1. Introduction
Although many different methods of global optimization have been developed, the efficiency of an optimization method is always determined by the specific nature of the particular problem. Parameter optimization of cellular dynamics models has become a field of particular interest, as the problem is widely applicable. Identification of the parameters of a nonlinear dynamic model is more difficult than the linear case since there are no general analytical results.
Nature-inspired metaheuristic methods still receive a great deal of attention nowadays [
1,
2,
3,
4,
5,
6,
7]. Among population-based nature-inspired metaheuristics are genetic algorithms (GA), ant colony optimization (ACO), firefly algorithm (FA), cuckoo search (CS), bat algorithm (BA), monarch butterfly optimization (MBO), whale optimization algorithm (WOA), salp swarm algorithm (SSA), emperor penguin optimizer (EPO), etc., all of which use multiple agents or “particles”.
Typically, metaheuristic methods can successfully solve complex identification problems. Even more effective behaviors can be achieved through a combination of different metaheuristic techniques, the so-called hybrid metaheuristics [
8].
The main purpose of hybrid algorithms is to exploit the advantages of different optimization strategies while avoiding their disadvantages. By choosing an appropriate combination of metaheuristic techniques, better algorithm performance can be achieved in solving hard optimization problems [
9]. There are many hybridization techniques that have been proven successful for various applications. In [
10], the authors propose a hybrid metaheuristic algorithm for clustering, which combines ant brood sorting with tabu search, with emphasis on flexibility and less specificity. Rosle et al. [
11] propose a hybrid of particle swarm optimization and harmony search, which outperforms the simulated annealing and the downhill simplex method. A novel hybrid integrating a support vector machine (SVM) model with GA is discussed in [
12]. The designed hybrid algorithm shows the smallest amount of outlier data and the best predictive stability. Another successful hybrid, combining GA and Variable Neighborhood Search, is proposed in [
13]. The algorithm’s feasibility and superiority are demonstrated in a real-world case.
An efficient combination of SVM and MBO is discussed in [
14]. Its application on an imbalanced disease classification problem demonstrates improvements using the developed hybrid method. Liu et al. [
15] propose a hybrid WOA enhanced with Lévy flight and differential evolution (DE). The experimental results of its application to solve a job shop scheduling problem show the superior performance of the hybrid. A combination of the advantages of the SSA and PSO algorithms are introduced in [
16] and applied to the optimal power flow problem. The proposed hybrid algorithm provides more efficient solutions even for conflict constraints. A hybrid metaheuristic optimization approach between EPO and ESA is proposed in [
17]. A test analysis on 53 benchmark functions shows the effectiveness and efficiency of the designed hybrid algorithm.
Following the literature, it is evident that many metaheuristic algorithms could be hybridized to improve the algorithms’ performance for a particular problem. Another example is the artificial bee colony (ABC) algorithm—one of the promising biologically inspired metaheuristic approaches introduced by Karaboga in 2005 [
18] for optimizing multi-variable and multimodal numerical functions. The known results indicate that ABC can be used efficiently for various optimization problems [
19,
20,
21,
22,
23].
Some hybrid algorithms involving ABC and its modifications that aim to improve the algorithm’s performance are the following: an ABC and quantum evolutionary algorithm hybrid is proposed in [
24] for solving continuous optimization problems. The authors use ABC to increase the local search capacity as well as the randomness of the populations. A hybrid between ABC and the monarch butterfly optimization for numerical optimization problems is proposed in [
25]. The hybrid is designed to boost the exploration versus exploitation balance of the original algorithms. Again, for numerical function optimization, a hybrid ABC algorithm with DE and free search is proposed in [
26]. The resulting hybrid shows excellent convergence and global searchability. A powerful hybrid algorithm that combines the features of both optimization algorithms ABC and BA are proposed in [
27]. An ABC algorithm with random location-updating is considered in [
28]. The designed algorithm shows stronger exploration ability, improved convergence speed and optimization precision. In [
29], the authors introduce an algorithm in which the crossover operator of GA is involved in improving the exploration of the canonical ABC. Crossover and mutation operators are introduced in a hybrid ABC to reduce the computational resources [
30].
This literature review shows that, as a result of the ABC hybridization, the algorithm’s performance is improved, and optimal solutions to different problems are achieved. This fact, and the known successful applications of GA, particularly for parameter identification of nonlinear models of cultivation processes, inspired the authors to work on the development of a new hybrid metaheuristic algorithm between ABC and GA.
The proposed hybrid algorithm in this paper, named ABC-GA, is designed to improve both the exploration and exploitation and thus present a powerful and efficient algorithm for real-world numerical optimization problems. As a result, the speed and precision of the algorithm’s convergence are also improved.
The performance of the proposed ABC-GA hybrid algorithm is examined with two different test groups, including classic benchmark test functions and a real nonlinear optimization problem. The ABC-GA is employed for a nonlinear model parameter identification problem. As a case study, an E. coli MC4110 fed-batch cultivation process model is considered. The optimization problem is solved using real experimental data.
The ABC-GA results were compared with different hybrid algorithms from the literature, between ACO, GA and FA, for each test group. The main contributions of the research can be summarized as below:
Combining the features of ABC and GA, a new hybrid algorithm ABC-GA is proposed and tested on classic benchmark test functions and a real-world problem.
The superiority of the designed hybrid ABC-GA is shown based on the comparison of the simulation results with other hybrid metaheuristic algorithms for the problems of the two different test groups.
Using the hybrid metaheuristic algorithm ABC-GA the optimal parameters’ values of a nonlinear mathematical model of an E. coli cultivation process are estimated.
The paper is organized as follows. In
Section 2, the background of the ABC-GA hybrid algorithm is given. The performance of the designed hybrid on different unconstrained optimization problems is studied in
Section 3. The considered real-world problem—a model parameter identification—is outlined in
Section 4, and the numerical results from the identification are presented and discussed. Concluding remarks are given in
Section 5.
2. ABC-GA Hybrid Algorithm
Based on the foraging behavior of the honey bees, the ABC algorithm was introduced by Karaboga for the purpose of numerical optimization problems [
18].
ABC operates on a set of food sources representing potential solutions to the problem under consideration. The initial set of solutions is generated during the initialization phase of the algorithm when the initial values of the algorithm’s parameters are set.
The ABC algorithm evolves in three phases on each iteration: employed bees’ phase, onlookers’ phase and scouts’ phase [
31], during which a new set of food sources is formed. The employed bees search for new food sources around those stored in their memory. Based on their evaluations, the newly found food sources may replace the old ones. Only certain food sources can be modified during the onlookers’ phase. They are selected based on the probability values associated with them. The last phase is when an abandoned food source is replaced by a new random one found by a scout.
These three phases are repeated for a predetermined number of cycles called maximum cycle number (MCN). The best food source represents a reasonable solution to the considered problem.
GA was developed to model adaptation processes using a recombination operator with a mutation in the background [
32].
GA maintains a population of individuals. Each individual represents a potential solution to the problem. Each solution is evaluated, so a new population is formed by selecting more fit individuals. In order to form new solutions, some elements of the new population undergo transformations by means of “genetic” operators. These are unary transformations (mutation type) which create new individuals by a small change in a single individual, and higher-order transformations (crossover type), which create new individuals by combining parts from several individuals.
After a certain number of generations, the algorithm converges and is expected to best represent a near-optimum (reasonable) solution to the problem under consideration.
The proposed hybrid algorithm is a collaborative combination between the ABC algorithm and GA. The aim is to avoid the poor convergence rate of the ABC algorithm, a disadvantage reported in [
26,
32,
33]. The population-based GA randomly generates an initial population that can be very far from the optimal solution and may require a lot of iterations to draw close to it. To overcome these limitations, ABC is applied to the generated initial solutions for a few iterations. The ABC outcomes are then used as an initial population of GA. Thus, GA starts with a population much closer to the optimal one compared to the randomly generated initial population. The best solution is obtained by the genetic evolution of the ABC, and the result uses fewer computational resources. This integration aims to provide a proper balance between exploration and exploitation (intensification and diversification).
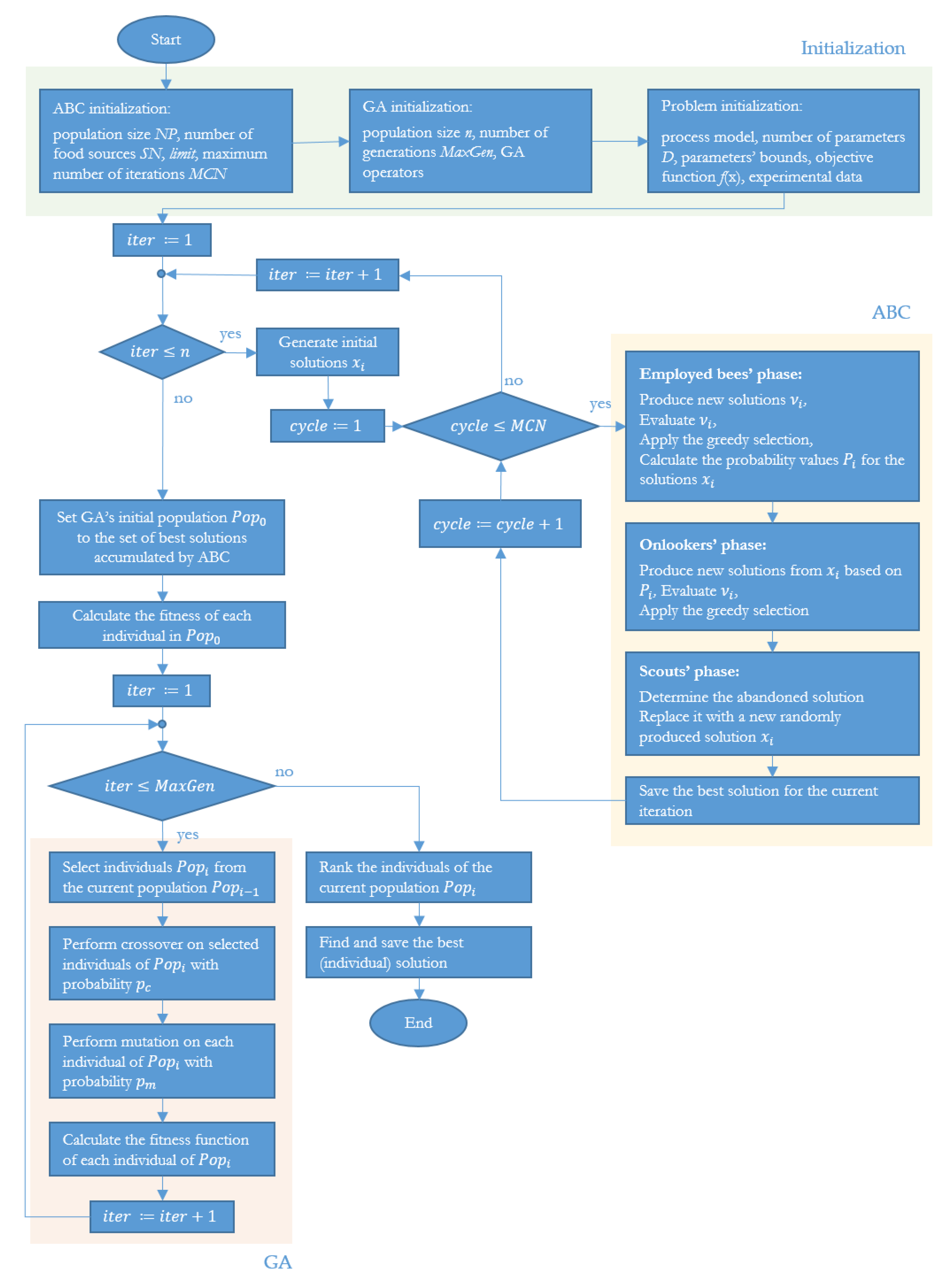
The main steps of the ABC-GA hybrid algorithm are presented in
Figure 1.
During the initialization phase of the algorithm, values of the input parameters of ABC and GA are set. The optimization problem is defined, as well as the problem parameters and their bounds.
The initial solutions are generated using Equation (1). Each solution is a D-dimensional vector
, limited by the lower and upper bounds of the corresponding parameter of the optimization problem:
where
i ∈ [1;
SN],
j ∈ [1;
D],
and
are the lower and the upper bounds of the dimension
j.
ABC utilizes this set of initial solutions in an attempt to get closer to finding the optimal solution to the optimization problem. ABC runs for a predetermined
MCN, and each cycle evolves in three phases, defined in [
31].
The search for new food sources (solutions) during the employed bees’ phase is based on Equation (2):
where
j is a random integer number in the range [1,
D];
k is randomly selected index;
k ∈ [1;
SN],
k ≠ i;
∈ [−1, 1] is a random number.
The onlookers’ search around certain food sources is based on the same Equation (3). The food sources are selected taking into account a probability value
associated with each food source, calculated by the roulette wheel selection:
where
is the fitness value of the solution
,
i ∈ [1;
SN].
During the greedy selection in the employed bees’ phase and the onlookers’ phase, the number of trials for each food source is updated when it is not replaced by a better one. When the trials exceed a predefined limit, the corresponding food source is abandoned and replaced by a new food source, randomly generated using Equation (1). This new food source represents a food source found by a scout.
The best solutions accumulated by ABC for n number of runs, where n is the population size of GA, are used as the initial population Pop0 of GA. GA iteratively improves the existing population by selecting and reproducing parents for a certain number of generations.
The selection of parents to produce next generations is performed based on the fitness of the individuals. The selection method for this particular case is the roulette wheel selection. Two genetic operators—crossover and mutation, are applied later to generate new individuals. The extended intermediate recombination and real value mutation employed here are defined in the following way.
The crossover operator combines the genetic material of two parents. The recombination process is unconditional. Let
x and
y be two
D-dimensional vectors denoting the parents from the current population. Let
be the result of the recombination. The elements of
z are generated using Equation (4).
where
j ∈ [1;
D];
∈ [−
δ; 1 +
δ] is chosen with uniform probability.
δ indicates to what degree an offspring can be generated out of the parents’ s scope; usually, the value of
δ is 0.25.
The mutation operator is applied to each element
with a probability inversely proportional to the number of dimensions
D. Let
be the result of the mutation. The elements of
z are evaluated using Equation (5).
where
j ∈ [1;
D];
sign ∈ {−1, +1}, chosen with equal probability;
,
∈ [0.1; 0.5
is the mutation range;
,
∈ {0, 1} from a Bernoulli probability distribution and
k ∈ {4, 5, …, 20} is the mutation precision related to the minimal step size and the distribution of mutation steps in the mutation range.
The fitness of each individual in the population is evaluated. Offspring with a better fitness evaluation move to the next generation until the maximum number of generations MaxGen is reached.
The optimal solution to the particular problem is the best individual from the last generation of GA. Since the initial population is not random but much closer to the optimal sought solution, GA converges to the optimal solution in much fewer iterations.
The hybrid ABC-GA proposed here applies the two algorithms, ABC and GA, sequentially. Therefore, the computational complexity of ABC and GA should be considered separately when the complexity of the hybrid is calculated.
For a given problem, let O (f) be the computational complexity of its fitness function evaluation. The complexity of the standard ABC can be expressed as O (MCN × NP × f). Since ABC is required to generate an initial population for GA, the algorithm is executed n number of times, where n is the population size of GA. The computational complexity of GA can be evaluated as O (MaxGen × n × f). The overall complexity of the ABC-GA hybrid algorithm can then be evaluated as O ((MCN × NP + MaxGen) × n × f).
It should be noted, however, that the values of the parameters MCN, NP and MaxGen used in the hybrid ABC-GA are far smaller than the values used in the standard case. The reason is that the ABC algorithm is used to give an initial push to GA, to trigger GA and the search for the optimal solution of the problem to start from a closed position rather than a random one. This way, GA needs far fewer steps to complete the search.
In order to improve the computation precision in the hybrid ABC-GA algorithm, the complexity of the algorithm is sacrificed.
3. ABC-GA Hybrid Algorithm Performance on Different Unconstrained Optimization Problems
The proposed algorithm ABC-GA is tested on nine well-known benchmark functions listed in
Table 1. All benchmark functions are considered with a dimension of 30 except for Wood’s function (dimension of 4).
To verify the performance of ABC-GA, the obtained results are compared with the hybrid algorithms ACO-GA and GA-ACO [
34] and ACO-FA [
35]. The common parameter settings of the hybrid algorithms used in all test sets are provided in
Table 2. The parameters were chosen to ensure enough configuration to find the optimal solution.
The performance of the four compared hybrid algorithms is evaluated by calculating the best value, the mean value and the standard deviation (SD). The obtained results are presented in
Table 3. The best and mean values for each problem are highlighted in bold.
According to
Table 3, ABC-GA produced the best value for all nine benchmark functions and the best mean value for eight of them. For Griewank, the best mean value was obtained by the hybrid ACO-GA.
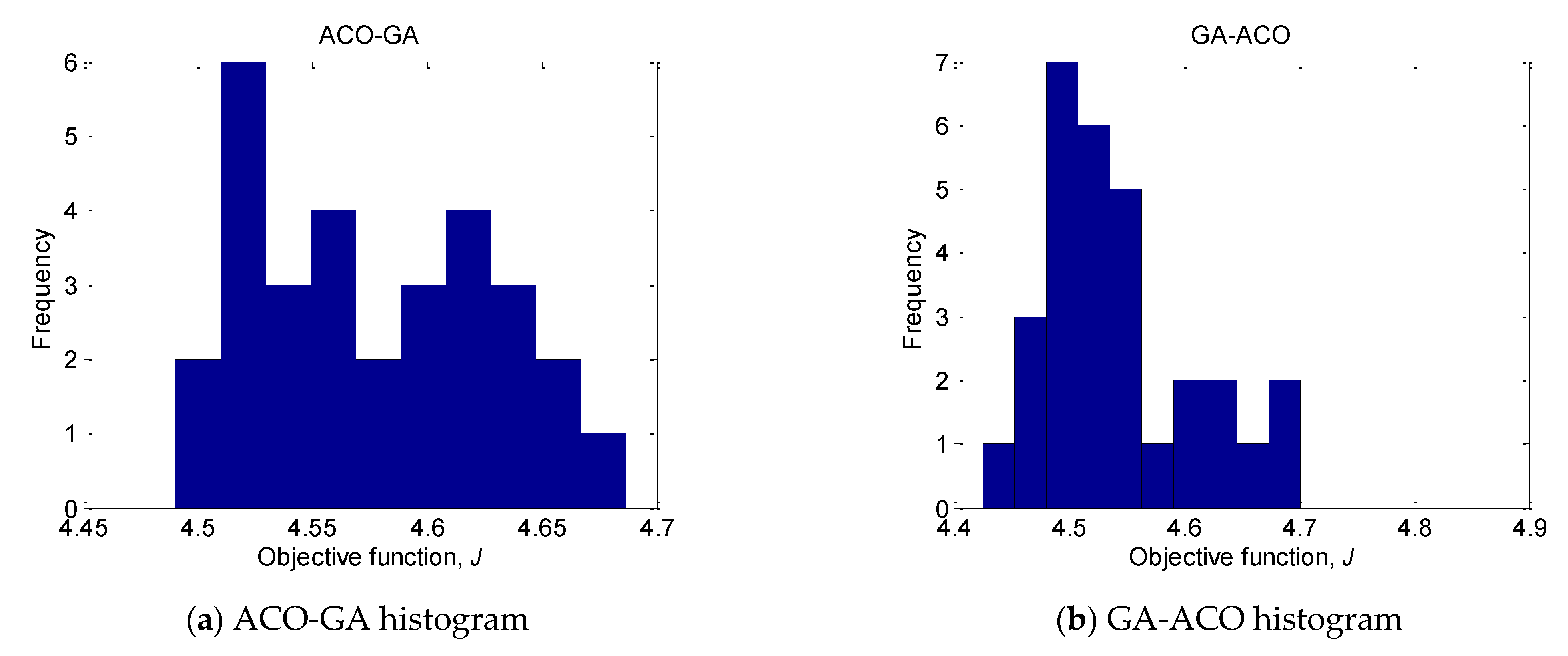
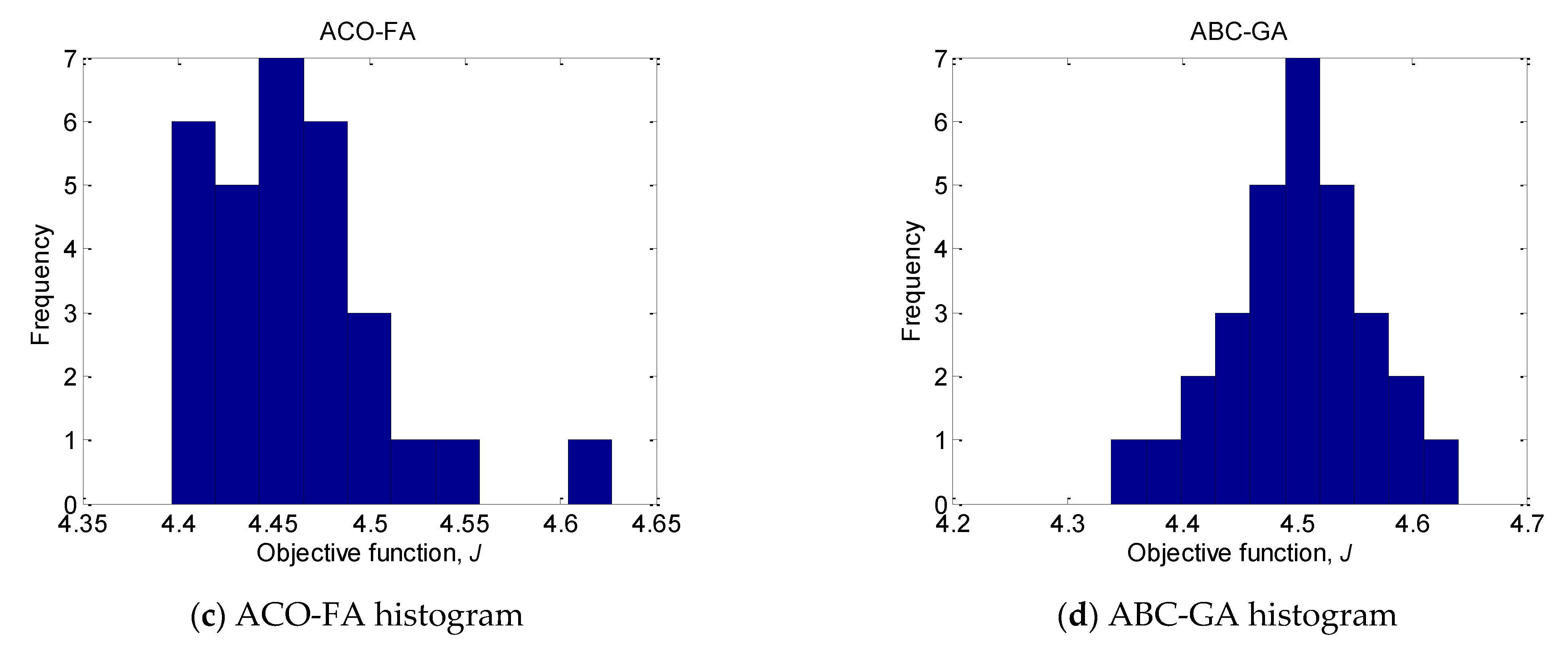
As can be seen for the unimodal Sphere function, the convergence accuracy and stability of ABC-GA are better than those of GA-ACO and ACO-FA. Only the results achieved by GA-ACO are close to the results of ABC-GA. The same results are obtained for the complex unimodal Rosenbrock. However, for the multimodal Rastrigin, Ackley, Griewank and unimodal Schaffer #1 mostly ACO-FA does not produce good results. In the case of Griewank, GA-ACO and ACO-FA results are significantly worse, too. The mean values of GA-ACO and ACO-FA are 9.71 × 10−3 and 8.70 × 10−7, respectively, compared to the mean ABC-GA value of 1.08 × 10−15. The ACO-FA shows the worse performance for all considered benchmark test functions. The results obtained by ABC-GA demonstrate the achieved good balance between algorithm’s exploration and exploitation.
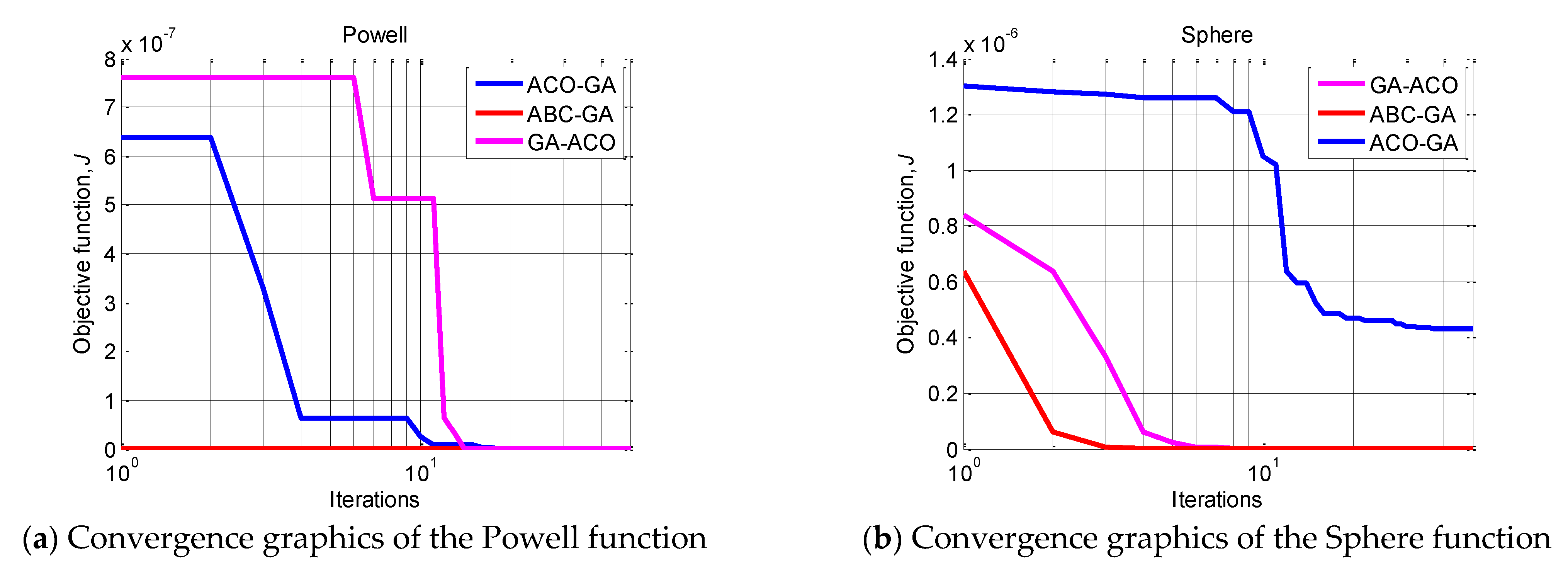
To illustrate the efficiency of ABC-GA, the convergence of the compared hybrid algorithms towards the optima for 30 runs is presented in
Figure 2. Two benchmark functions, Sphere and Powell, are chosen as the most indicative. For the rest of the functions, the superiority of ABC-GA is very clearly shown by the results in
Table 3. ACO-FA is not included in the chart because it produces much worse results.
The plots show the comparative performance of the ABC-GA hybrid algorithm. In terms of convergence, it can be noticed that ABC-GA has a relatively fast convergence toward its final optimal value compared to the other hybrid algorithms.
5. Conclusions
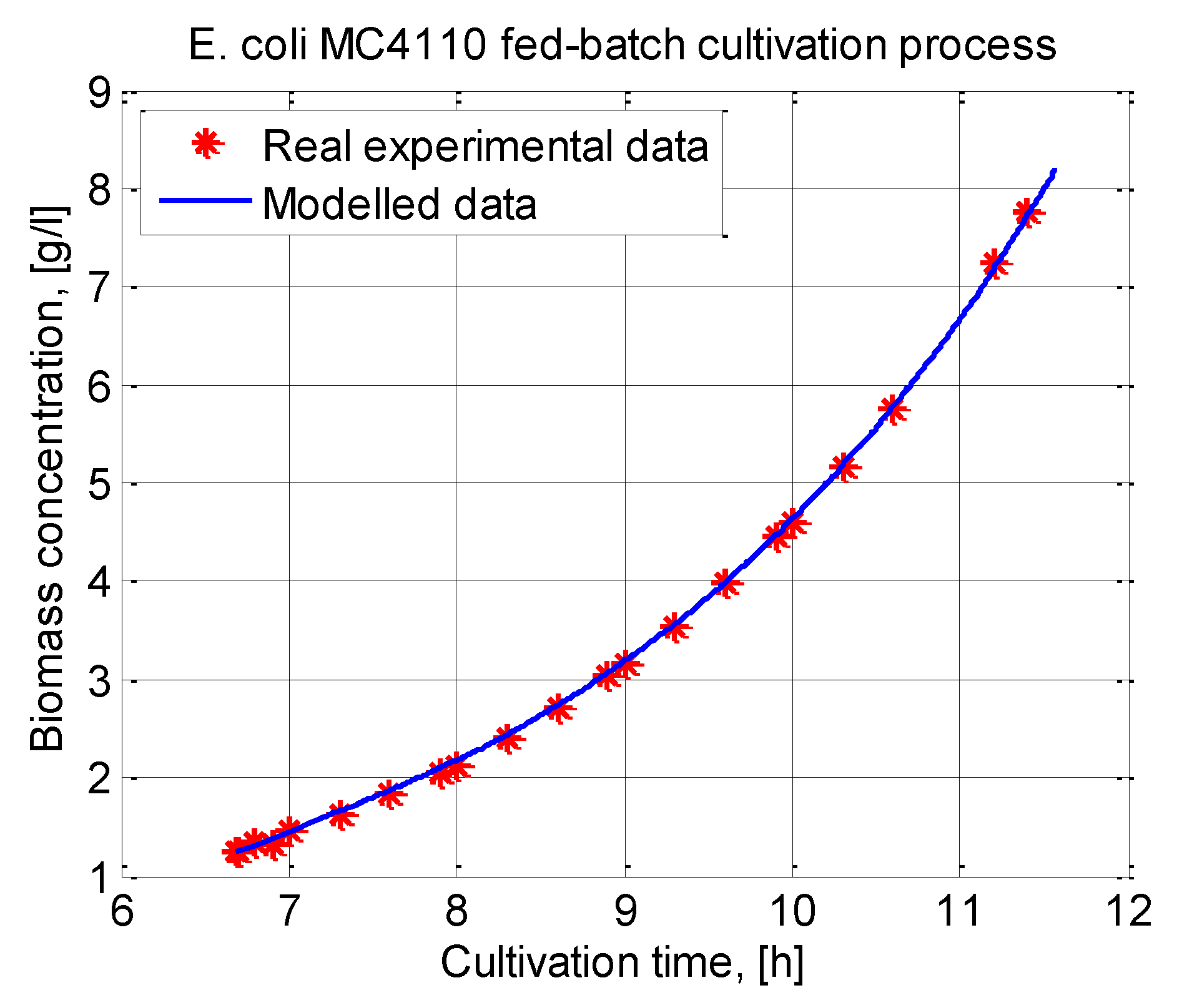
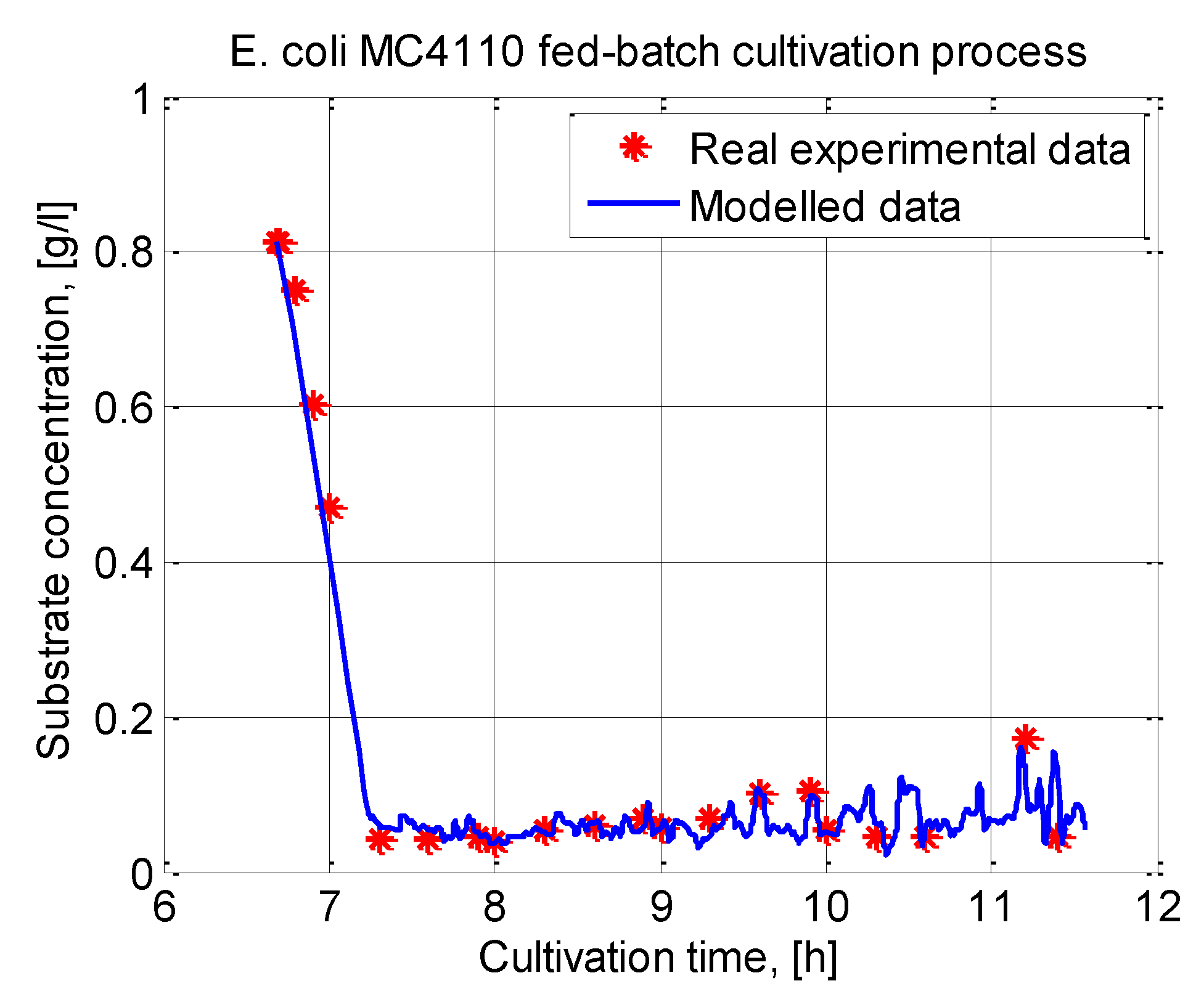
In this paper, an ABC-GA hybrid algorithm is designed and applied to the parameter identification of a cultivation model. A system of nonlinear ordinary differential equations is used to model bacteria growth and substrate utilization. Model parameter identification is performed using a real experimental data set from an E. coli MC4110 fed-batch cultivation process.
The ABC-GA algorithm uses, in the beginning, a small ABC population size (10 individuals) and obtains the GA initial set of solutions for only 25 iterations. Next, for only 50 generations, GA converges to the final solution.
The proposed hybrid algorithm is further compared to other nature-inspired population-based hybrid metaheuristics known in the literature. As competing algorithms, GA, FA and ACO hybrids are chosen and applied to the same parameter identification problem. It is shown that the ABC-GA hybrid algorithm outperforms the other competitor algorithms.
Since the hybrid uses a low number of individuals, the known dependence of sampling capability of GAs on the population size could be exhibited. For example, in the case when ABC-GA obtains the worst results (
Table 7). Further improvements to the designed ABC-GA hybrid will be directed to overcoming this dependence. Another direction for improving the algorithm is the algorithm’s parameters setting. A more in-depth study on the influence of the control parameters (both for ABC and GA) on the hybrid algorithm’s performance could be made.
The good results observed here for the ACO-FA hybrid algorithm and the results in [
47] could be the basis for possible future work where hybridization between ABC and ACO, ABC and FA or ABC and CS algorithm could be investigated.