The Effect of a Peptide Substrate Containing an Unnatural Branched Amino Acid on Chymotrypsin Activity
Abstract
1. Introduction
2. Materials and Methods
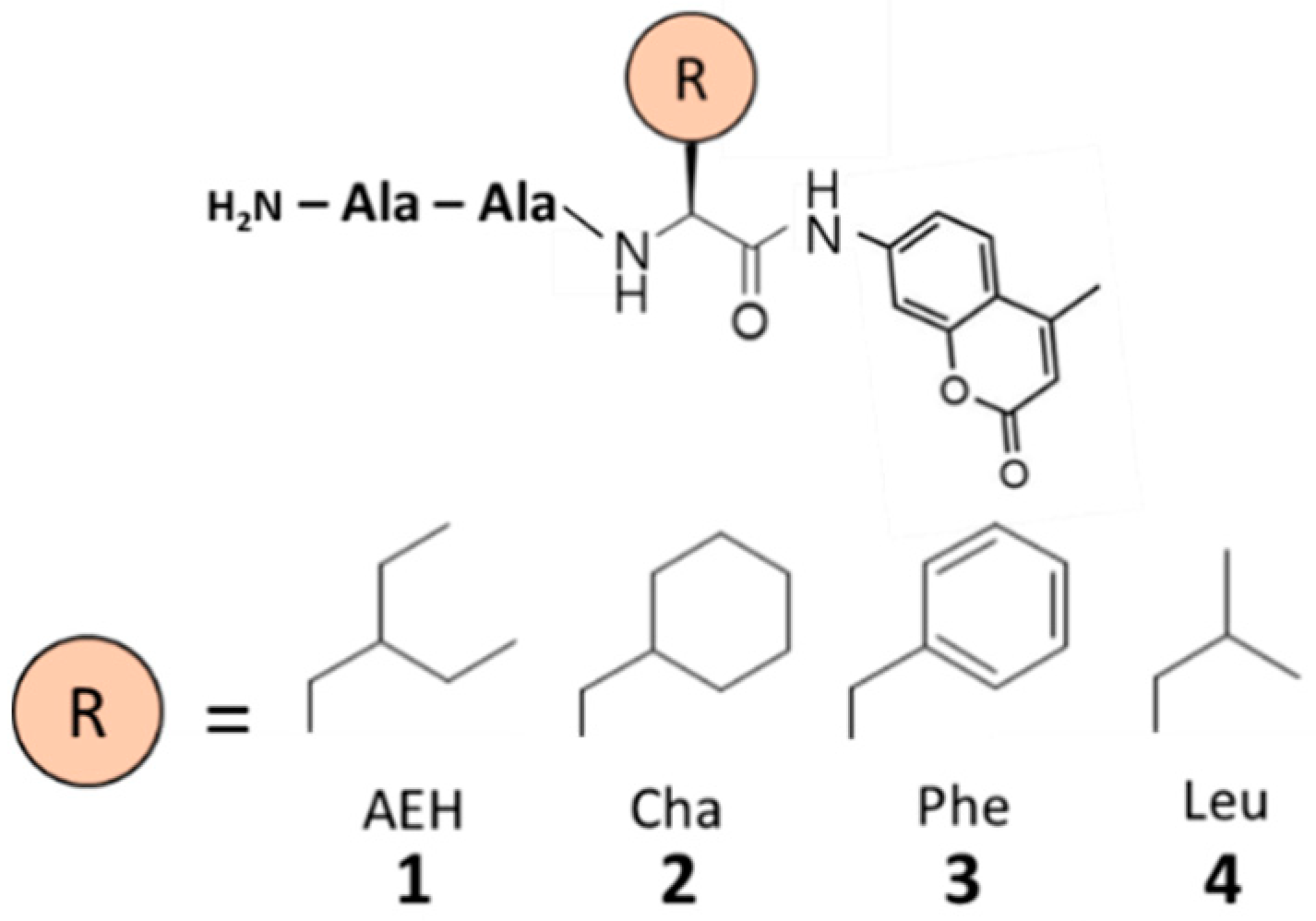
2.1. Design of Substrates
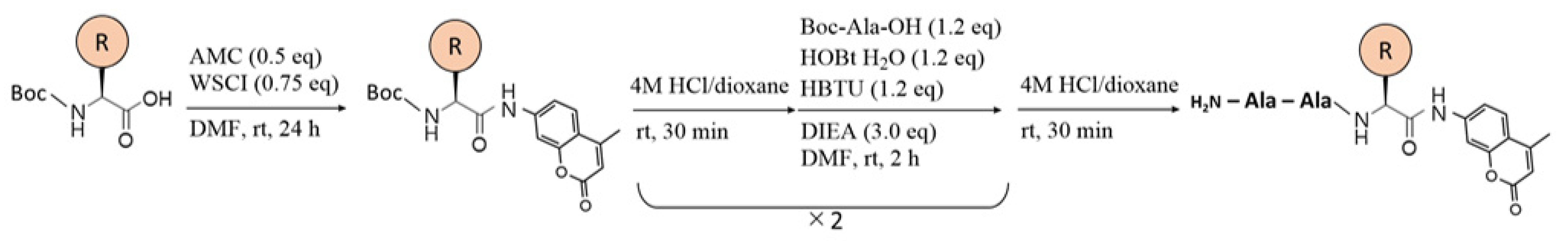
2.2. Synthesis of Boc-AEH-OH
2.3. Synthesis of Peptide-MCA Substrates
2.4. Chymotrypsin Activity Assay
3. Results and Discussion
3.1. Hydrophobicity of the Peptide Substrates
3.2. Chymotrypsin Activity of the Peptide Substrates
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Turk, V.; Stoka, V.; Vasiljeva, O.; Renko, M.; Sun, T.; Turk, B.; Turk, D. Cysteine Cathepsins: From Structure, Function and Regulation to New Frontiers. Biochim. Biophys. Acta Proteins Proteom. 2012, 1824, 68–88. [Google Scholar] [CrossRef] [PubMed]
- Figueiredo, J.-L.; Aikawa, M.; Zheng, C.; Aaron, J.; Lax, L.; Libby, P.; Filho, J.L.d.L.; Gruener, S.; Fingerle, J.; Haap, W.; et al. Selective Cathepsin S Inhibition Attenuates Atherosclerosis in Apolipoprotein E-Deficient Mice with Chronic Renal Disease. Am. J. Pathol. 2015, 185, 1156–1166. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Yu, W.; Kovalski, K.; Ossowski, L. Requirement for Specific Proteases in Cancer Cell Intravasation as Revealed by a Novel Semiquantitative PCR-Based Assay. Cell 1998, 94, 353–362. [Google Scholar] [CrossRef]
- Pham, C.T.N.; Ley, T.J. Dipeptidyl Peptidase I Is Required for the Processing and Activation of Granzymes A and B In Vivo. Proc. Natl. Acad. Sci. USA 1999, 96, 8627–8632. [Google Scholar] [CrossRef]
- Hart, T.C.; Harta, P.S.; Bowden, D.W.; Michalec, M.D.; Callison, S.A.; Walker, S.J.; Zhang, Y.; Firatli, E. Mutations of the Cathepsin C Gene Are Responsible for Papillon-Lefèvre Syndrome. J. Med. Genet. 1999, 36, 881–887. [Google Scholar]
- Hart, T.C.; Hart, P.S.; Michalec, M.D.; Zhang, Y.; Firatli, E.; Dyke, T.E.V.; Stabholz, A.; Zlorogorski, A.; Shapira, L.; Soskolne, W.A. Haim-Munk Syndrome and Papillon-Lefèvre Syndrome Are Allelic Mutations in Cathepsin C. J. Med. Genet. 2000, 37, 88–94. [Google Scholar] [CrossRef]
- Gocheva, V.; Joyce, J.A. Cysteine Cathepsins and the Cutting Edge of Cancer Invasion. Cell Cycle 2007, 6, 60–64. [Google Scholar] [CrossRef]
- Adkison, A.M.; Raptis, S.Z.; Kelley, D.G.; Pham, C.T.N. Dipeptidyl Peptidase I Activates Neutrophil-Derived Serine Proteases and Regulates the Development of Acute Experimental Arthritis. J. Clin. Investig. 2002, 109, 363–371. [Google Scholar] [CrossRef]
- Yang, Y.; Hong, H.; Zhang, Y.; Cai, W. Molecular Imaging of Proteases in Cancer. Cancer Growth Metastasis 2009, 17, 13–27. [Google Scholar] [CrossRef]
- McKerrow, J.H.; Bhargava, V.; Hansell, E.; Huling, S.; Kuwahara, T.; Matley, M.; Coussens, L.; Warren, R. A Functional Proteomics Screen of Proteases in Colorectal Carcinoma. Mol. Med. 2000, 6, 450–460. [Google Scholar] [CrossRef]
- Klezovitch, O.; Chevillet, J.; Mirosevich, J.; Roberts, R.L.; Matusik, R.J.; Vasioukhin, V. Hepsin Promotes Prostate Cancer Progression and Metastasis. Cancer Cell 2004, 6, 185–195. [Google Scholar] [CrossRef] [PubMed]
- Łęgowska, M.; Hamon, Y.; Wojtysiak, A.; Grzywa, R.; Sieńczyk, M.; Burster, T.; Korkmaz, B.; Lesner, A. Development of the First Internally-Quenched Fluorescent Substrates of Human Cathepsin C: The Application in the Enzyme Detection in Biological Samples. Arch. Biochem. Biophys. 2016, 612, 91–102. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, S.; Sakabe, M.; Ishizawa, T.; Hasegawa, K.; Urano, Y.; Kokudo, N. Visualization of the Leakage of Pancreatic Juice Using a Chymotrypsin-Activated Fluorescent Probe. Br. J. Surg. 2013, 100, 1220–1228. [Google Scholar] [CrossRef] [PubMed]
- Poreba, M.; Mihelic, M.; Krai, P.; Rajkovic, J.; Krezel, A.; Pawelczak, M.; Klemba, M.; Turk, D.; Turk, B.; Latajka, R.; et al. Unnatural Amino Acids Increase Activity and Specificity of Synthetic Substrates for Human and Malarial Cathepsin C. Amino Acids 2014, 46, 931–943. [Google Scholar] [CrossRef] [PubMed]
- Imamura, T.; Matsushita, K.; Travis, J.; Potempa, J. Inhibition of Trypsin-Like Cysteine Proteinases (Gingipains) from Porphyromonas gingivalis by Tetracycline and Its Analogues. Antimicrob. Agents Chemother. 2001, 45, 2871–2876. [Google Scholar] [CrossRef][Green Version]
- Mittoo, S.; Sundstrom, L.E.; Bradley, M. Synthesis and Evaluation of Fluorescent Probes for the Detection of Calpain Activity. Anal. Biochem. 2003, 319, 234–238. [Google Scholar] [CrossRef]
- Steimle, A.; Kalbacher, H.; Maurer, A.; Beifuss, B.; Bender, A.; Schäfer, A.; Müller, R.; Autenrieth, I.B.; Frick, J.-S. A Novel Approach for Reliable Detection of Cathepsin S Activities in Mouse Antigen Presenting Cells. J. Immunol. Methods 2016, 432, 87–94. [Google Scholar] [CrossRef]
- Tokmina-Roszyk, M.; Tokmina-Roszyk, D.; Bhowmick, M.; Fields, G.B. Development of a Förster Resonance Energy Transfer Assay for Monitoring Bacterial Collagenase Triple-Helical Peptidase Activity. Anal. Biochem. 2014, 453, 61–69. [Google Scholar] [CrossRef]
- Sato, D.; Kato, T. Novel Fluorescent Substrates for Detection of Trypsin Activity and Inhibitor Screening by Self-Quenching. Bioorganic Med. Chem. Lett. 2016, 26, 5736–5740. [Google Scholar] [CrossRef]
- Zimmerman, M.; Yurewicz, E.; Patel, G. A New Fluorogenic Substrate for Chymotrypsin. Anal. Biochem. 1976, 70, 258–262. [Google Scholar] [CrossRef]
- Tran, T.V.; Ellis, K.A.; Kam, C.-M.; Hudig, D.; Powers, J.C. Dipeptidyl Peptidase I: Importance of Progranzyme Activation Sequences, Other Dipeptide Sequences, and the N-Terminal Amino Group of Synthetic Substrates for Enzyme Activity. Arch. Biochem. Biophys. 2002, 403, 160–170. [Google Scholar] [CrossRef]
- Kraft, M.; Radke, D.; Wieland, G.D.; Zipfel, P.F.; Horn, U. A Fluorogenic Substrate as Quantitative In Vivo Reporter to Determine Protein Expression and Folding of Tobacco Etch Virus Protease in Escherichia coli. Protein Expr. Purif. 2007, 52, 478–484. [Google Scholar] [CrossRef] [PubMed]
- Schulze, A.; Wermann, M.; Demuth, H.-U.; Yoshimoto, T.; Ramsbeck, D.; Schlenzig, D.; Schilling, S. Continuous Assays for Meprin Alpha and Beta Using Prolyl Tripeptidyl Aminopeptidase (PtP) from Porphyromonas gingivalis. Anal. Biochem. 2018, 559, 11–16. [Google Scholar] [CrossRef] [PubMed]
- Bergmann, B.M.; Fruton, J.S. On Proteolytic Enzymes: XIII. Synthetic Substrates for Chymotrypsin. J. Biol. Chem. 1937, 118, 405–415. [Google Scholar] [CrossRef]
- Fruton, J.S.; Bergmann, M. The Multiple Specificity of Chymotrypsin. J. Biol. Chem. 1942, 145, 253–265. [Google Scholar] [CrossRef]
- Hein, G.E.; Jones, J.B.; Niemann, C. The Kinetics of the α-Chymotrypsin-Catalyzed Hydrolysis of Acetyl-L-leucine Methyl Ester. Biochim. Biophys. Acta 1962, 65, 353–355. [Google Scholar] [CrossRef]
- Waite, H.R.; Niemann, C. The α-Chymotrypsin-Catalyzed Hydrolysis of a Series of Acylated-L-valine Esters. Biochemistry 1962, 1, 250–253. [Google Scholar] [CrossRef]
- Spreti, N.; Mancini, M.V.; Germani, R.; Profio, P.D.; Savelli, G. Substrate Effect on α-Chymotrypsin Activity in Aqueous Solutions of “Big-Head” Ammonium Salts. J. Mol. Catal. 2008, 50, 1–6. [Google Scholar] [CrossRef]
- Watanabe, L.A.; Jose, B.; Kato, T.; Nishino, N.; Yoshida, M. Synthesis of l-α-amino-ω-bromoalkanoic Acid for Side Chain Modification. Tetrahedron Lett. 2004, 45, 491–494. [Google Scholar] [CrossRef]
- DelMar, E.G.; Largman, C.; Brodrick, J.W.; Geokas, M.C. A Sensitive New Substrate for Chymotrypsin. Anal. Biochem. 1979, 99, 316–320. [Google Scholar] [CrossRef]

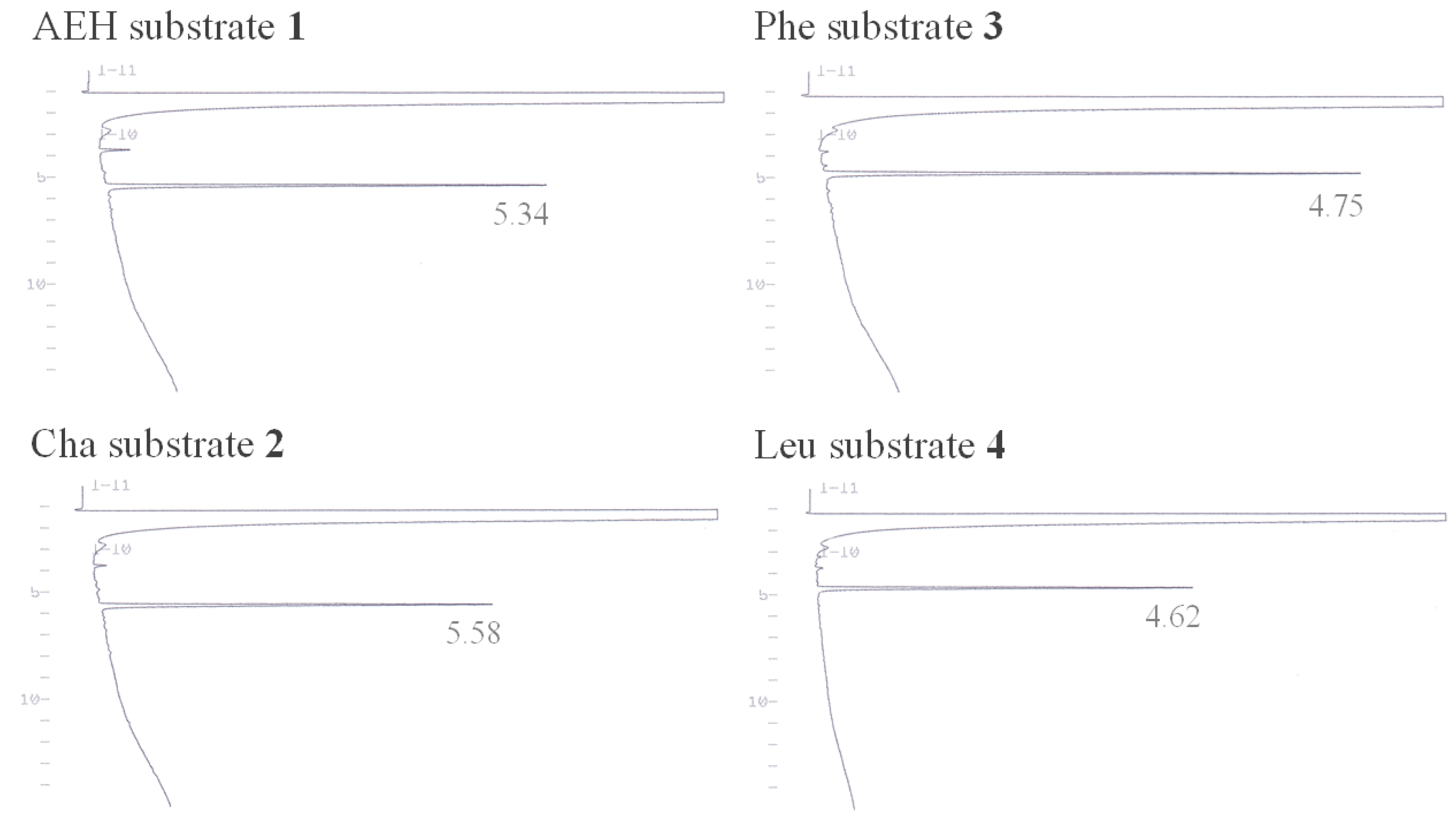
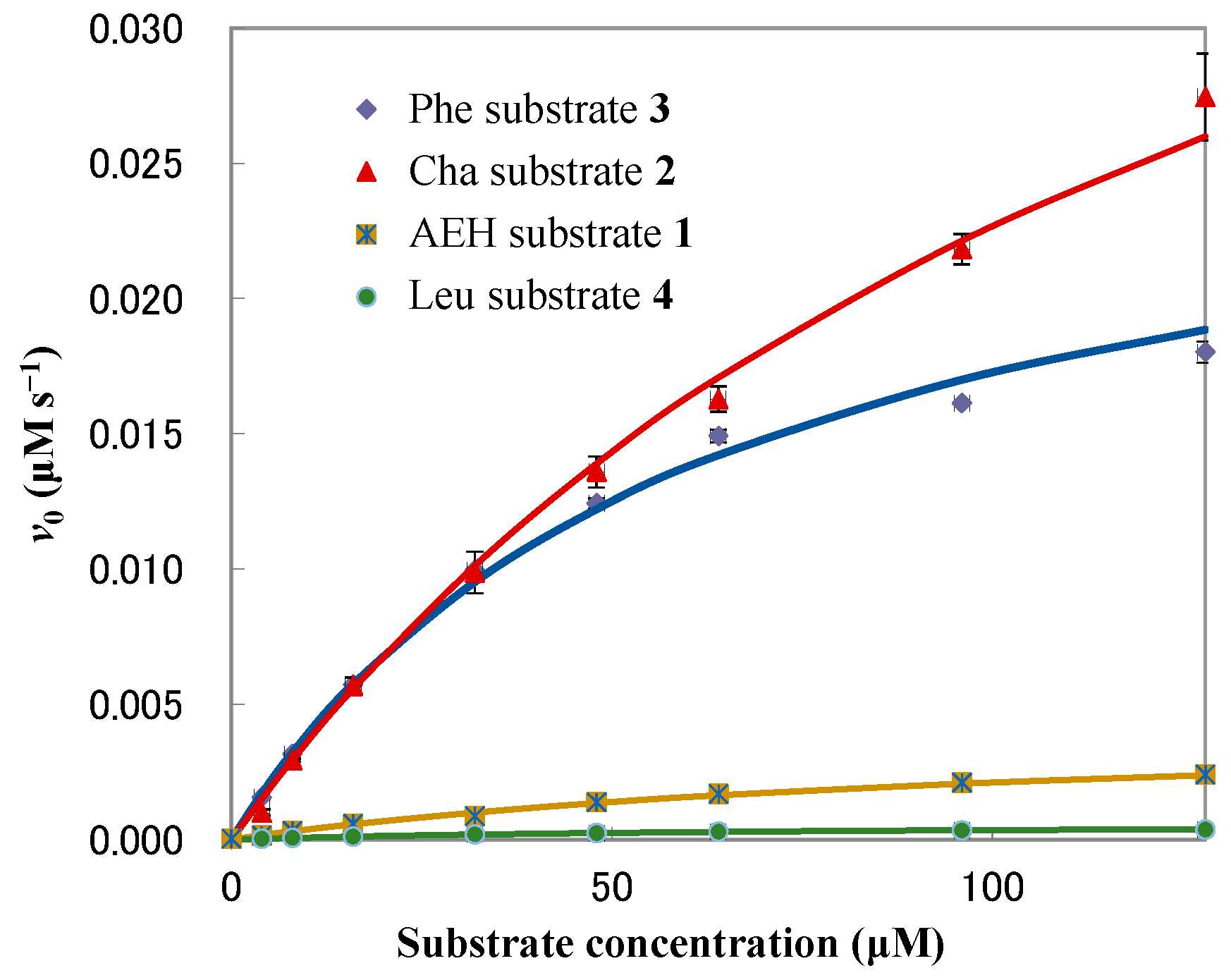
| Sample | HPLC Retention Time (min) | Vmax (nM s−1) | Km (μM) | kcat (s−1) | kcat/Km (M−1 s−1) |
|---|---|---|---|---|---|
| Leu substrate 4 | 4.62 | 0.60 ± 0.01 | 72.83 ± 1.22 | (0.60 ± 0.01) × 10−3 | 8.21 ± 0.03 |
| AEH substrate 1 | 5.34 | 4.36 ± 0.09 | 107.79 ± 3.12 | (4.36 ± 0.09) × 10−3 | 40.67 ± 0.43 |
| Cha substrate 2 | 5.58 | 54.54 ± 1.62 | 140.48 ± 5.65 | (5.45 ± 0.16) × 10−2 | 388.32 ± 4.54 |
| Phe substrate 3 | 4.75 | 27.96 ± 0.71 | 61.97 ± 2.40 | (2.80 ± 0.07) × 10−2 | 452.35 ± 6.62 |
| Suc-F-pNA a | 720 | 0.011 | 15 | ||
| A-A-F-AMC a | 500 | 0.83 | 2000 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yamawaki, Y.; Yufu, T.; Kato, T. The Effect of a Peptide Substrate Containing an Unnatural Branched Amino Acid on Chymotrypsin Activity. Processes 2021, 9, 242. https://doi.org/10.3390/pr9020242
Yamawaki Y, Yufu T, Kato T. The Effect of a Peptide Substrate Containing an Unnatural Branched Amino Acid on Chymotrypsin Activity. Processes. 2021; 9(2):242. https://doi.org/10.3390/pr9020242
Chicago/Turabian StyleYamawaki, Yuuki, Tomoki Yufu, and Tamaki Kato. 2021. "The Effect of a Peptide Substrate Containing an Unnatural Branched Amino Acid on Chymotrypsin Activity" Processes 9, no. 2: 242. https://doi.org/10.3390/pr9020242
APA StyleYamawaki, Y., Yufu, T., & Kato, T. (2021). The Effect of a Peptide Substrate Containing an Unnatural Branched Amino Acid on Chymotrypsin Activity. Processes, 9(2), 242. https://doi.org/10.3390/pr9020242





