Biological Indicators for Fecal Pollution Detection and Source Tracking: A Review
Abstract
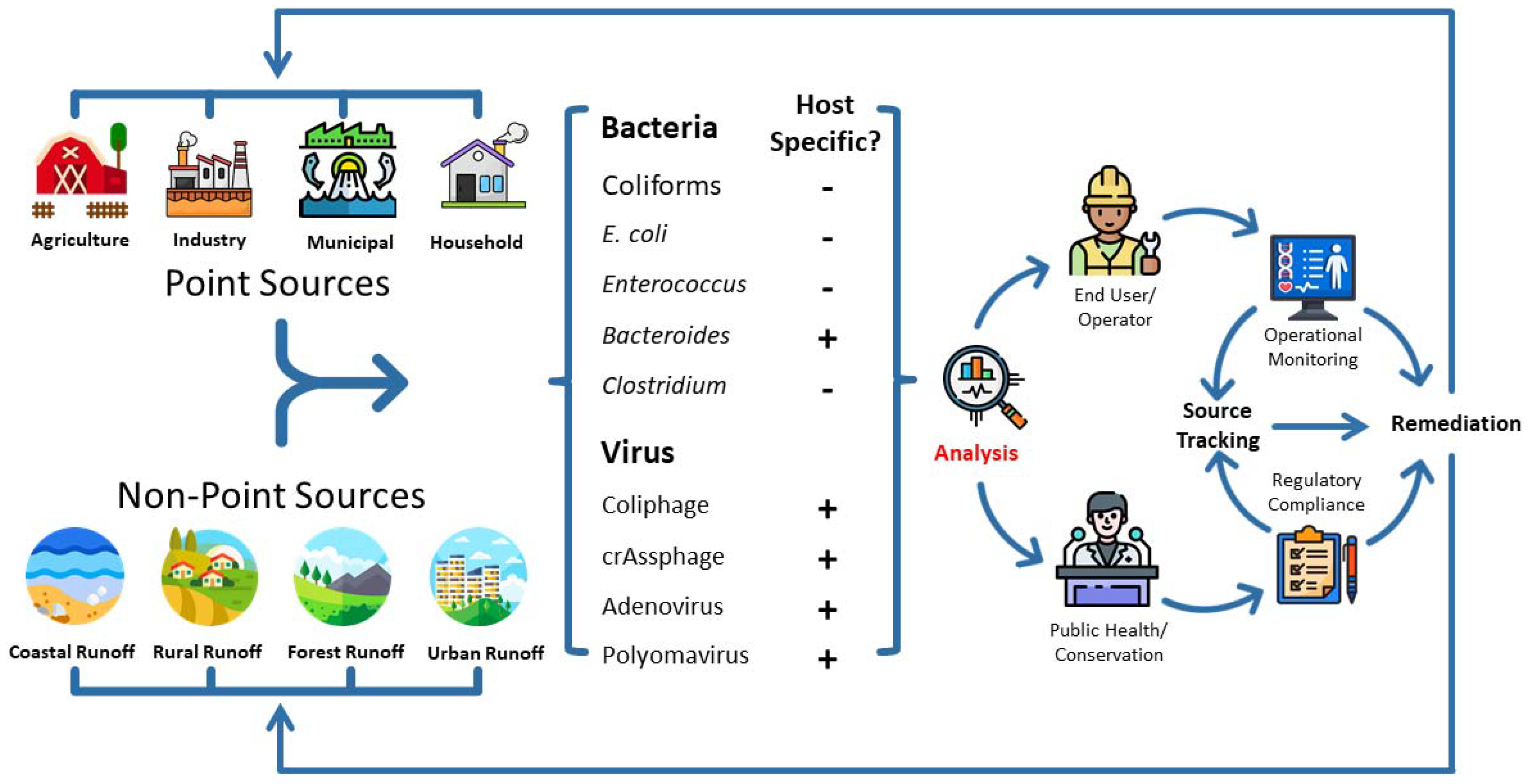
1. Introduction
2. Fecal Pollution Detection Using Biological Markers
2.1. Bacterial Markers
- (1)
- It should be detected only in feces-polluted environments and should not be detected in non-fecal-polluted environments; i.e., it should have a high association with fecal pollution with little or no background noise;
- (2)
- It should be detectable in the same host as the pathogens, and its abundance should reflect pathogen abundance both inside and outside the host; i.e., it should have tight correlations with pathogens/diseases;
- (3)
- It should be detectable in the host intestine, feces, and also fecal-polluted environmental waters so that it can be assessed using inexpensive and simple methods; i.e., it should have good detection sensitivity;
- (4)
- It should not multiply outside the host intestine but should be viable or in a detectable form for some time comparable to fecal pathogens;
- (5)
- It should be safe to measure for field and lab workers, not causing severe human diseases.
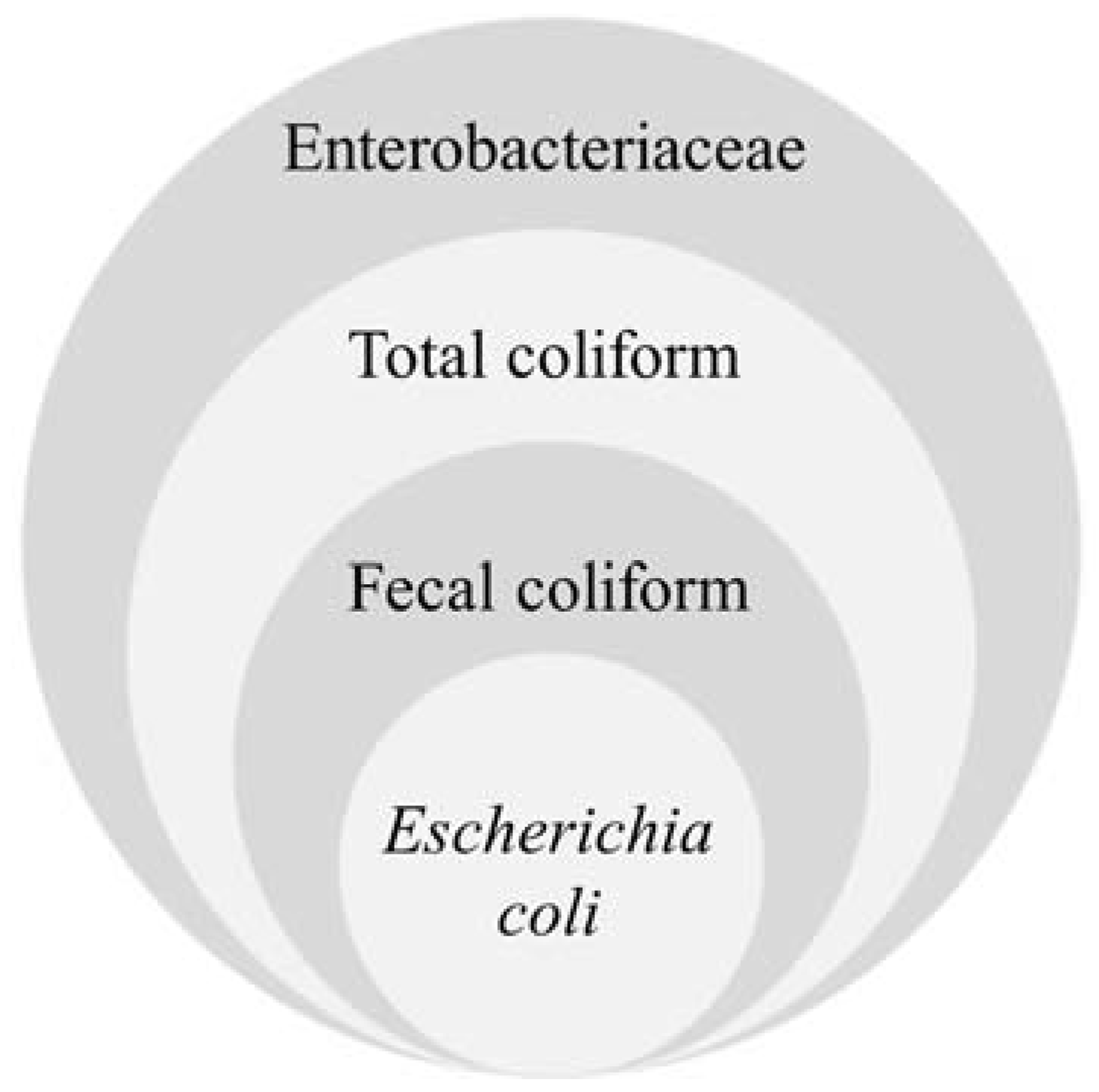
2.1.1. Coliforms
2.1.2. Escherichia coli
2.1.3. Enterococcus
2.1.4. Clostridium perfringens
2.1.5. Bacteroides
2.2. Viral Markers
2.2.1. Coliphage
2.2.2. CrAssphage
2.2.3. Human Adenovirus (HAdV)
2.2.4. Human Polyomavirus (HPyV)
3. Emerging Bioengineering Enumeration Methods
3.1. A Time-Lapse Coherent Imaging Platform
3.2. An Online Culture-Independent Imaging Platform
3.3. An On-Site Microflow Cytometry Free of Fluorescence Staining
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Glassmeyer, S.T.; Furlong, E.T.; Kolpin, D.W.; Cahill, J.D.; Zaugg, S.D.; Werner, S.L.; Meyer, M.T.; Kryak, D.D. Transport of chemical and microbial compounds from known wastewater discharges: Potential for use as indicators of human fecal contamination. Environ. Sci. Technol. 2005, 39, 5157–5169. [Google Scholar] [CrossRef]
- Karkman, A.; Pärnänen, K.; Larsson, D.J. Fecal pollution can explain antibiotic resistance gene abundances in anthropogenically impacted environments. Nat. Commun. 2019, 10, 80. [Google Scholar] [CrossRef]
- Cabral, J.P. Water microbiology. Bacterial pathogens and water. Int. J. Environ. Res. Public Health 2010, 7, 3657–3703. [Google Scholar] [CrossRef] [PubMed]
- Paruch, L.; Paruch, A.M.; Eiken, H.G.; Sorheim, R. Faecal pollution affects abundance and diversity of aquatic microbial community in anthropo-zoogenically influenced lotic ecosystems. Sci. Rep. 2019, 9, 19469. [Google Scholar] [CrossRef] [PubMed]
- Anderson, K.L.; Whitlock, J.E.; Harwood, V.J. Persistence and differential survival of fecal indicator bacteria in subtropical waters and sediments. Appl. Environ. Microbiol. 2005, 71, 3041–3048. [Google Scholar] [CrossRef]
- Porter, K.D.H.; Quilliam, R.S.; Reaney, S.M.; Oliver, D.M. High resolution characterisation of E. coli proliferation profiles in livestock faeces. Waste Manag. 2019, 87, 537–545. [Google Scholar] [CrossRef] [PubMed]
- He, Q.; Qiu, S.; Jiang, Y.; Wu, Z.; Liu, Z. Land-use change caused microbial pollution in a karst underground river, Chongqing, China. Environ. Earth Sci. 2016, 75, 709. [Google Scholar] [CrossRef]
- Rehman, F.; Subyani, A.M.; Cheema, T.; Harbi, H.M.; Azeem, T.; Naseem, A.A.; Ullah, M.F.; Riaz, O.; Ur Rehman, S. Contribution to the comparative study of treated and untreated wastewater: A case study (Wadi Fatima and Al Misk Lake) in Saudi Arabia. Arab. J. Geosci. 2020, 13, 842. [Google Scholar] [CrossRef]
- Abd-Elhamid, H.F.; Abd-Elmoneem, S.M.; Abdelaal, G.M.; Zelenakova, M.; Vranayova, Z.; Abd-Elaty, I. Investigating and Managing the Impact of Using Untreated Wastewater for Irrigation on the Groundwater Quality in Arid and Semi-Arid Regions. Int. J. Environ. Res. Public Health 2021, 18, 7485. [Google Scholar] [CrossRef]
- Kuroda, K.; Murakami, M.; Oguma, K.; Muramatsu, Y.; Takada, H.; Taldzawa, S. Assessment of Groundwater Pollution in Tokyo Using PPCPs as Sewage Markers. Environ. Sci. Technol. 2012, 46, 1455–1464. [Google Scholar] [CrossRef]
- World Health Organization & the United Nations Children’s Fund. Progress on Drinking Water, Sanitation and Hygiene: 2017 Update and SDG Baselines; WHO: Geneva, Switzerland, 2017; Licence: CC BY-NC-SA 3.0 IGO. [Google Scholar]
- Troeger, C.; Blacker, B.F.; Khalil, I.A.; Rao, P.C.; Cao, S.; Zimsen, S.R.; Albertson, S.B.; Stanaway, J.D.; Deshpande, A.; Abebe, Z. Estimates of the global, regional, and national morbidity, mortality, and aetiologies of diarrhoea in 195 countries: A systematic analysis for the Global Burden of Disease Study 2016. Lancet Infect. Dis. 2018, 18, 1211–1228. [Google Scholar] [CrossRef]
- Fuhrmeister, E.R.; Ercumen, A.; Pickering, A.J.; Jeanis, K.M.; Ahmed, M.; Brown, S.; Arnold, B.F.; Hubbard, A.E.; Alam, M.; Sen, D.; et al. Predictors of Enteric Pathogens in the Domestic Environment from Human and Animal Sources in Rural Bangladesh. Environ. Sci. Technol. 2019, 53, 10023–10033. [Google Scholar] [CrossRef]
- Wanjugi, P.; Sivaganesan, M.; Korajkic, A.; McMinn, B.; Kelty, C.A.; Rhodes, E.; Cyterski, M.; Zepp, R.; Oshima, K.; Stachler, E.; et al. Incidence of somatic and F+ coliphage in Great Lake Basin recreational waters. Water Res. 2018, 140, 200–210. [Google Scholar] [CrossRef]
- Jagai, J.S.; Castronovo, D.A.; Monchak, J.; Naumova, E.N. Seasonality of cryptosporidiosis: A meta-analysis approach. Environ. Res. 2009, 109, 465–478. [Google Scholar] [CrossRef]
- Niendorf, S.; Ebner, W.; Marques, A.M.; Bierbaum, S.; Babikir, R.; Huzly, D.; Maaßen, S.; Grundmann, H.; Panning, M. Rotavirus outbreak among adults in a university hospital in Germany. J. Clin. Virol. 2020, 129, 104532. [Google Scholar] [CrossRef] [PubMed]
- Owusu-Ansah, E.d.-G.J.; Sampson, A.; Amponsah, S.K.; Abaidoo, R.C.; Dalsgaard, A.; Hald, T. Probabilistic quantitative microbial risk assessment model of norovirus from wastewater irrigated vegetables in Ghana using genome copies and fecal indicator ratio conversion for estimating exposure dose. Sci. Total Environ. 2017, 601, 1712–1719. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.H.; Huang, A.S.; Liao, Y.S.; Liu, Y.L.; Chiou, C.S. A large outbreak of salmonellosis associated with sandwiches contaminated with multiple bacterial pathogens purchased via an online shopping service. Foodborne Pathog. Dis. 2014, 11, 230–233. [Google Scholar] [CrossRef] [PubMed]
- Minnesota Department of Health. Salmonellosis. 2010. Available online: https://www.health.state.mn.us/diseases/reportable/dcn/sum10/salmonellosis.html (accessed on 15 August 2021).
- U.S. Centers for Disease Control and Prevention. Multistate Outbreak of Multidrug-Resistant Salmonella Heidelberg Infections Linked to Foster Farms Brand Chicken (Final Update). Available online: https://www.cdc.gov/salmonella/heidelberg-10-13/ (accessed on 21 August 2021).
- Hopkins, J.; Hague, H.; Hudgin, G.; Ross, L.; Moore, D. An Outbreak of Cryptosporidium at a Recreational Water Park in Niagara Region, Canada. J. Environ. Health 2013, 75, 28–33. [Google Scholar]
- Waldron, L.S.; Ferrari, B.C.; Cheung-Kwok-Sang, C.; Beggs, P.J.; Stephens, N.; Power, M.L. Molecular epidemiology and spatial distribution of a waterborne cryptosporidiosis outbreak in Australia. Appl. Environ. Microbiol. 2011, 77, 7766–7771. [Google Scholar] [CrossRef]
- Chen, T.; Leung, R.K.; Zhou, Z.; Liu, R.; Zhang, X.; Zhang, L. Investigation of key interventions for shigellosis outbreak control in China. PLoS ONE 2014, 9, e95006. [Google Scholar] [CrossRef]
- Topal, S.; Çelik, H.; Yilmaz, Ş.; Özoğlu, E.; Karaoğlanoğlu, O.; Temel, F.; Şirin, H. Outbreak of Shigella sonnei infection in Terme City, Turkey, September 2012. Turk Hij. Deney. Biyol. Derg. 2019, 76, 141–148. [Google Scholar] [CrossRef]
- Gill, A.; Huszczynski, G. Enumeration of Escherichia coli O157:H7 in Outbreak-Associated Beef Patties. J. Food Prot. 2016, 79, 1266–1268. [Google Scholar] [CrossRef]
- Asia Pacific Emerging Infections Network, University of Washington, Seattle, USA. Japan: E. coli O157:H7 Outbreak, Pickled Cabbage. 2012. Available online: https://blogs.uw.edu/apecein/2012/08/31/japan-e-coli-o157h7-outbreak-pickled-cabbage/ (accessed on 11 August 2021).
- Kenyon, J.; Inns, T.; Aird, H.; Swift, C.; Astbury, J.; Forester, E.; Decraene, V. Campylobacter outbreak associated with raw drinking milk, North West England, 2016. Epidemiol. Infect. 2020, 148, e13. [Google Scholar] [CrossRef] [PubMed]
- Institute of Environmental Science and Research Funded by Ministry of Health, New Zealand. Annual Summary of Outbreaks in New Zealand. 2016. Available online: https://surv.esr.cri.nz/surveillance/annual_outbreak.php?we_objectID=4741 (accessed on 2 September 2021).
- Viray, M.; Hofmeister, M.; Johnston, D.; Krishnasamy, V.; Nichols, C.; Foster, M.; Balajadia, R.; Wise, M.; Manuzak, A.; Lin, Y. Public health investigation and response to a hepatitis A outbreak from imported scallops consumed raw—Hawaii, 2016. Epidemiol. Infect. 2019, 147, E28. [Google Scholar] [CrossRef] [PubMed]
- Rakesh, P.; Mainu, T.C.R.; Raj, A.; Babu, D.; Rajiv, M.; Mohandas, K.S.; Das, A.; Balasubramanian, A. Investigating a community wide outbreak of hepatitis A in Kerala, India. J. Fam. Med. Prim. Care 2018, 7, 1537. [Google Scholar]
- Maguire, J.E.; Glasgow, K.; Glass, K.; Roczo-Farkas, S.; Bines, J.E.; Sheppeard, V.; Macartney, K.; Quinn, H.E. Rotavirus epidemiology and monovalent rotavirus vaccine effectiveness in Australia: 2010–2017. Pediatrics 2019, 144, e20191024. [Google Scholar] [CrossRef]
- Kujawski, S.A.; Lu, X.; Schneider, E.; Blythe, D.; Boktor, S.; Farrehi, J.; Haupt, T.; McBride, D.; Stephens, E.; Sakthivel, S.K.; et al. Outbreaks of Adenovirus-associated Respiratory Illness on 5 College Campuses in the United States, 2018–2019. Clin. Infect. Dis. 2021, 72, 1992–1999. [Google Scholar] [CrossRef]
- Dai, M.; Wu, Y.; Tan, H.; Deng, J.; Hou, M.; Peng, W.; Chen, G.; Li, Y.; Li, H.; Pan, P. Cross-infection of adenovirus among medical staff: A warning from the intensive care unit in a tertiary care teaching hospital in China. Int. J. Infect. Dis. 2020, 98, 390–397. [Google Scholar] [CrossRef]
- Belluz, J. The Dreaded Norovirus Has Hit the Winter Olympics. at Least 199 People Are Sick. Vox. 2018. Available online: https://www.vox.com/science-and-health/2018/2/6/16980028/winter-olympics-norovirus (accessed on 15 September 2021).
- Fumian, T.M.; Ferreira, F.C.; de Andrade, J.d.S.R.; Canal, N.; Silva Gomes, G.; Teixeira, L.B.; Miagostovich, M.P. Norovirus Foodborne Outbreak Associated with the Consumption of Ice Pop, Southern Brazil, 2020. Food Environ. Virol. 2021, 1–7. [Google Scholar] [CrossRef]
- Rizzo, L.; Manaia, C.; Merlin, C.; Schwartz, T.; Dagot, C.; Ploy, M.; Michael, I.; Fatta-Kassinos, D. Urban wastewater treatment plants as hotspots for antibiotic resistant bacteria and genes spread into the environment: A review. Sci. Total Environ. 2013, 447, 345–360. [Google Scholar] [CrossRef]
- Palacios, O.A.; Contreras, C.A.; Muñoz-Castellanos, L.N.; González-Rangel, M.O.; Rubio-Arias, H.; Palacios-Espinosa, A.; Nevárez-Moorillón, G.V. Monitoring of indicator and multidrug resistant bacteria in agricultural soils under different irrigation patterns. Agric. Water Manag. 2017, 184, 19–27. [Google Scholar] [CrossRef]
- Proia, L.; Anzil, A.; Subirats, J.; Borrego, C.; Farre, M.; Llorca, M.; Balcazar, J.L.; Servais, P. Antibiotic resistance along an urban river impacted by treated wastewaters. Sci. Total Environ. 2018, 628–629, 453–466. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Li, X.; Zheng, S.; Kai, Z.; Jin, T.; Shi, R.; Huang, H.; Zheng, X. Effects of wastewater treatment and manure application on the dissemination of antimicrobial resistance around swine feedlots. J. Clean. Prod. 2021, 280, 123794. [Google Scholar] [CrossRef]
- Luczkiewicz, A.; Jankowska, K.; Fudala-Ksiazek, S.; Olanczuk-Neyman, K. Antimicrobial resistance of fecal indicators in municipal wastewater treatment plant. Water Res. 2010, 44, 5089–5097. [Google Scholar] [CrossRef]
- Andreev, N.; Ronteltap, M.; Boincean, B.; Wernli, M.; Zubcov, E.; Bagrin, N.; Borodin, N.; Lens, P.N.L. Lactic acid fermentation of human urine to improve its fertilizing value and reduce odour emissions. J. Environ. Manag. 2017, 198, 63–69. [Google Scholar] [CrossRef]
- Shen, Y.; Ren, L.; Li, G.; Chen, T.; Guo, R. Influence of aeration on CH4, N2O and NH3 emissions during aerobic composting of a chicken manure and high C/N waste mixture. Waste Manag. 2011, 31, 33–38. [Google Scholar] [CrossRef]
- Han, Z.; Sun, D.; Wang, H.; Li, R.; Bao, Z.; Qi, F. Effects of ambient temperature and aeration frequency on emissions of ammonia and greenhouse gases from a sewage sludge aerobic composting plant. Bioresour. Technol. 2018, 270, 457–466. [Google Scholar] [CrossRef]
- Petkuviene, J.; Vaiciute, D.; Katarzyte, M.; Gecaite, I.; Rossato, G.; Vybernaite-Lubiene, I.; Bartoli, M. Feces from piscivorous and herbivorous birds stimulate differentially phytoplankton growth. Water 2019, 11, 2567. [Google Scholar] [CrossRef]
- Wang, Y.; Gu, X.; Zeng, Q.; Mao, Z.; Wang, W. Contrasting response of a plankton community to two filter-feeding fish and their feces: An in situ enclosure experiment. Aquaculture 2016, 465, 330–340. [Google Scholar] [CrossRef]
- Börjesson, G.; Kätterer, T. Soil fertility effects of repeated application of sewage sludge in two 30-year-old field experiments. Nutr. Cycl. Agroecosyst. 2018, 112, 369–385. [Google Scholar] [CrossRef]
- Pradhan, S.K.; Cofie, O.; Nikiema, J.; Heinonen-Tanski, H. Fecal sludge derived products as fertilizer for lettuce cultivation in urban agriculture. Sustainability 2019, 11, 7101. [Google Scholar] [CrossRef]
- Bartelme, R.P.; Oyserman, B.O.; Blom, J.E.; Sepulveda-Villet, O.J.; Newton, R.J. Stripping Away the Soil: Plant Growth Promoting Microbiology Opportunities in Aquaponics. Front. Microbiol. 2018, 9, 8. [Google Scholar] [CrossRef] [PubMed]
- Pradhan, S.K.; Nikiema, J.; Cofie, O.O.; Heinonen-Tanski, H.; Drechsel, P. Fecal sludge-derived pellet fertilizer in maize cultivation. J. Water Sanit. Hyg. Dev. 2016, 6, 474–481. [Google Scholar] [CrossRef]
- Hashemi, S.; Han, M. Optimizing source-separated feces degradation and fertility using nitrifying microorganisms. J. Environ. Manag. 2018, 206, 540–546. [Google Scholar] [CrossRef]
- Yoshizawa, N.; Usui, M.; Fukuda, A.; Asai, T.; Higuchi, H.; Okamoto, E.; Seki, K.; Takada, H.; Tamura, Y. Manure compost is a potential source of tetracycline-resistant Escherichia coli and tetracycline resistance genes in Japanese farms. Antibiotics 2020, 9, 76. [Google Scholar] [CrossRef]
- Korajkic, A.; McMinn, B.R.; Harwood, V.J. Relationships between microbial indicators and pathogens in recreational water settings. Int. J. Environ. Res. Public Health 2018, 15, 2842. [Google Scholar] [CrossRef]
- U.S. Environmental Protection Agency. Basic Information about Nonpoint Source (NPS) Pollution. 2020. Available online: https://www.epa.gov/nps/basic-information-about-nonpoint-source-nps-pollution (accessed on 5 August 2021).
- Edberg, S.C.; Rice, E.W.; Karlin, R.J.; Allen, M.J. Escherichia coli: The best biological drinking water indicator for public health protection. Symp. Ser. Soc. Appl. Microbiol. 2000, 88, 106S–116S. [Google Scholar] [CrossRef]
- Rochelle-Newall, E.; Nguyen, T.M.; Le, T.P.; Sengtaheuanghoung, O.; Ribolzi, O. A short review of fecal indicator bacteria in tropical aquatic ecosystems: Knowledge gaps and future directions. Front. Microbiol. 2015, 6, 308. [Google Scholar] [CrossRef]
- Ahmed, W.; Gyawali, P.; Feng, S.; McLellan, S.L. Host Specificity and Sensitivity of Established and Novel Sewage-Associated Marker Genes in Human and Nonhuman Fecal Samples. Appl. Environ. Microbiol. 2019, 85, e00641-19. [Google Scholar] [CrossRef]
- Tran, N.H.; Gin, K.Y.; Ngo, H.H. Fecal pollution source tracking toolbox for identification, evaluation and characterization of fecal contamination in receiving urban surface waters and groundwater. Sci. Total Environ. 2015, 538, 38–57. [Google Scholar] [CrossRef]
- Ahmed, W.; Goonetilleke, A.; Gardner, T. Human and bovine adenoviruses for the detection of source-specific fecal pollution in coastal waters in Australia. Water Res. 2010, 44, 4662–4673. [Google Scholar] [CrossRef]
- Hundesa, A.; Bofill-Mas, S.; Maluquer de Motes, C.; Rodriguez-Manzano, J.; Bach, A.; Casas, M.; Girones, R. Development of a quantitative PCR assay for the quantitation of bovine polyomavirus as a microbial source-tracking tool. J. Virol. Methods 2010, 163, 385–389. [Google Scholar] [CrossRef] [PubMed]
- Harrach, B.; Tarján, Z.L.; Benkő, M. Adenoviruses across the animal kingdom: A walk in the zoo. FEBS Lett. 2019, 593, 3660–3673. [Google Scholar] [CrossRef] [PubMed]
- Robinson, R.K. Encyclopedia of Food Microbiology; Academic Press: New York, NY, USA, 2014. [Google Scholar]
- Masiello, S.N.; Martin, N.H.; Trmcic, A.; Wiedmann, M.; Boor, K.J. Identification and characterization of psychrotolerant coliform bacteria isolated from pasteurized fluid milk. J. Dairy Sci. 2016, 99, 130–140. [Google Scholar] [CrossRef]
- Martin, N.H.; Trmčić, A.; Hsieh, T.-H.; Boor, K.J.; Wiedmann, M. The evolving role of coliforms as indicators of unhygienic processing conditions in dairy foods. Front. Microbiol. 2016, 7, 1549. [Google Scholar] [CrossRef]
- Craun, G.F.; Berger, P.S.; Calderon, R.L. Coliform bacteria and waterborne disease outbreaks. J.-Am. Water Work. Assoc. 1997, 89, 96–104. [Google Scholar] [CrossRef]
- U.S. Environmental Protection Agency. National Interim Primary Drinking Water Regulations; U.S. Environmental Protection Agency: Washington, DC, USA, 1976.
- U.S. Environmental Protection Agency. National Primary Drinking Water Regulations Total Coliforms Final Rule; U.S. Environmental Protection Agency: Washington, DC, USA, 1989.
- Das, S.K.; Das, S.K. Influence of phosphorus and organic matter on microbial transformation of arsenic. Environ. Technol. Innov. 2020, 19, 100930. [Google Scholar] [CrossRef]
- Rosenblueth, M.; Martinez, L.; Silva, J.; Martinez-Romero, E. Klebsiella variicola, a novel species with clinical and plant-associated isolates. Syst. Appl. Microbiol. 2004, 27, 27–35. [Google Scholar] [CrossRef]
- Kabler, P.W.; Clark, H.F. Coliform group and fecal coliform organisms as indicators of pollution in drinking water. J.-Am. Water Work. Assoc. 1960, 52, 1577–1579. [Google Scholar] [CrossRef]
- Coliform Bacteria in Drinking Water Supplies; New York State Department of Health, Center for Environmental Health: New York, NY, USA, 2017.
- Florini, S.; Shahsavari, E.; Ngo, T.; Aburto-Medina, A.; Smith, D.J.; Ball, A.S. Factors Influencing the Concentration of Fecal Coliforms in Oysters in the River Blackwater Estuary, UK. Water 2020, 12, 1086. [Google Scholar] [CrossRef]
- Bartram, J.; Ballance, R. Water Quality Monitoring: A Practical Guide to the Design and Implementation of Freshwater Quality Studies and Monitoring Programmes; CRC Press: Boca Raton, FL, USA, 1996. [Google Scholar]
- Leclercq, A.; Wanegue, C.; Baylac, P. Comparison of fecal coliform agar and violet red bile lactose agar for fecal coliform enumeration in foods. Appl. Environ. Microbiol. 2002, 68, 1631–1638. [Google Scholar] [CrossRef] [PubMed]
- Alonso, J.; Soriano, A.; Amoros, I.; Ferrus, M. Quantitative determination of E. coli, and fecal coliforms in water using a chromogenic medium. J. Environ. Sci. Health Part A 1998, 33, 1229–1248. [Google Scholar] [CrossRef]
- Evans, T.M.; LeChevallier, M.W.; Waarvick, C.E.; Seidler, R.J. Coliform species recovered from untreated surface water and drinking water by the membrane filter, standard, and modified most-probable-number techniques. Appl. Environ. Microbiol. 1981, 41, 657–663. [Google Scholar] [CrossRef]
- Dunling, W.; Fiessel, W. Evaluation of media for simultaneous enumeration of total coliform and Escherichia coli in drinking water supplies by membrane filtration techniques. J. Environ. Sci. 2008, 20, 273–277. [Google Scholar]
- Tominaga, T. Rapid quantification of coliforms in ready-to-eat foods using lateral-flow immunochromatographic assay. J. Food Saf. 2020, 40, e12835. [Google Scholar] [CrossRef]
- Hu, S.; Yu, Y.; Li, R.; Xia, X.; Xiao, X.; Li, X. Real-time TaqMan PCR for rapid detection and quantification of coliforms in chilled meat. Food Anal. Methods 2016, 9, 813–822. [Google Scholar] [CrossRef]
- Martin, M.C.; Martinez, N.; del Rio, B.; Ladero, V.; Fernandez, M.; Alvarez, M.A. A novel real-time polymerase chain reaction-based method for the detection and quantification of lactose-fermenting Enterobacteriaceae in the dairy and other food industries. J. Dairy Sci. 2010, 93, 860–867. [Google Scholar] [CrossRef]
- Li, X.; Harwood, V.J.; Nayak, B.; Staley, C.; Sadowsky, M.J.; Weidhaas, J. A novel microbial source tracking microarray for pathogen detection and fecal source identification in environmental systems. Environ. Sci. Technol. 2015, 49, 7319–7329. [Google Scholar] [CrossRef] [PubMed]
- Cheswick, R.; Cartmell, E.; Lee, S.; Upton, A.; Weir, P.; Moore, G.; Nocker, A.; Jefferson, B.; Jarvis, P. Comparing flow cytometry with culture-based methods for microbial monitoring and as a diagnostic tool for assessing drinking water treatment processes. Environ. Int. 2019, 130, 104893. [Google Scholar] [CrossRef]
- Xia, Y.; Li, A.-D.; Deng, Y.; Jiang, X.-T.; Li, L.-G.; Zhang, T. MinION nanopore sequencing enables correlation between resistome phenotype and genotype of coliform bacteria in municipal sewage. Front. Microbiol. 2017, 8, 2105. [Google Scholar] [CrossRef]
- Moore, A.C.; Herwaldt, B.L.; Craun, G.F.; Calderon, R.L.; Highsmith, A.K.; Juranek, D.D. Waterborne disease in the United States, 1991 and 1992. J.-Am. Water Work. Assoc. 1994, 86, 87–97. [Google Scholar] [CrossRef]
- U.S. Environmental Protection Agency. Test Methods for Escherichia coli and Enterococci in Water by the Membrane Filter Procedure; (Method #1103.1); U.S. Environmental Protection Agency: Washington, DC, USA, 1985.
- U.S. Environmental Protection Agency. Ambient Water Quality Criteria for Bacteria—1986; U.S. Environmental Protection Agency: Washington, DC, USA, 1986.
- Ishii, S.; Sadowsky, M.J. Escherichia coli in the Environment: Implications for Water Quality and Human Health. Microbes. Environ. 2008, 23, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Souid, F.; Agoubi, B.; Hamdi, M.; Telahigue, F.; Kharroubi, A. Groundwater chemical and fecal contamination assessment of the Jerba unconfined aquifer, southeast of Tunisia. Arab. J. Geosci. 2017, 10, 231. [Google Scholar] [CrossRef]
- Dheenan, P.S.; Jha, D.K.; Das, A.K.; Vinithkumar, N.V.; Devi, M.P.; Kirubagaran, R. Geographic information systems and multivariate analysis to evaluate fecal bacterial pollution in coastal waters of Andaman, India. Environ. Pollut. 2016, 214, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Byappanahalli, M.N.; Whitman, R.L.; Shively, D.A.; Sadowsky, M.J.; Ishii, S. Population structure, persistence, and seasonality of autochthonous Escherichia coli in temperate, coastal forest soil from a Great Lakes watershed. Environ. Microbiol. 2006, 8, 504–513. [Google Scholar] [CrossRef] [PubMed]
- Power, M.L.; Littlefield-Wyer, J.; Gordon, D.M.; Veal, D.A.; Slade, M.B. Phenotypic and genotypic characterization of encapsulated Escherichia coli isolated from blooms in two Australian lakes. Environ. Microbiol. 2005, 7, 631–640. [Google Scholar] [CrossRef]
- Escobar-Paramo, P.; Le Menac’h, A.; Le Gall, T.; Amorin, C.; Gouriou, S.; Picard, B.; Skurnik, D.; Denamur, E. Identification of forces shaping the commensal Escherichia coli genetic structure by comparing animal and human isolates. Environ. Microbiol. 2006, 8, 1975–1984. [Google Scholar] [CrossRef]
- Gordon, D.M.; Cowling, A. The distribution and genetic structure of Escherichia coli in Australian vertebrates: Host and geographic effects. Microbiology 2003, 149, 3575–3586. [Google Scholar] [CrossRef]
- Clermont, O.; Lescat, M.; O’Brien, C.L.; Gordon, D.M.; Tenaillon, O.; Denamur, E. Evidence for a human-specific Escherichia coli clone. Environ. Microbiol. 2008, 10, 1000–1006. [Google Scholar] [CrossRef]
- Williams, A.; Avery, L.; Killham, K.; Jones, D.L. Persistence of Escherichia coli O157 on farm surfaces under different environmental conditions. J. Appl. Microbiol. 2005, 98, 1075–1083. [Google Scholar] [CrossRef]
- Sinton, L.W.; Hall, C.H.; Lynch, P.A.; Davies-Colley, R.J. Sunlight inactivation of fecal indicator bacteria and bacteriophages from waste stabilization pond effluent in fresh and saline waters. Appl. Environ. Microbiol. 2002, 68, 1122–1131. [Google Scholar] [CrossRef]
- Brennan, F.P.; Grant, J.; Botting, C.H.; O’Flaherty, V.; Richards, K.G.; Abram, F. Insights into the low-temperature adaptation and nutritional flexibility of a soil-persistent Escherichia coli. FEMS Microbiol. Ecol. 2013, 84, 75–85. [Google Scholar] [CrossRef]
- Vital, M.; Hammes, F.; Egli, T. Competition of Escherichia coli O157 with a drinking water bacterial community at low nutrient concentrations. Water Res. 2012, 46, 6279–6290. [Google Scholar] [CrossRef]
- Haller, L.; Poté, J.; Loizeau, J.-L.; Wildi, W. Distribution and survival of faecal indicator bacteria in the sediments of the Bay of Vidy, Lake Geneva, Switzerland. Ecol. Indic. 2009, 9, 540–547. [Google Scholar] [CrossRef]
- Schwering, M.; Song, J.; Louie, M.; Turner, R.J.; Ceri, H. Multi-species biofilms defined from drinking water microorganisms provide increased protection against chlorine disinfection. Biofouling 2013, 29, 917–928. [Google Scholar] [CrossRef] [PubMed]
- Mellefont, L.A.; McMeekin, T.A.; Ross, T. Performance evaluation of a model describing the effects of temperature, water activity, pH and lactic acid concentration on the growth of Escherichia coli. Int. J. Food Microbiol. 2003, 82, 45–58. [Google Scholar] [CrossRef]
- Abia, A.L.; Ubomba-Jaswa, E.; Momba, M.N. Impact of seasonal variation on Escherichia coli concentrations in the riverbed sediments in the Apies River, South Africa. Sci. Total Environ. 2015, 537, 462–469. [Google Scholar] [CrossRef] [PubMed]
- Stocker, M.D.; Pachepsky, Y.A.; Hill, R.L.; Shelton, D.R. Depth-Dependent Survival of Escherichia coli and Enterococci in Soil after Manure Application and Simulated Rainfall. Appl. Environ. Microbiol. 2015, 81, 4801–4808. [Google Scholar] [CrossRef]
- Kleinheinz, G.T.; McDermott, C.M.; Hughes, S.; Brown, A. Effects of rainfall on E. coli concentrations at Door County, Wisconsin beaches. Int. J. Microbiol. 2009, 2009, 876050. [Google Scholar] [CrossRef]
- Mugler, C.; Ribolzi, O.; Viguier, M.; Janeau, J.L.; Jarde, E.; Latsachack, K.; Henry-Des-Tureaux, T.; Thammahacksa, C.; Valentin, C.; Sengtaheuanghoung, O.; et al. Experimental and modelling evidence of splash effects on manure borne Escherichia coli washoff. Environ. Sci. Pollut. Res. Int. 2021, 28, 33009–33020. [Google Scholar] [CrossRef]
- Stumpf, C.H.; Piehler, M.F.; Thompson, S.; Noble, R.T. Loading of fecal indicator bacteria in North Carolina tidal creek headwaters: Hydrographic patterns and terrestrial runoff relationships. Water Res. 2010, 44, 4704–4715. [Google Scholar] [CrossRef]
- Abia, A.L.; James, C.; Ubomba-Jaswa, E.; Benteke Momba, M.N. Microbial Remobilisation on Riverbed Sediment Disturbance in Experimental Flumes and a Human-Impacted River: Implication for Water Resource Management and Public Health in Developing Sub-Saharan African Countries. Int. J. Environ. Res. Public Health 2017, 14, 306. [Google Scholar] [CrossRef]
- Ducarmon, Q.R.; Zwittink, R.D.; Hornung, B.V.H.; van Schaik, W.; Young, V.B.; Kuijper, E.J. Gut Microbiota and Colonization Resistance against Bacterial Enteric Infection. Microbiol. Mol. Biol. Rev. 2019, 83, e00007-19. [Google Scholar] [CrossRef] [PubMed]
- Momose, Y.; Hirayama, K.; Itoh, K. Competition for proline between indigenous Escherichia coli and E. coli O157: H7 in gnotobiotic mice associated with infant intestinal microbiota and its contribution to the colonization resistance against E. coli O157: H7. Antonie Van Leeuwenhoek 2008, 94, 165–171. [Google Scholar] [CrossRef] [PubMed]
- Panhwar, S.; Aftab, A.; Keerio, H.A.; Sarmadivaleh, M.; Tamer, U. A Novel Approach for Real-Time Enumeration of Escherichia coli ATCC 47076 in Water through High Multi-Functional Engineered Nano-Dispersible Electrode. J. Electrochem. Soc. 2021, 168, 037514. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention. National Shiga Toxin-Producing Escherichia coli (STEC) Surveillance Annual Report, 2016; CDC: Atlanta, GA, USA, 2018. [Google Scholar]
- Wong, S.Y.; Paschos, A.; Gupta, R.S.; Schellhorn, H.E. Insertion/deletion-based approach for the detection of Escherichia coli O157: H7 in freshwater environments. Environ. Sci. Technol. 2014, 48, 11462–11470. [Google Scholar] [CrossRef]
- Cox, C.R.; Gilmore, M.S. Native microbial colonization of Drosophila melanogaster and its use as a model of Enterococcus faecalis pathogenesis. Infect. Immun. 2007, 75, 1565–1576. [Google Scholar] [CrossRef]
- Ghosh, A.; Kukanich, K.; Brown, C.E.; Zurek, L. Resident Cats in Small Animal Veterinary Hospitals Carry Multi-Drug Resistant Enterococci and are Likely Involved in Cross-Contamination of the Hospital Environment. Front. Microbiol. 2012, 3, 62. [Google Scholar] [CrossRef]
- Cai, Y. Identification and characterization of Enterococcus species isolated from forage crops and their influence on silage fermentation. J. Dairy Sci. 1999, 82, 2466–2471. [Google Scholar] [CrossRef]
- Napier, M.D.; Haugland, R.; Poole, C.; Dufour, A.P.; Stewart, J.R.; Weber, D.J.; Varma, M.; Lavender, J.S.; Wade, T.J. Exposure to human-associated fecal indicators and self-reported illness among swimmers at recreational beaches: A cohort study. Environ. Health 2017, 16, 103. [Google Scholar] [CrossRef]
- Wade, T.J.; Sams, E.; Brenner, K.P.; Haugland, R.; Chern, E.; Beach, M.; Wymer, L.; Rankin, C.C.; Love, D.; Li, Q. Rapidly measured indicators of recreational water quality and swimming-associated illness at marine beaches: A prospective cohort study. Environ. Health 2010, 9, 66. [Google Scholar] [CrossRef] [PubMed]
- Wade, T.J.; Calderon, R.L.; Brenner, K.P.; Sams, E.; Beach, M.; Haugland, R.; Wymer, L.; Dufour, A.P. High sensitivity of children to swimming-associated gastrointestinal illness: Results using a rapid assay of recreational water quality. Epidemiology 2008, 19, 375–383. [Google Scholar] [CrossRef]
- U.S. Environmental Protection Agency. Recreational Water Quality Criteria; U.S. Environmental Protection Agency: Washington, DC, USA, 2012.
- Wymer, L.J.; Brenner, K.P.; Martinson, J.W.; Stutts, W.R.; Schaub, S.; Dufour, A.P. The EMPACT Beaches Project: Results from a Study on Microbiological Monitoring in Recreational Waters; USEPA: Washington, DC, USA, 2005.
- Health and Welfare Canada. Guidelines for Canadian Recreational Water Quality, 3rd ed.; Healthy Environments and Consumer Safety Branch, Health Canada: Ottawa, ON, Canada, 2012. [Google Scholar]
- Soller, J.A.; Schoen, M.E.; Bartrand, T.; Ravenscroft, J.E.; Ashbolt, N.J. Estimated human health risks from exposure to recreational waters impacted by human and non-human sources of faecal contamination. Water Res. 2010, 44, 4674–4691. [Google Scholar] [CrossRef] [PubMed]
- Miller, W.R.; Munita, J.M.; Arias, C.A. Mechanisms of antibiotic resistance in enterococci. Expert Rev. Anti-Infect. 2014, 12, 1221–1236. [Google Scholar] [CrossRef] [PubMed]
- Cheng, W.; Chen, J.-C. Effect of cultivation broth pH, temperature and NaCl concentration on virulence of an Enterococcus-like bacterium to the giant freshwater prawn Macrobrachium rosenbergii. Dis. Aquat. Org. 1999, 36, 233–237. [Google Scholar] [CrossRef]
- Dubin, K.; Pamer, E.G. Enterococci and their interactions with the intestinal microbiome. Microbiol. Spectr. 2017, 5, 5–6. [Google Scholar] [CrossRef]
- Roslev, P.; Iversen, L.; Sonderbo, H.L.; Iversen, N.; Bastholm, S. Uptake and persistence of human associated Enterococcus in the mussel Mytilus edulis: Relevance for faecal pollution source tracking. J. Appl. Microbiol. 2009, 107, 944–953. [Google Scholar] [CrossRef]
- Nishiyama, M.; Iguchi, A.; Suzuki, Y. Identification of Enterococcus faecium and Enterococcus faecalis as vanC-type vancomycin-resistant enterococci (VRE) from sewage and river water in the provincial city of Miyazaki, Japan. J. Environ. Sci. Health Part A 2015, 50, 16–25. [Google Scholar] [CrossRef]
- Piggot, A.M.; Klaus, J.S.; Johnson, S.; Phillips, M.C.; Solo-Gabriele, H.M. Relationship between enterococcal levels and sediment biofilms at recreational beaches in South Florida. Appl. Environ. Microbiol. 2012, 78, 5973–5982. [Google Scholar] [CrossRef]
- Wiegner, T.N.; Mead, L.H.; Molloy, S.L. A comparison of water quality between low-and high-flow river conditions in a tropical estuary, Hilo Bay, Hawaii. Estuaries Coasts 2013, 36, 319–333. [Google Scholar] [CrossRef]
- Deller, S.; Mascher, F.; Platzer, S.; Reinthaler, F.F.; Marth, E. Effect of solar radiation on survival of indicator bacteria in bathing waters. Cent. Eur. J. Public Health 2006, 14, 133–137. [Google Scholar] [CrossRef]
- Heim, S.; Del Mar Lleo, M.; Bonato, B.; Guzman, C.A.; Canepari, P. The viable but nonculturable state and starvation are different stress responses of Enterococcus faecalis, as determined by proteome analysis. J. Bacteriol. 2002, 184, 6739–6745. [Google Scholar] [CrossRef]
- Dorevitch, S.; Doi, M.; Hsu, F.C.; Lin, K.T.; Roberts, J.D.; Liu, L.C.; Gladding, R.; Vannoy, E.; Li, H.; Javor, M.; et al. A comparison of rapid and conventional measures of indicator bacteria as predictors of waterborne protozoan pathogen presence and density. J. Environ. Monit. 2011, 13, 2427–2435. [Google Scholar] [CrossRef]
- Mansilha, C.R.; Coelho, C.A.; Reinas, A.; Moutinho, A.; Ferreira, S.; Pizarro, C.; Tavares, A. Salmonella: The forgotten pathogen: Health hazards of compliance with European Bathing Water Legislation. Mar. Pollut. Bull. 2010, 60, 819–826. [Google Scholar] [CrossRef]
- Wang, H.; Naghavi, M.; Allen, C.; Barber, R.M.; Bhutta, Z.A.; Carter, A.; Casey, D.C.; Charlson, F.J.; Chen, A.Z.; Coates, M.M. Global, regional, and national life expectancy, all-cause mortality, and cause-specific mortality for 249 causes of death, 1980–2015: A systematic analysis for the Global Burden of Disease Study 2015. Lancet 2016, 388, 1459–1544. [Google Scholar] [CrossRef]
- Koo, B.S.; Hwang, E.H.; Kim, G.; Park, J.Y.; Oh, H.; Lim, K.S.; Kang, P.; Lee, H.Y.; Jeong, K.J.; Mo, I.; et al. Prevalence and characterization of Clostridium perfringens isolated from feces of captive cynomolgus monkeys (Macaca fascicularis). Anaerobe 2020, 64, 102236. [Google Scholar] [CrossRef]
- Matches, J.R.; Liston, J.; Curran, D. Clostridium perfringens in the environment. Appl. Microbiol. 1974, 28, 655–660. [Google Scholar] [CrossRef] [PubMed]
- Sabry, M.; Abd El-Moein, K.; Hamza, E.; Abdel Kader, F. Occurrence of Clostridium perfringens Types A, E, and C in Fresh Fish and Its Public Health Significance. J. Food Prot. 2016, 79, 994–1000. [Google Scholar] [CrossRef] [PubMed]
- Scott, A.; Tien, Y.C.; Drury, C.F.; Reynolds, W.D.; Topp, E. Enrichment of antibiotic resistance genes in soil receiving composts derived from swine manure, yard wastes, or food wastes, and evidence for multiyear persistence of swine Clostridium spp. Can. J. Microbiol. 2018, 64, 201–208. [Google Scholar] [CrossRef] [PubMed]
- Milton, A.A.P.; Sanjukta, R.; Gogoi, A.P.; Momin, K.M.; Priya, G.B.; Das, S.; Ghatak, S.; Sen, A.; Kandpal, B.K. Prevalence, molecular typing and antibiotic resistance of Clostridium perfringens in free range ducks in Northeast India. Anaerobe 2020, 64, 102242. [Google Scholar] [CrossRef]
- Fujioka, R.S. Monitoring coastal marine waters for spore-forming bacteria of faecal and soil origin to determine point from non-point source pollution. Water Sci. Technol. 2001, 44, 181–188. [Google Scholar] [CrossRef] [PubMed]
- Skanavis, C.; Yanko, W.A. Clostridium perfringens as a potential indicator for the presence of sewage solids in marine sediments. Mar. Pollut. Bull. 2001, 42, 31–35. [Google Scholar] [CrossRef]
- Paredes-Sabja, D.; Sarker, N.; Setlow, B.; Setlow, P.; Sarker, M.R. Roles of DacB and spm proteins in clostridium perfringens spore resistance to moist heat, chemicals, and UV radiation. Appl. Environ. Microbiol. 2008, 74, 3730–3738. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Paredes-Sabja, D.; Sarker, M.; Green, C.; Setlow, P.; Li, Y.q. Effects of wet heat treatment on the germination of individual spores of C lostridium perfringens. J. Appl. Microbiol. 2012, 113, 824–836. [Google Scholar] [CrossRef] [PubMed]
- Savichtcheva, O.; Okabe, S. Alternative indicators of fecal pollution: Relations with pathogens and conventional indicators, current methodologies for direct pathogen monitoring and future application perspectives. Water Res. 2006, 40, 2463–2476. [Google Scholar] [CrossRef]
- Vijayavel, K.; Kashian, D. Evaluation of Clostridium perfringens as a tracer of sewage contamination in sediments by two enumeration methods. Environ. Monit. Assess. 2014, 186, 5617–5624. [Google Scholar] [CrossRef]
- Sartory, D. Membrane filtration enumeration of faecal clostridia and Clostridium perfringens in water. Water Res. 1986, 20, 1255–1260. [Google Scholar] [CrossRef]
- Sartory, D.P.; Field, M.; Curbishley, S.M.; Pritchard, A.M. Evaluation of two media for the membrane filtration enumeration of Clostridium perfringens from water. Lett. Appl. Microbiol. 1998, 27, 323–327. [Google Scholar] [CrossRef]
- Marshall, R.S.; Steenbergen, J.F.; McClung, L. Rapid technique for the enumeration of Clostridium perfringens. Appl. Microbiol. 1965, 13, 559–563. [Google Scholar] [CrossRef]
- Karpowicz, E.; Novinscak, A.; Barlocher, F.; Filion, M. qPCR quantification and genetic characterization of Clostridium perfringens populations in biosolids composted for 2 years. J. Appl. Microbiol. 2010, 108, 571–581. [Google Scholar] [CrossRef]
- Milton, A.A.P.; Momin, K.M.; Priya, G.B.; Ghatak, S.; Gandhale, P.N.; Angappan, M.; Das, S.; Sen, A. A novel in situ methodology for visual detection of Clostridium perfringens in pork harnessing saltatory rolling circle amplification. Anaerobe 2021, 69, 102324. [Google Scholar] [CrossRef]
- Bernhard, A.E.; Field, K.G. Identification of nonpoint sources of fecal pollution in coastal waters by using host-specific 16S ribosomal DNA genetic markers from fecal anaerobes. Appl. Environ. Microbiol. 2000, 66, 1587–1594. [Google Scholar] [CrossRef] [PubMed]
- Pilla, R.; Suchodolski, J.S. The Role of the Canine Gut Microbiome and Metabolome in Health and Gastrointestinal Disease. Front. Vet. Sci. 2019, 6, 498. [Google Scholar] [CrossRef]
- Bergamaschi, M.; Tiezzi, F.; Howard, J.; Huang, Y.J.; Gray, K.A.; Schillebeeckx, C.; McNulty, N.P.; Maltecca, C. Gut microbiome composition differences among breeds impact feed efficiency in swine. Microbiome 2020, 8, 110. [Google Scholar] [CrossRef] [PubMed]
- Kabiri, L.; Alum, A.; Rock, C.; McLain, J.E.; Abbaszadegan, M. Isolation of Bacteroides from fish and human fecal samples for identification of unique molecular markers. Can. J. Microbiol. 2013, 59, 771–777. [Google Scholar] [CrossRef] [PubMed]
- Ko, H.Y.; Cho, K.; Park, S.; Kim, J.H.; Kang, J.H.; Jeong, Y.S.; Choi, J.D.; Sin, Y.; Lee, C.; Ko, G. Host-Specific Bacteroides Markers-Based Microbial Source Tracking in Aquaculture Areas. Microbes Environ. 2018, 33, 151–161. [Google Scholar] [CrossRef]
- Kildare, B.J.; Leutenegger, C.M.; McSwain, B.S.; Bambic, D.G.; Rajal, V.B.; Wuertz, S. 16S rRNA-based assays for quantitative detection of universal, human-, cow-, and dog-specific fecal Bacteroidales: A Bayesian approach. Water Res. 2007, 41, 3701–3715. [Google Scholar] [CrossRef]
- Kobayashi, A.; Sano, D.; Hatori, J.; Ishii, S.; Okabe, S. Chicken- and duck-associated Bacteroides-Prevotella genetic markers for detecting fecal contamination in environmental water. Appl. Microbiol. Biotechnol. 2013, 97, 7427–7437. [Google Scholar] [CrossRef]
- Bernhard, A.E.; Field, K.G. A PCR assay To discriminate human and ruminant feces on the basis of host differences in Bacteroides-Prevotella genes encoding 16S rRNA. Appl. Environ. Microbiol. 2000, 66, 4571–4574. [Google Scholar] [CrossRef]
- Layton, A.; McKay, L.; Williams, D.; Garrett, V.; Gentry, R.; Sayler, G. Development of Bacteroides 16S rRNA gene TaqMan-based real-time PCR assays for estimation of total, human, and bovine fecal pollution in water. Appl. Environ. Microbiol. 2006, 72, 4214–4224. [Google Scholar] [CrossRef]
- Reischer, G.H.; Kasper, D.C.; Steinborn, R.; Farnleitner, A.H.; Mach, R.L. A quantitative real-time PCR assay for the highly sensitive and specific detection of human faecal influence in spring water from a large alpine catchment area. Lett. Appl. Microbiol. 2007, 44, 351–356. [Google Scholar] [CrossRef]
- Okabe, S.; Okayama, N.; Savichtcheva, O.; Ito, T. Quantification of host-specific Bacteroides-Prevotella 16S rRNA genetic markers for assessment of fecal pollution in freshwater. Appl. Microbiol. Biotechnol. 2007, 74, 890–901. [Google Scholar] [CrossRef]
- Dick, L.K.; Bernhard, A.E.; Brodeur, T.J.; Santo Domingo, J.W.; Simpson, J.M.; Walters, S.P.; Field, K.G. Host distributions of uncultivated fecal Bacteroidales bacteria reveal genetic markers for fecal source identification. Appl. Environ. Microbiol. 2005, 71, 3184–3191. [Google Scholar] [CrossRef]
- Bacic, M.K.; Smith, C.J. Laboratory maintenance and cultivation of bacteroides species. Curr. Protoc. Microbiol. 2008, 9, 13C-1. [Google Scholar] [CrossRef]
- Livingston, S.J.; Kominos, S.D.; Yee, R.B. New medium for selection and presumptive identification of the Bacteroides fragilis group. J. Clin. Microbiol. 1978, 7, 448–453. [Google Scholar] [CrossRef] [PubMed]
- Ho, P.L.; Ho, L.Y.; Yau, C.Y.; Tong, M.K.; Chow, K.H. A Novel Selective Medium for Isolation of Bacteroides fragilis from Clinical Specimens. J. Clin. Microbiol. 2017, 55, 384–390. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, S.A.; Drucker, D.B.; Keaney, M.G.; Ganguli, L.A. Evaluation of the RAPID ID 32A system for the identification of Bacteroides fragilis and related organisms. J. Appl. Bacteriol. 1991, 71, 360–365. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, W.; Hughes, B.; Harwood, V.J. Current status of marker genes of Bacteroides and related taxa for identifying sewage pollution in environmental waters. Water 2016, 8, 231. [Google Scholar] [CrossRef]
- Kirs, M.; Caffaro-Filho, R.A.; Wong, M.; Harwood, V.J.; Moravcik, P.; Fujioka, R.S. Human-associated Bacteroides spp. and human polyomaviruses as microbial source tracking markers in Hawaii. Appl. Environ. Microbiol. 2016, 82, 6757–6767. [Google Scholar] [CrossRef]
- Boehm, A.B.; Soller, J.A. Refined ambient water quality thresholds for human-associated fecal indicator HF183 for recreational waters with and without co-occurring gull fecal contamination. Microb. Risk Anal. 2020, 16, 100139. [Google Scholar] [CrossRef]
- Shanks, O.C.; Kelty, C.A.; Sivaganesan, M.; Varma, M.; Haugland, R.A. Quantitative PCR for genetic markers of human fecal pollution. Appl. Environ. Microbiol. 2009, 75, 5507–5513. [Google Scholar] [CrossRef]
- Toledo-Hernandez, C.; Ryu, H.; Gonzalez-Nieves, J.; Huertas, E.; Toranzos, G.A.; Santo Domingo, J.W. Tracking the primary sources of fecal pollution in a tropical watershed in a one-year study. Appl. Environ. Microbiol. 2013, 79, 1689–1696. [Google Scholar] [CrossRef] [PubMed]
- Zimmer-Faust, A.G.; Thulsiraj, V.; Marambio-Jones, C.; Cao, Y.; Griffith, J.F.; Holden, P.A.; Jay, J.A. Effect of freshwater sediment characteristics on the persistence of fecal indicator bacteria and genetic markers within a Southern California watershed. Water Res. 2017, 119, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Stachler, E.; Kelty, C.; Sivaganesan, M.; Li, X.; Bibby, K.; Shanks, O.C. Quantitative CrAssphage PCR Assays for Human Fecal Pollution Measurement. Environ. Sci. Technol. 2017, 51, 9146–9154. [Google Scholar] [CrossRef] [PubMed]
- Okoh, A.I.; Sibanda, T.; Gusha, S.S. Inadequately treated wastewater as a source of human enteric viruses in the environment. Int. J. Environ. Res. Public Health 2010, 7, 2620–2637. [Google Scholar] [CrossRef]
- Liu, Y.; Gilchrist, A.; Zhang, J.; Li, X.F. Detection of viable but nonculturable Escherichia coli O157:H7 bacteria in drinking water and river water. Appl. Environ. Microbiol. 2008, 74, 1502–1507. [Google Scholar] [CrossRef] [PubMed]
- Kitajima, M.; Sassi, H.P.; Torrey, J.R. Pepper mild mottle virus as a water quality indicator. NPJ Clean Water 2018, 1, 19. [Google Scholar] [CrossRef]
- Dutilh, B.E.; Cassman, N.; McNair, K.; Sanchez, S.E.; Silva, G.G.; Boling, L.; Barr, J.J.; Speth, D.R.; Seguritan, V.; Aziz, R.K.; et al. A highly abundant bacteriophage discovered in the unknown sequences of human faecal metagenomes. Nat. Commun. 2014, 5, 4498. [Google Scholar] [CrossRef]
- McMinn, B.R.; Huff, E.M.; Rhodes, E.R.; Korajkic, A. Concentration and quantification of somatic and F+ coliphages from recreational waters. J. Virol. Methods 2017, 249, 58–65. [Google Scholar] [CrossRef]
- McMinn, B.R.; Rhodes, E.R.; Huff, E.M.; Korajkic, A. Decay of infectious adenovirus and coliphages in freshwater habitats is differentially affected by ambient sunlight and the presence of indigenous protozoa communities. Virol. J. 2020, 17, 1. [Google Scholar] [CrossRef]
- Tomat, D.; Balagué, C.; Casabonne, C.; Verdini, R.; Quiberoni, A. Resistance of foodborne pathogen coliphages to thermal and physicochemical treatments applied in food manufacture. Innov. Food Sci. Emerg. Technol. 2015, 30, 184–191. [Google Scholar] [CrossRef]
- Guan, D.; Kniel, K.; Calci, K.R.; Hicks, D.T.; Pivarnik, L.F.; Hoover, D.G. Response of four types of coliphages to high hydrostatic pressure. Food Microbiol. 2006, 23, 546–551. [Google Scholar] [CrossRef] [PubMed]
- Vergara, G.; Goh, S.; Rezaeinejad, S.; Chang, S.; Sobsey, M.; Gin, K. Evaluation of FRNA coliphages as indicators of human enteric viruses in a tropical urban freshwater catchment. Water Res. 2015, 79, 39–47. [Google Scholar] [CrossRef] [PubMed]
- Muniesa, M.; Mocé-Llivina, L.; Katayama, H.; Jofre, J. Bacterial host strains that support replication of somatic coliphages. Antonie Van Leeuwenhoek 2003, 83, 305–315. [Google Scholar] [CrossRef]
- U.S. Environmental Protection Agency. Male-Specific (F+) and Somatic Coliphage in Water by Two-Step Enrichment Procedure; U.S. Environmental Protection Agency: Wahington, DC, USA, 2001.
- U.S. Environmental Protection Agency. Method 1602: Male-Specific (F+) and Somatic Coliphage in Water by Single Agar Layer (SAL) Procedure; U.S. Environmental Protection Agency: Wahington, DC, USA, 2001.
- Muniesa, M.; Ballesté, E.; Imamovic, L.; Pascual-Benito, M.; Toribio-Avedillo, D.; Lucena, F.; Blanch, A.; Jofre, J. Bluephage: A rapid method for the detection of somatic coliphages used as indicators of fecal pollution in water. Water Res. 2018, 128, 10–19. [Google Scholar] [CrossRef]
- Mendez, J.; Toribio-Avedillo, D.; Mangas-Casas, R.; Martinez-Gonzalez, J. Bluephage, a method for efficient detection of somatic coliphages in one hundred milliliter water samples. Sci. Rep. 2020, 10, 2977. [Google Scholar] [CrossRef]
- Haramoto, E.; Yamada, K.; Nishida, K. Prevalence of protozoa, viruses, coliphages and indicator bacteria in groundwater and river water in the Kathmandu Valley, Nepal. Trans. R. Soc. Trop. Med. Hyg. 2011, 105, 711–716. [Google Scholar] [CrossRef]
- Lee, J.E.; Lee, H.; Cho, Y.H.; Hur, H.G.; Ko, G. F+ RNA coliphage-based microbial source tracking in water resources of South Korea. Sci. Total Environ. 2011, 412, 127–131. [Google Scholar] [CrossRef]
- Toribio-Avedillo, D.; Martin-Diaz, J.; Blanco-Picazo, P.; Blanch, A.R.; Muniesa, M. F-specific coliphage detection by the Bluephage method. Water Res. 2020, 184, 116215. [Google Scholar] [CrossRef]
- Agullo-Barcelo, M.; Galofre, B.; Sala, L.; Garcia-Aljaro, C.; Lucena, F.; Jofre, J. Simultaneous detection of somatic and F-specific coliphages in different settings by Escherichia coli strain CB390. FEMS Microbiol. Lett. 2016, 363. [Google Scholar] [CrossRef][Green Version]
- Benjamin-Chung, J.; Arnold, B.F.; Wade, T.J.; Schiff, K.; Griffith, J.F.; Dufour, A.P.; Weisberg, S.B.; Colford, J.M., Jr. Coliphages and Gastrointestinal Illness in Recreational Waters: Pooled Analysis of Six Coastal Beach Cohorts. Epidemiology 2017, 28, 644–652. [Google Scholar] [CrossRef]
- Shkoporov, A.N.; Khokhlova, E.V.; Fitzgerald, C.B.; Stockdale, S.R.; Draper, L.A.; Ross, R.P.; Hill, C. ΦCrAss001 represents the most abundant bacteriophage family in the human gut and infects Bacteroides intestinalis. Nat. Commun. 2018, 9, 4781. [Google Scholar] [CrossRef]
- Edwards, R.A.; Vega, A.A.; Norman, H.M.; Ohaeri, M.; Levi, K.; Dinsdale, E.A.; Cinek, O.; Aziz, R.K.; McNair, K.; Barr, J.J.; et al. Global phylogeography and ancient evolution of the widespread human gut virus crAssphage. Nat. Microbiol. 2019, 4, 1727–1736. [Google Scholar] [CrossRef]
- Kongprajug, A.; Mongkolsuk, S.; Sirikanchana, K. CrAssphage as a potential human sewage marker for microbial source tracking in Southeast Asia. Environ. Sci. Technol. Lett. 2019, 6, 159–164. [Google Scholar] [CrossRef]
- Stachler, E.; Crank, K.; Bibby, K. Co-occurrence of crAssphage with antibiotic resistance genes in an impacted urban watershed. Environ. Sci. Technol. Lett. 2019, 6, 216–221. [Google Scholar] [CrossRef]
- Ahmed, W.; Payyappat, S.; Cassidy, M.; Besley, C. A duplex PCR assay for the simultaneous quantification of Bacteroides HF183 and crAssphage CPQ_056 marker genes in untreated sewage and stormwater. Environ. Int. 2019, 126, 252–259. [Google Scholar] [CrossRef] [PubMed]
- Stachler, E.; Akyon, B.; de Carvalho, N.A.; Ference, C.; Bibby, K. Correlation of crAssphage qPCR Markers with Culturable and Molecular Indicators of Human Fecal Pollution in an Impacted Urban Watershed. Environ. Sci. Technol. 2018, 52, 7505–7512. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Greaves, J.; Arp, L.; Stone, D.; Bibby, K. Comparative fate of CrAssphage with culturable and molecular fecal pollution indicators during activated sludge wastewater treatment. Environ. Int. 2020, 136, 105452. [Google Scholar] [CrossRef]
- Wolf, S.; Hewitt, J.; Greening, G.E. Viral multiplex quantitative PCR assays for tracking sources of fecal contamination. Appl. Environ. Microbiol. 2010, 76, 1388–1394. [Google Scholar] [CrossRef] [PubMed]
- McQuaig, S.M.; Scott, T.M.; Lukasik, J.O.; Paul, J.H.; Harwood, V.J. Quantification of human polyomaviruses JC virus and BK virus by TaqMan quantitative PCR and comparison to other water quality indicators in water and fecal samples. Appl. Environ. Microbiol. 2009, 75, 3379–3388. [Google Scholar] [CrossRef] [PubMed]
- Pal, A.; Sirota, L.; Maudru, T.; Peden, K.; Lewis, A.M., Jr. Real-time, quantitative PCR assays for the detection of virus-specific DNA in samples with mixed populations of polyomaviruses. J. Virol. Methods 2006, 135, 32–42. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, W.; Zhang, Q.; Kozak, S.; Beale, D.; Gyawali, P.; Sadowsky, M.J.; Simpson, S. Comparative decay of sewage-associated marker genes in beach water and sediment in a subtropical region. Water Res. 2019, 149, 511–521. [Google Scholar] [CrossRef] [PubMed]
- Rowe, W.P.; Huebner, R.J.; Gilmore, L.K.; Parrott, R.H.; Ward, T.G. Isolation of a cytopathogenic agent from human adenoids undergoing spontaneous degeneration in tissue culture. Proc. Soc. Exp. Biol. Med. 1953, 84, 570–573. [Google Scholar] [CrossRef]
- Parks, R.J. Adenovirus protein IX: A new look at an old protein. Mol. Ther. 2005, 11, 19–25. [Google Scholar] [CrossRef]
- Matthews, D.A. Adenovirus protein V induces redistribution of nucleolin and B23 from nucleolus to cytoplasm. J. Virol. 2001, 75, 1031–1038. [Google Scholar] [CrossRef]
- Noble, R.T.; Allen, S.M.; Blackwood, A.D.; Chu, W.; Jiang, S.C.; Lovelace, G.L.; Sobsey, M.D.; Stewart, J.R.; Wait, D.A. Use of viral pathogens and indicators to differentiate between human and non-human fecal contamination in a microbial source tracking comparison study. J. Water Health 2003, 1, 195–207. [Google Scholar] [CrossRef][Green Version]
- Hundesa, A.; De Motes, C.M.; Albinana-Gimenez, N.; Rodriguez-Manzano, J.; Bofill-Mas, S.; Sunen, E.; Girones, R.R. Development of a qPCR assay for the quantification of porcine adenoviruses as an MST tool for swine fecal contamination in the environment. J. Virol. Methods 2009, 158, 130–135. [Google Scholar] [CrossRef]
- Ogorzaly, L.; Bertrand, I.; Paris, M.; Maul, A.; Gantzer, C. Occurrence, survival, and persistence of human adenoviruses and F-specific RNA phages in raw groundwater. Appl. Environ. Microbiol. 2010, 76, 8019–8025. [Google Scholar] [CrossRef] [PubMed]
- Hewitt, J.; Greening, G.E.; Leonard, M.; Lewis, G.D. Evaluation of human adenovirus and human polyomavirus as indicators of human sewage contamination in the aquatic environment. Water Res. 2013, 47, 6750–6761. [Google Scholar] [CrossRef]
- Li, D.; He, M.; Jiang, S.C. Detection of infectious adenoviruses in environmental waters by fluorescence-activated cell sorting assay. Appl. Environ. Microbiol. 2010, 76, 1442–1448. [Google Scholar] [CrossRef]
- Gerba, C.P.; Gramos, D.M.; Nwachuku, N. Comparative inactivation of enteroviruses and adenovirus 2 by UV light. Appl. Environ. Microbiol. 2002, 68, 5167–5169. [Google Scholar] [CrossRef]
- Eischeid, A.C.; Thurston, J.A.; Linden, K.G. UV disinfection of adenovirus: Present state of the research and future directions. Crit. Rev. Environ. Sci. Technol. 2011, 41, 1375–1396. [Google Scholar] [CrossRef]
- Flint, S.J.; Enquist, L.W.; Racaniello, V.R.; Skalka, A.M. Principles of Virology: Molecular Biology, Pathogenesis, and Control; ASM Press: Washington, DC, USA, 2000. [Google Scholar]
- Sassoubre, L.M.; Love, D.C.; Silverman, A.I.; Nelson, K.L.; Boehm, A.B. Comparison of enterovirus and adenovirus concentration and enumeration methods in seawater from Southern California, USA and Baja Malibu, Mexico. J. Water Health 2012, 10, 419–430. [Google Scholar] [CrossRef] [PubMed]
- Lipp, E.K.; Lukasik, J.; Rose, J.B. Human enteric viruses and parasites in the marine environment. Methods Microbiol. 2001, 30, 559–588. [Google Scholar]
- Lachiewicz, A.; Cianciolo, R.; Miller, M.; Derebail, V. Adenovirus causing fever, upper respiratory infection, and allograft nephritis complicated by persistent asymptomatic viremia. Transpl. Infect. Dis. 2014, 16, 648–652. [Google Scholar] [CrossRef] [PubMed]
- Demoliner, M.; Gularte, J.S.; Girardi, V.; Eisen, A.K.A.; de Souza, F.G.; Staggemeier, R.; Henzel, A.; Spilki, F.R. Microbial Source Tracking in Small Farms: Use of Different Methods for Adenovirus Detection. Water Air Soil Pollut. 2021, 232, 63. [Google Scholar] [CrossRef]
- Schoch, C.L.; Ciufo, S.; Domrachev, M.; Hotton, C.L.; Kannan, S.; Khovanskaya, R.; Leipe, D.; Mcveigh, R.; O’Neill, K.; Robbertse, B. NCBI Taxonomy: A comprehensive update on curation, resources and tools. Database 2020, 2020, baaa062. [Google Scholar] [CrossRef] [PubMed]
- Padgett, B.; Zurhein, G.; Walker, D.; Eckroade, R.; Dessel, B. Cultivation of papova-like virus from human brain with progressive multifocal leucoencephalopathy. Lancet 1971, 297, 1257–1260. [Google Scholar] [CrossRef]
- Bofill-Mas, S.; Rusiñol, M.; Fernandez-Cassi, X.; Carratalà, A.; Hundesa, A.; Girones, R. Quantification of human and animal viruses to differentiate the origin of the fecal contamination present in environmental samples. BioMed Res. Int. 2013, 2013, 192089. [Google Scholar] [CrossRef]
- Bofill-Mas, S.; Pina, S.; Girones, R. Documenting the epidemiologic patterns of polyomaviruses in human populations by studying their presence in urban sewage. Appl. Environ. Microbiol. 2000, 66, 238–245. [Google Scholar] [CrossRef]
- McQuaig, S.; Griffith, J.; Harwood, V.J. Association of fecal indicator bacteria with human viruses and microbial source tracking markers at coastal beaches impacted by nonpoint source pollution. Appl. Environ. Microbiol. 2012, 78, 6423–6432. [Google Scholar] [CrossRef]
- Albinana-Gimenez, N.; Clemente-Casares, P.; Calgua, B.; Huguet, J.M.; Courtois, S.; Girones, R. Comparison of methods for concentrating human adenoviruses, polyomavirus JC and noroviruses in source waters and drinking water using quantitative PCR. J. Virol. Methods 2009, 158, 104–109. [Google Scholar] [CrossRef]
- Bofill-Mas, S.; Albinana-Gimenez, N.; Clemente-Casares, P.; Hundesa, A.; Rodriguez-Manzano, J.; Allard, A.; Calvo, M.; Girones, R. Quantification and stability of human adenoviruses and polyomavirus JCPyV in wastewater matrices. Appl. Environ. Microbiol. 2006, 72, 7894–7896. [Google Scholar] [CrossRef] [PubMed]
- Templeton, M.R.; Andrews, R.C.; Hofmann, R. Particle-associated viruses in water: Impacts on disinfection processes. Crit. Rev. Environ. Sci. Technol. 2008, 38, 137–164. [Google Scholar] [CrossRef]
- Haramoto, E.; Katayama, H. Application of acidic elution to virus concentration using electropositive filters. Food Environ. Virol. 2013, 5, 77–80. [Google Scholar] [CrossRef]
- Ahmed, W.; Harwood, V.; Gyawali, P.; Sidhu, J.P.; Toze, S. Comparison of concentration methods for quantitative detection of sewage-associated viral markers in environmental waters. Appl. Environ. Microbiol. 2015, 81, 2042–2049. [Google Scholar] [CrossRef]
- Calgua, B.; Barardi, C.R.M.; Bofill-Mas, S.; Rodriguez-Manzano, J.; Girones, R. Detection and quantitation of infectious human adenoviruses and JC polyomaviruses in water by immunofluorescence assay. J. Virol. Methods 2011, 171, 1–7. [Google Scholar] [CrossRef]
- Wong, M.V.; Hashsham, S.A.; Gulari, E.; Rouillard, J.-M.; Aw, T.G.; Rose, J.B. Detection and characterization of human pathogenic viruses circulating in community wastewater using multi target microarrays and polymerase chain reaction. J. Water Health 2013, 11, 659–670. [Google Scholar] [CrossRef] [PubMed]
- Hewitt, J.; Leonard, M.; Greening, G.E.; Lewis, G.D. Influence of wastewater treatment process and the population size on human virus profiles in wastewater. Water Res. 2011, 45, 6267–6276. [Google Scholar] [CrossRef] [PubMed]
- Carini, P.; Marsden, P.J.; Leff, J.W.; Morgan, E.E.; Strickland, M.S.; Fierer, N. Relic DNA is abundant in soil and obscures estimates of soil microbial diversity. Nat. Microbiol. 2016, 2, 16242. [Google Scholar] [CrossRef]
- Wang, H.; Koydemir, H.C.; Qiu, Y.; Bai, B.; Zhang, Y.; Jin, Y.; Tok, S.; Yilmaz, E.C.; Gumustekin, E.; Rivenson, Y. Early detection and classification of live bacteria using time-lapse coherent imaging and deep learning. Light Sci. Appl. 2020, 9, 118. [Google Scholar] [CrossRef]
- Højris, B.; Christensen, S.C.B.; Albrechtsen, H.-J.; Smith, C.; Dahlqvist, M. A novel, optical, on-line bacteria sensor for monitoring drinking water quality. Sci. Rep. 2016, 6, 23935. [Google Scholar] [CrossRef] [PubMed]
- Simões, J.; Yang, Z.; Dong, T. An ultrasensitive fluorimetric sensor for pre-screening of water microbial contamination risk. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2021, 258, 119805. [Google Scholar] [CrossRef] [PubMed]
- Guo, T.; Wei, Y.; Xu, C.; Watts, B.R.; Zhang, Z.; Fang, Q.; Zhang, H.; Selvaganapathy, P.R.; Deen, M.J. Counting of Escherichia coli by a microflow cytometer based on a photonic–microfluidic integrated device. Electrophoresis 2015, 36, 298–304. [Google Scholar] [CrossRef] [PubMed]
| Leading Pathogen | Diseases | Year | Country | No. of Cases | Citation |
|---|---|---|---|---|---|
| Salmonella spp. | Salmonellosis | 2010 | China | 324 | [18] |
| Salmonellosis | 2010 | USA | 695 | [19] | |
| Gastroenteritis | 2013 | USA | 634 | [20] | |
| Cryptosporidium | Cryptosporidiosis | 2010 | Canada | 12 | [21] |
| Cryptosporidiosis | 2011 | Australia | 1141 | [22] | |
| Shigella sonnei | Shigellosis | 2012 | China | 134 | [23] |
| Shigellosis | 2012 | Turkey | 4239 | [24] | |
| E. coli O157:H7 | Diarrhea/Vomiting | 2012 | Canada | 5 | [25] |
| Diarrhea/Vomiting | 2012 | Japan | 115 | [26] | |
| Campylobacter | Campylobacteriosis | 2016 | UK | 69 | [27] |
| Campylobacteriosis | 2016 | New Zelaland | 967 | [28] | |
| Hepatitis A virus | Hepatitis A | 2016 | USA | 292 | [29] |
| Jaundice | 2016 | India | 233 | [30] | |
| Rotavirus G2P | Gastroenteritis | 2017 | Germany | 32 | [16] |
| Rotavirus G3P | Gastroenteritis | 2017 | Australia | 2319 | [31] |
| Adenovirus | Respiratory illness | 2018 | USA | 168 | [32] |
| Fever | 2019 | China | 27 | [33] | |
| Norovirus | Diarrhea/Vomiting | 2018 | South Korea | 199 | [34] |
| Gastroenteritis | 2020 | Brazil | 176 | [35] |
| Host | Primer Set | Sequence (5′ → 3′) | Citation |
|---|---|---|---|
| Human | HF183F/Bac708R | F: ATCATGAGTTCACATGTCCG | [157] |
| R: CAATCGGAGTTCTTCGTG | |||
| HF134F/HF654R | F: GCCGTCTACTCTTGGCC | [157] | |
| R: CCTGCCTCTACTGTACTC | |||
| HuBac566F/HuBac 692R | F: GGGTTTAAAGGGAGCGTAGG | [158] | |
| R: CTACACCACGAATTCCGCCT | |||
| BacHF/BacHR | F: CTTGGCCAGCCTTCTGAAAG | [159] | |
| R: CCCCATCGTCTACCGAAAATAC | |||
| Cattle | CF128F/Bac708R | F: CCAACYTTCCCGWTACTC | [157] |
| R: See above | |||
| CS406F/Bac581R | F: GAAGGATGAAGGTTCTATGGATTGT | [160] | |
| R: CGCTCCCTTTAAACCCAATAAA | |||
| CS621F/Bac725R | F: AACCACAGCCCGCGATT | [160] | |
| R: CAATCGGAGTTCTTCGTGATATCTA | |||
| Swine | Bac41F/PS183R | F: TACAGGCTTAACACATGCAAGTCG | [160] |
| R: CTCATACGGTATTAATCCGCCTTT | |||
| PigBac1F/PigBac1R | F: CGGGTTGTAAACTGCTTTTATGAAG | [160] | |
| R: CGCTCCCTTTAAACCCAATAAA | |||
| PF163F/Bac708R | F: GCGGATTAATACCGTATGA | [161] | |
| R: See above | |||
| Dog | BacCan-545F/Bac725R | F: GGAGCGCAGACGGGTTTT | [155] |
| R: See above | |||
| Chicken | C160F-HU/Bac265R-HU | F: AAGGGAGATTAATACCCGATGATG | [156] |
| R: CCGTTACCCCGCCTACTAC | |||
| Duck | Bac366F-HU/Duck474R-HU | F: TTGGTCAATGGGCGGAAG | [156] |
| R: GCACATTCCCACACGTGAGA |
| Target Virus | qPCR Assay | Primer or Probe Sequence (5′ → 3′) | Target Gene | Citation |
|---|---|---|---|---|
| CrAssphage | CPQ_056 | F: CAGAAGTACAAACTCCTAAAAAACGTAGAG | Predicted coding region | [172] |
| R: GATGACCAATAAACAAGCCATTAGC | ||||
| Probe: [FAM]-AATAACGATTTACGTGATGTAAC-[MGB] | ||||
| CPQ_064 | F: TGTATAGATGCTGCTGCAACTGTACTC | Predicted coding region | [172] | |
| R: CGTTGTTTTCATCTTTATCTTGTCCAT | ||||
| Probe: [FAM]-CTGAAATTGTTCATAAGCAA-[MGB] | ||||
| HAdV | VTB-1-HAdVF | F: GCCTGGGGAACAAGTTCAGA | Hexon | [199] |
| R: GCGTAAAGCGCACTTTGTAAG | ||||
| Probe: [Quasar]-CAGTCGCTGYGACCTGTCTGTGGTT-[BHQ2] | ||||
| VTB-2-HAdVC | F: GAGACGTACTTCAGCCTGAAT | Hexon | [199] | |
| R: GATGAACCGCAGCGTCAA | ||||
| Probe: [FAM]-CCTACGCACGACGTGACCACAGA-[BHQ1] | ||||
| HPyV | BKPyV and JCPyV | SM2: AGTCTTTAGGGTCTTCTACCTTT | Conserved T-antigen | [200] |
| P6: GGTGCCAACCTATGGAACAG | ||||
| KGJ3 probe: [FAM]-TCATCACTGGCAAACAT-[MGB] | ||||
| HPyV-2 | F: ATGTTTGCCAGTGATGATGAAAA | JVGP5 | [201] | |
| R: GGAAAGTCTTTAGGGTCTTCTACCTTT | ||||
| Probe: [FAM]-AGGATCCCAACACTCTACCCCACCTAAAAAGA-[MGB] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, E.; Saleem, F.; Edge, T.A.; Schellhorn, H.E. Biological Indicators for Fecal Pollution Detection and Source Tracking: A Review. Processes 2021, 9, 2058. https://doi.org/10.3390/pr9112058
Li E, Saleem F, Edge TA, Schellhorn HE. Biological Indicators for Fecal Pollution Detection and Source Tracking: A Review. Processes. 2021; 9(11):2058. https://doi.org/10.3390/pr9112058
Chicago/Turabian StyleLi, Enze, Faizan Saleem, Thomas A. Edge, and Herb E. Schellhorn. 2021. "Biological Indicators for Fecal Pollution Detection and Source Tracking: A Review" Processes 9, no. 11: 2058. https://doi.org/10.3390/pr9112058
APA StyleLi, E., Saleem, F., Edge, T. A., & Schellhorn, H. E. (2021). Biological Indicators for Fecal Pollution Detection and Source Tracking: A Review. Processes, 9(11), 2058. https://doi.org/10.3390/pr9112058






