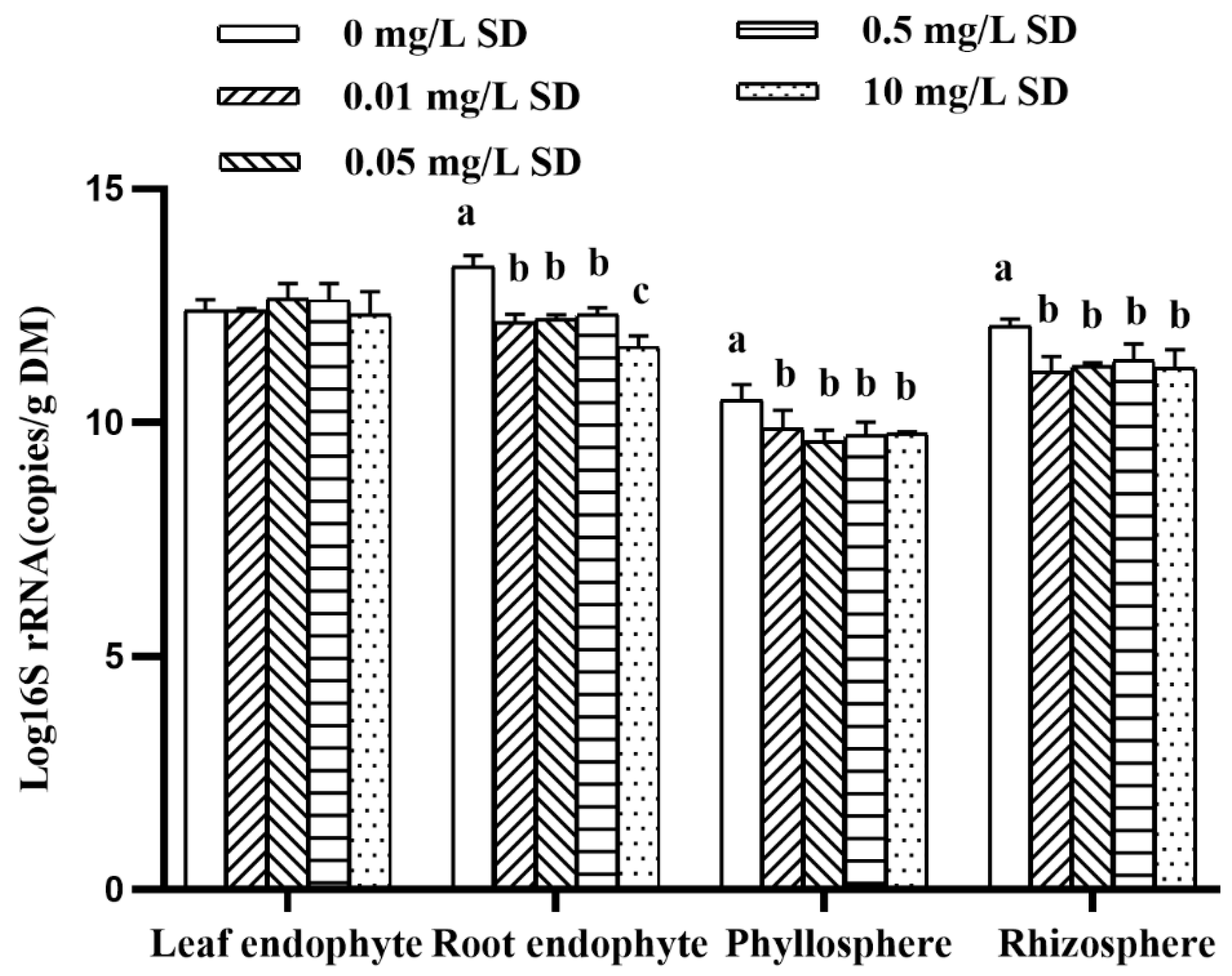
3.3. Antibiotic Resistomes in Plant Microbiomes
3.4. Membrane Permeability of the Roots
3.5. The Ultrastructural Responses of Lettuce to the SD Stress Culture Environment
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mathew, A.G.; Cissell, R.; Liamthong, S. Antibiotic resistance in bacteria associated with food animals: A United States perspective of livestock production. Foodborne Pathog. Dis. 2007, 4, 115–133. [Google Scholar] [CrossRef]
- Micinski, J.; Pogorzelska, J.; Slyamowa, A.; Kobzhassarov, T.; Bermagambetova, N.; Dzik, S.; Kowalski, P.M.; Zaborowska-Sapeta, K.; Kowalski, I.M. Hazards to Humans and Animals Associated with Antibiotic Misuse. J. Elementol. 2015, 20, 1077–1086. [Google Scholar] [CrossRef][Green Version]
- Sarmah, A.K.; Meyer, M.T.; Boxall, A.B. A global perspective on the use, sales, exposure pathways, occurrence, fate and effects of veterinary antibiotics (VAs) in the environment. Chemosphere 2006, 65, 725–759. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Wang, J.; Lu, C.; Liao, Q.H.; Gudda, F.O.; Ling, W.T. Antibiotics in animal manure and manure-based fertilizers: Occurrence and ecological risk assessment. Chemosphere 2020, 255, 127006. [Google Scholar] [CrossRef] [PubMed]
- Menz, J.; Olsson, O.; Kummerer, K. Antibiotic residues in livestock manure: Does the EU risk assessment sufficiently protect against microbial toxicity and selection of resistant bacteria in the environment? J. Hazard. Mater. 2019, 379, 120807. [Google Scholar] [CrossRef] [PubMed]
- Lyu, J.; Yang, L.S.; Zhang, L.; Ye, B.X.; Wang, L. Antibiotics in soil and water in China-a systematic review and source analysis. Environ. Pollut. 2020, 266, 115147. [Google Scholar] [CrossRef]
- Li, Y.W.; Wu, X.L.; Mo, C.H.; Tai, Y.P.; Huang, X.P.; Xiang, L. Investigation of Sulfonamide, Tetracycline, and Quinolone Antibiotics in Vegetable Farmland Soil in the Pearl River Delta Area, Southern China. J. Agric. Food Chem. 2011, 59, 7268–7276. [Google Scholar] [CrossRef] [PubMed]
- Peltzer, P.M.; Lajmanovich, R.C.; Attademo, A.M.; Junges, C.M.; Teglia, C.M.; Martinuzzi, C.; Curi, L.; Culzoni, M.J.; Goicoechea, H.C. Ecotoxicity of veterinary enrofloxacin and ciprofloxacin antibiotics on anuran amphibian larvae. Environ. Toxicol. Pharmacol. 2017, 51, 114–123. [Google Scholar] [CrossRef]
- Nasri, A.; Hannachi, A.; Allouche, M.; Barhoumi, B.; Saidi, I.; Dallali, M.; Harrath, A.H.; Mansour, L.; Mahmoudi, E.; Beyrem, H.; et al. Chronic ecotoxicity of ciprofloxacin exposure on taxonomic diversity of a meiobenthic nematode community in microcosm experiments. J. King Saud Univ. Sci. 2020, 32, 1470–1475. [Google Scholar] [CrossRef]
- Xie, Z.X.; Tang, J.; Wu, X.W.; Fan, S.S.; Cheng, H.M.; Li, X.D.; Hua, R.M. Bioconcentration and ecotoxicity of sulfadiazine in the aquatic midge Chironomus riparius. Environ. Toxicol. Pharmacol. 2019, 66, 69–74. [Google Scholar] [CrossRef]
- Pino, M.R.; Val, J.; Mainar, A.M.; Zuriaga, E.; Espanol, C.; Langa, E. Acute toxicological effects on the earthworm Eisenia fetida of 18 common pharmaceuticals in artificial soil. Sci. Total Environ. 2015, 518, 225–237. [Google Scholar] [CrossRef]
- Pan, M.; Chu, L.M. Transfer of antibiotics from wastewater or animal manure to soil and edible crops. Environ. Pollut. 2017, 231, 829–836. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.J.; Hu, H.W.; Gou, M.; Wang, J.T.; Chen, D.; He, J.Z. Temporal succession of soil antibiotic resistance genes following application of swine, cattle and poultry manures spiked with or without antibiotics. Environ. Pollut. 2017, 231, 1621–1632. [Google Scholar] [CrossRef] [PubMed]
- Xie, W.Y.; McGrath, S.P.; Su, J.Q.; Hirsch, P.R.; Clark, I.M.; Shen, Q.; Zhu, Y.G.; Zhao, F.J. Long-Term Impact of Field Applications of Sewage Sludge on Soil Antibiotic Resistome. Environ. Sci. Technol. 2016, 50, 12602–12611. [Google Scholar] [CrossRef]
- Chen, Y.; Su, J.Q.; Zhang, J.; Li, P.; Chen, H.; Zhang, B.; Gin, K.Y.; He, Y. High-throughput profiling of antibiotic resistance gene dynamic in a drinking water river-reservoir system. Water Res. 2019, 149, 179–189. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.L.; Cui, H.L.; Su, J.Q.; Penuelas, J.; Zhu, Y.G. Antibiotic Resistomes in Plant Microbiomes. Trends Plant Sci. 2019, 24, 530–541. [Google Scholar] [CrossRef]
- Yazdi, M.; Kolahi, M.; Kazemi, E.M.; Barnaby, A.G. Study of the contamination rate and change in growth features of lettuce (Lactuca sativa Linn.) in response to cadmium and a survey of its phytochelatin synthase gene. Ecotoxicol. Environ. Saf. 2019, 180, 295–308. [Google Scholar] [CrossRef]
- Hayes, K.L.; Mui, J.; Song, B.; Sani, E.S.; Eisenman, S.W.; Sheffield, J.B.; Kim, B. Effects, uptake, and translocation of aluminum oxide nanoparticles in lettuce: A comparison study to phytotoxic aluminum ions. Sci. Total Environ. 2020, 719, 137393. [Google Scholar] [CrossRef]
- Song, Y.; Zhang, F.L.; Li, H.P.; Qiu, B.; Gao, Y.; Cui, D.; Yang, Z.G. Antioxidant defense system in lettuces tissues upon various As species exposure. J. Hazard. Mater. 2020, 399, 123003. [Google Scholar] [CrossRef]
- Xu, D.M.; Pan, H.; Yao, J.C.; Feng, Y.X.; Wu, P.P.; Shao, K. Stress responses and biological residues of sulfanilamide antibiotics in Arabidopsis thaliana. Ecotoxicol. Environ. Saf. 2020, 199, 110727. [Google Scholar] [CrossRef]
- Berendonk, T.U.; Manaia, C.M.; Merlin, C.; Fatta-Kassinos, D.; Cytryn, E.; Walsh, F.; Burgmann, H.; Sorum, H.; Norstrom, M.; Pons, M.N.; et al. Tackling antibiotic resistance: The environmental framework. Nat. Rev. Microbiol. 2015, 13, 310–317. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.X.; Ren, S.W.; Niu, T.Q.; Guo, Y.H.; Qi, S.Y.; Han, X.K.; Liu, D.; Pan, F. Distribution of antibiotic-resistant bacteria in chicken manure and manure-fertilized vegetables. Environ. Sci. Pollut. Res. 2014, 21, 1231–1241. [Google Scholar] [CrossRef]
- Bellino, A.; Lofrano, G.; Carotenuto, M.; Libralato, G.; Baldantoni, D. Antibiotic effects on seed germination and root development of tomato (Solanum lycopersicum L.). Ecotoxicol. Environ. Saf. 2018, 148, 135–141. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.G.; Yu, W.T.; Ma, Q.; Wang, J.; Zhou, H.; Jiang, C.M. The combined effect of sulfadiazine and copper on soil microbial activity and community structure. Ecotoxicol. Environ. Saf. 2016, 134, 43–52. [Google Scholar] [CrossRef] [PubMed]
- Hemkemeyer, M.; Christensen, B.T.; Martens, R.; Tebbe, C.C. Soil particle size fractions harbour distinct microbial communities and differ in potential for microbial mineralisation of organic pollutants. Soil Biol. Biochem. 2015, 90, 255–265. [Google Scholar] [CrossRef]
- Rafraf, I.D.; Lekunberri, I.; Sanchez-Melsio, A.; Aouni, M.; Borrego, C.M.; Balcazar, J.L. Abundance of antibiotic resistance genes in five municipal wastewater treatment plants in the Monastir Governorate, Tunisia. Environ. Pollut. 2016, 219, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Pei, R.T.; Kim, S.C.; Carlson, K.H.; Pruden, A. Effect of River Landscape on the sediment concentrations of antibiotics and corresponding antibiotic resistance genes (ARG). Water Res. 2006, 40, 2427–2435. [Google Scholar] [CrossRef]
- Liao, H.; Lu, X.; Rensing, C.; Friman, V.P.; Geisen, S.; Chen, Z.; Yu, Z.; Wei, Z.; Zhou, S.; Zhu, Y. Hyperthermophilic Composting Accelerates the Removal of Antibiotic Resistance Genes and Mobile Genetic Elements in Sewage Sludge. Environ. Sci. Technol. 2018, 52, 266–276. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.G.; Johnson, T.A.; Su, J.Q.; Qiao, M.; Guo, G.X.; Stedtfeld, R.D.; Hashsham, S.A.; Tiedje, J.M. Diverse and abundant antibiotic resistance genes in Chinese swine farms. Proc. Natl. Acad. Sci. USA 2013, 110, 3435–3440. [Google Scholar] [CrossRef] [PubMed]
- Lv, Y.; Li, Y.Y.; Liu, X.H.; Xu, K. The tolerance mechanism and accumulation characteristics of Phragmites australis to sulfamethoxazole and ofloxacin. Chemosphere 2020, 253, 126695. [Google Scholar] [CrossRef]
- Yang, L.; Feng, Y.X.; Zhang, H.; Yu, X.Z. Estimating the synergistic and antagonistic effects of dual antibiotics on plants through root elongation test. Ecotoxicology 2020, 1–12. [Google Scholar] [CrossRef]
- Eraslan, F.; Inal, A.; Savasturk, O.; Gunes, A. Changes in antioxidative system and membrane damage of lettuce in response to salinity and boron toxicity. Sci. Hortic Amst. 2007, 114, 5–10. [Google Scholar] [CrossRef]
- United States Environmental Protection Agency. Development Document for the Proposed Revisions to the National Pollutant Discharge Elimination System Regulation and the Effluent Guidelines for Concentrated Animal Feeding Operations by Engineering and Analysis Division, Office of Science and Technology; United States Environmental Protection Agency: Washington, DC, USA, 2001.
- Ahmed, M.B.M.; Rajapaksha, A.U.; Lim, J.E.; Vu, N.T.; Kim, I.S.; King, H.M.; Lee, S.S.; Ok, Y.S. Distribution and Accumulative Pattern of Tetracyclines and Sulfonamides in Edible Vegetables of Cucumber, Tomato, and Lettuce. J. Agric. Food Chem. 2015, 63, 398–405. [Google Scholar] [CrossRef]
- Compant, S.; Clement, C.; Sessitsch, A. Plant growth-promoting bacteria in the rhizo- and endosphere of plants: Their role, colonization, mechanisms involved and prospects for utilization. Soil Biol. Biochem. 2010, 42, 669–678. [Google Scholar] [CrossRef]
- Zhang, Y.J.; Hu, H.W.; Chen, Q.L.; Singh, B.K.; Yan, H.; Chen, D.L.; He, J.Z. Transfer of antibiotic resistance from manure-amended soils to vegetable microbiomes. Environ. Int. 2019, 130, 104912. [Google Scholar] [CrossRef]
- Wang, F.H.; Qiao, M.; Chen, Z.; Su, J.Q.; Zhu, Y.G. Antibiotic resistance genes in manure-amended soil and vegetables at harvest. J. Hazard. Mater. 2015, 299, 215–221. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.; Chen, Q.; Chen, S.; Zhu, Y.G. Does organically produced lettuce harbor higher abundance of antibiotic resistance genes than conventionally produced? Environ. Int. 2017, 98, 152–159. [Google Scholar] [CrossRef]
- Migliore, L.; Cozzolino, S.; Fiori, M. Phytotoxicity to and uptake of enrofloxacin in crop plants. Chemosphere 2003, 52, 1233–1244. [Google Scholar] [CrossRef]
- Liu, X.N.; Lv, Y.; Gao, S.; Xu, K. Ofloxacin induces etiolation in Welsh onion leaves. Chemosphere 2021, 267, 128918. [Google Scholar] [CrossRef]
- Fu, L.; Huang, T.; Wang, S.; Wang, X.H.; Su, L.M.; Li, C.; Zhao, Y.H. Toxicity of 13 different antibiotics towards freshwater green algae Pseudokirchneriella subcapitata and their modes of action. Chemosphere 2017, 168, 217–222. [Google Scholar] [CrossRef]
- Davi, V.; Tanimoto, H.; Ershov, D.; Haupt, A.; De Belly, H.; Le Borgne, R.; Couturier, E.; Boudaoud, A.; Minc, N. Mechanosensation Dynamically Coordinates Polar Growth and Cell Wall Assembly to Promote Cell Survival. Dev. Cell 2018, 45, 170–182. [Google Scholar] [CrossRef] [PubMed]





| Target Gene | Primer Pair a | Sequence (5′→3′) | Annealing Temp. (°C) | Reference |
|---|---|---|---|---|
| 16s rRNA | 16s rRNA-F | GTGSTGCAYGGYTGTCGTCA | 60 | [26] |
| 16s rRNA-R | ACGTCRTCCMCACCTTCCTC | |||
| sul1 | sul1-F | CGCACCGGAAACATCGCTGCAC | 62 | [27] |
| sul1-R | TGAAGTTCCGCCGCAAGGCTCG | |||
| sul2 | sul2-F | CTCCGATGGAGGCCGGTAT | 60 | [28] |
| sul2-R | GGGAATGCCATCTGCCTTGA | |||
| tnpA | tnpA-F | CCGATCACGGAAAGCTCAAG | 60 | [29] |
| tnpA-R | GGCTCGCATGACTTCGAATC |
| Treatment | Leaf | Root |
|---|---|---|
| Control | Not detectable | Not detectable |
| 0.01 mg/L SD | Not detectable | Not detectable |
| 0.05 mg/L SD | Not detectable | Not detectable |
| 0.5 mg/L SD | 3.90 ± 0.91 c | 6.48 ± 1.62 b |
| 2 mg/LSD | 9.55 ± 1.73 b | 22.86 ± 2.41 c |
| 10 mg/L SD | 13.06 ± 1.76 a | 39.80 ± 2.34 b |
| 50 mg/L SD | 16.74 ± 1.88 a | 120.87 ± 17.33 a |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mei, L.; Chen, Y.-X.; Wang, C.; Chen, J.-H.; Zhang, Z.-J.; Zhou, M.-Y.; Feng, J.-T.; Wang, Y. Antibiotic Resistance Gene Transformation and Ultrastructural Alterations of Lettuce (Lactuca sativa L.) Resulting from Sulfadiazine Accumulation in Culture Solution. Processes 2021, 9, 1451. https://doi.org/10.3390/pr9081451
Mei L, Chen Y-X, Wang C, Chen J-H, Zhang Z-J, Zhou M-Y, Feng J-T, Wang Y. Antibiotic Resistance Gene Transformation and Ultrastructural Alterations of Lettuce (Lactuca sativa L.) Resulting from Sulfadiazine Accumulation in Culture Solution. Processes. 2021; 9(8):1451. https://doi.org/10.3390/pr9081451
Chicago/Turabian StyleMei, Liang, Ying-Xin Chen, Chao Wang, Jia-Hua Chen, Zhi-Jin Zhang, Min-Yao Zhou, Jin-Tao Feng, and Yan Wang. 2021. "Antibiotic Resistance Gene Transformation and Ultrastructural Alterations of Lettuce (Lactuca sativa L.) Resulting from Sulfadiazine Accumulation in Culture Solution" Processes 9, no. 8: 1451. https://doi.org/10.3390/pr9081451
APA StyleMei, L., Chen, Y.-X., Wang, C., Chen, J.-H., Zhang, Z.-J., Zhou, M.-Y., Feng, J.-T., & Wang, Y. (2021). Antibiotic Resistance Gene Transformation and Ultrastructural Alterations of Lettuce (Lactuca sativa L.) Resulting from Sulfadiazine Accumulation in Culture Solution. Processes, 9(8), 1451. https://doi.org/10.3390/pr9081451





