Abstract
Gemcitabine (GEM) is the first-line chemotherapy drug for patients with advanced pancreatic cancer. Moringa oleifera (MO) exhibited various biological activities, including anticancer effects. Nevertheless, the effectiveness of their combination against pancreatic cancer has not yet been explored. This study evaluates the effect of MO and GEM against pancreatic cancer through network pharmacology. TCMSP, TCMID, and PubMed were used to identify and screen MO bioactive compounds. MO and GEM genes were predicted through DGIdb, CTD, and DrugBank. Pancreatic cancer genes were retrieved from OMIM and MalaCards. Protein–protein interaction (PPI) and compound-target-pathway network were established via STRING and Cytoscape. Gene ontology (GO) and pathway enrichment analysis were conducted using DAVID Bioinformatic Tools. Catechin, kaempferol, quercetin, and epicatechin that met the drug screening requirements, and three additional compounds, glucomoringin, glucoraphanin, and moringinine, were identified as bioactive compounds in MO. Catechin was found to be the main hub compound in MO. TP53, AKT1, VEGFA, and CCND1 from PPI network were discovered as hub genes to have biological importance in pancreatic cancer. GO and pathway analysis revealed that MO and GEM combination was mainly associated with cancer, including pancreatic cancer, through regulation of apoptosis. Combination therapy between MO and GEM might provide insight in pancreatic cancer treatment.
1. Introduction
According to the World Health Organization, cancer is defined as a disease that occurs in any organ or tissue of the body when the abnormal cells grow and divide uncontrollably beyond the usual boundaries and invade or spread to the other parts of the body or organs [1]. Pancreatic cancer is one of the most lethal human cancers, which appears to be a significant unresolved health issue in the world. It is often diagnosed at an advanced stage due to the distinctive anatomical location of the pancreas, which is associated with a very poor prognosis. Based on GLOBOCAN 2020, the estimated incidence of pancreatic cancer was 495,773 new cases and 466,003 deaths per year, making this cancer ranked as the 12th most common cancer and the 7th leading cause of cancer death in the world, respectively [2].
Gemcitabine (GEM), or also known as 2′,2′-difluorodeoxycytidine (dFdC), is a golden-standard treatment for locally advanced and metastatic pancreatic cancer, since it was proven to be superior to fluorouracil (5-FU) in terms of overall survival, performance status, and pain control in 1997 [3,4]. Although it has been accepted widely, the chemoresistance is one of the major issues associated with this drug. Currently, studies on drug combination have been well recognized in the cancer research community, with the aim to reduce toxicity, minimize the induction of drug resistance, as well as to achieve additional therapeutic effect [5]. Thus, the focus had been drawn towards traditional herbal medicine, which could be an alternative approach to overcome the limitation of gemcitabine against pancreatic cancer.
Moringa oleifera (MO), also known as drumstick or horseradish tree, is native to Africa and South Asia and is most widely cultivated in northwestern India [6]. MO has been demonstrated to possess pharmacological and biological activities, such as antitumor, antioxidant, antimicrobial, hypoglycemic, hypotensive, hepatoprotective and immunomodulatory activities [7]. These could be attributed to the presence of numerous bioactive compounds, including flavonoids, phenolic compounds, vitamins, isothiocyanates, saponins, and tannins, that have been reported in MO studies for their therapeutic effect [6].
Many efforts have been made to study the synergistic effects of combination therapy between conventional and traditional herbal medicines. Network pharmacology is one of those, which offers a systematic approach and a novel perspective to present the dynamic relation between drugs, potential targets, and associated pathways by constructing the interactions among drug–targets–diseases [8]. Bioinformatics, a cost-effective drug development approach, also provides a powerful platform to study network-based drug discovery to address the complexity of the multi-target mechanism [9]. Hence, in this study, a network pharmacology approach was used to predict the target genes and evaluate the effect of MO in combination with GEM against pancreatic cancer.
2. Materials and Methods
2.1. Identification of Chemical Compounds of MO
Identification of chemical compounds of MO were performed by using Traditional Chinese Medicine Systems Pharmacology, TCMSP (https://www.tcmspw.com/tcmsp.php, (accessed on 2 December 2020)) [10], a unique Chinese herbal medicine pharmacology database system, which integrates the association between drugs, targets, and disease networks, and Traditional Chinese Medicine Integrated Database, TCMID (http://bidd.group/TCMID/, (accessed on 2 December 2020)) [11]. The Latin name of MO was searched in these two databases to retrieve the composition of chemical and pharmacokinetic properties of each chemical compound. However, the constituents of MO cannot be found in the databases.
Alternatively, studies related to MO were searched through the electronic literature database, PubMed, (https://pubmed.ncbi.nlm.nih.gov/, (accessed on 7 December 2020)) [12], to determine the biochemical compounds. Huang et al., 2020 [13] studied the effect of antiinsulin resistance of MO, and listed the compounds found in MO in their article. The 3D structures of the selected compounds were downloaded from PubChem (https://pubchem.ncbi.nlm.nih.gov/, (accessed on 26 August 2021)) [14].
2.2. The Screening of Bioactive Compounds in MO
The absorption, distribution, metabolism, and excretion (ADME) properties have been regarded as essential indicators for orally administered herbal medicine. The evaluation of three parameters, including the oral bioavailability (OB), drug-likeness (DL), and Caco-2 permeability, were applied to predict the potential bioactive compounds of MO. The three parameters were evaluated using an in silico integrative ADME model through the TCMSP database, with the threshold of OB ≥ 30%, DL ≥ 0.18, and Caco-2 permeability ≥ −0.4. Compounds listed in Huang et al., 2020 [13] that met the criteria of ADME properties were considered as bioactive compounds. The bioavailability details of some MO compounds that cannot be found in the TCMSP database were searched through online published literature using PubMed.
2.3. Target Genes Prediction of MO Bioactive Compounds and GEM
The target genes of MO bioactive compounds and GEM were retrieved from the three databases, which are Drugbank (https://go.drugbank.com/, (accessed on 17 December 2020)) [15], The Drug Gene Interaction Database, DgIdb (https://www.dgidb.org/, (accessed on 17 December 2020)) [16], and Comparative Toxicogenomics Database, CTD (http://ctdbase.org/, (accessed on 17 December 2020)) [17]. All the target genes were limited to Homo sapiens only, and duplicated target genes were removed from the list.
2.4. Prediction of Pancreatic Cancer Target Genes
The target genes related to pancreatic cancer were derived from two databases, which are Online Mendelian Inheritance in Man, OMIM (https://www.omim.org/, (accessed on 17 December 2020)) [18] and Malacards (http://www.malacards.org/, (accessed on 17 December 2020)) [19], with the keyword “pancreatic cancer”.
2.5. Construction of Venn Diagram
All the predicted target genes for MO, GEM, and pancreatic cancer were imported into the Bioinformatics and Evolutionary Genomics (http://bioinformatics.psb.ugent.be/webtools/Venn/, (accessed on 17 December 2020)) to identify the pancreatic cancer related target genes of MO and GEM. The Venn diagram constructed from the tool represents the intersection of potential target genes between drug and disease.
2.6. PPI Network Analysis
A protein–protein interaction (PPI) network was constructed via The Search Tool for the Retrieval of Interacting Genes, STRING database, version 11.0 (https://string-db.org/, (accessed on 18 December 2020)) [20] by inputting the target genes of MO and GEM against pancreatic cancer for a better understanding of the protein interactions. The PPI networks were set as “homo sapiens” organism, highest confidence (0.900) for the minimum required interaction score and excluded the disconnected node in the network. The PPI were imported into Cytoscape software, version 3.8.2 (https://cytoscape.org/ (accessed on 4 January 2021)) [21], an open-source software tool for visualizing and integrating complex interaction networks with any kind of attribute data [22] for the identification of hub genes. The hub genes were identified through Cytoscape plug-in cytoHubba by first selecting the top 10 genes under the, i.e., “Degree”, “Closeness”, and “Betweenness” parameters to calculate the topological features of each node in the network. The shared top 10 hub genes by the three parameters were determined using the Venn diagram tool.
2.7. GO and Pathway Enrichment Analysis
The Database for Annotation, Visualization, and Integrated Discovery, DAVID, version 6.8 (https://david.ncifcrf.gov/home.jsp, (accessed on 27 January 2021)) [23], an online software that offers comprehensive data for high-throughput gene functional analysis in the context of clarifying the biological characteristics, was used to perform the Gene Ontology (GO) enrichment analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis. The enrichment analysis was conducted to reveal the underlying mechanism of the combination between MO and GEM against pancreatic cancer through the biological process, cellular components, molecular functions, and key signaling pathways.
2.8. Network Construction
Compound-target-pathway network construction was established by using Cytoscape software, version 3.8.2 to better demonstrate the pharmacological mechanism of MO combined with GEM against pancreatic cancer. Every node in the network was analyzed and ranked according to their “degree” value, and nodes with higher than average node of degree were considered as hub compounds.
3. Results
3.1. Identification and Screening of MO Compound
According to the literature of Huang studies, a total of 32 compounds have been identified in MO (Supplementary Table S1). However, only seven compounds were found to be listed in the TCMSP database, which are catechin, epicatechin, quercetin, kaempferol, caffeic acid, p-coumaric acid, and ferulic acid. Four compounds, which are catechin, epicatechin, quercetin, and kaempferol, were selected as the bioactive compound that met the requirement of the drug screening for further analyses (Table 1).

Table 1.
The ADME properties of the bioactive compounds of Moringa oleifera found in Traditional Chinese Medicine Systems Pharmacology (TCMSP).
The screening of the remaining 25 compounds of MO were conducted using DgIdb, CTD, and Drugbank databases to search for the target genes. Target genes were found only for 3 compounds out of the 25, i.e., glucomoringin, glucoraphanin, and moringinine. The bioavailability of these compounds was determined through literature search since these compounds were not found in the TCMSP database. Glucomoringin and glucoraphanin were reported to have a good bioavailability, while the solubility of moringinine can be improved by the formation of benzylamine salts with benzoic acid derivatives [24,25,26,27,28,29,30]. These three compounds were also regarded as bioactive compounds, as they were reported to exhibit anticancer activity in several types of cancers [30,31,32]. The chemical structures of the selected bioactive compounds in MO are summarized in Table 2. Catechin, epicatechin, quercetin, and kaempferol are categorized in the flavonoids group. Each of the compounds have similar structures with different positions of functional groups. Glucomoringin and glucoraphanin are classified in the glucosinolates group, while moringinine is categorized in the benzylamine group.

Table 2.
Chemical structures of the selected bioactive compounds in MO.
3.2. Target Gene Prediction of MO, GEM, and Pancreatic Cancer
In MO, a total of 1092 potential pancreatic cancer target genes of the seven bioactive compounds were found in Drugbank, DgIdb, and CTD databases (Supplementary Table S2). There are 352 (Supplementary Table S3) and 421 target genes (Supplementary Table S4) for GEM and pancreatic cancer, respectively, that were retrieved from several databases.
3.3. Screening of Potential Pancreatic Cancer Related Target Genes of MO and GEM
The potential pancreatic cancer related target genes of MO and GEM were represented by the Venn diagram (Figure 1). The diagram revealed 81 MO intersection, 89 GEM intersection, and 137 MO + GEM intersection target genes against pancreatic cancer. The online databases identified 33 shared biotargets of MO and GEM against pancreatic cancer.
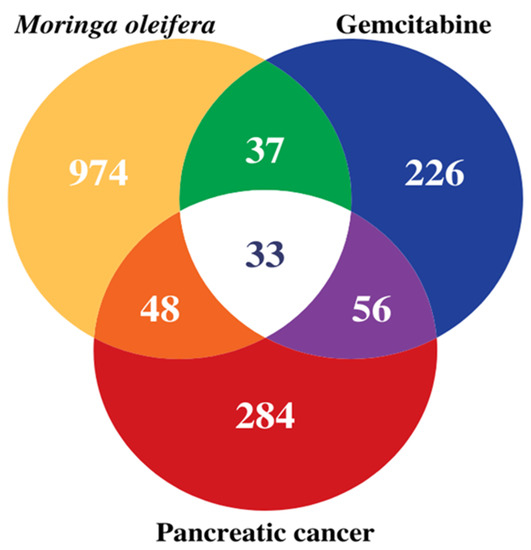
Figure 1.
The relation of target genes via Venn diagram between 7 compounds of MO and GEM against pancreatic cancer. There are 137 target genes in MO + GEM intersection (orange, white, and purple) against pancreatic cancer. Biotargets that are shared by MO and GEM against pancreatic cancer are 33 (white) in total. MO and GEM intersections have 81 (orange and white) and 89 (purple and white) target genes against pancreatic cancer, respectively.
3.4. PPI Network
In this study, we investigated four intersections. The PPI network of the MO + GEM intersection (Figure 2a), shared biotargets intersection (Figure 2b), MO intersection (Figure 2c), and GEM intersection (Figure 2d) targets against pancreatic cancer were constructed using STRING database. There were 136 nodes, with one disconnected node (MIR24-2), and 878 edges of MO + GEM intersection, 33 nodes and 122 edges of shared biotargets intersection, 81 nodes and 373 edges of MO intersection, and 88 nodes, with one disconnected node (MIR24-2), and 425 edges of GEM intersection targets against pancreatic cancer. The networks showed that these target genes have complex interactions between each other. Several potential pancreatic cancer related target genes of MO and GEM were also identified in the network and were considered as the hub genes based on the degree, closeness, and betweenness values. All the hub genes for each intersection are shown in Figure 3. TP53, AKT1, VEGFA, and CCND1 were identified as hub genes that overlapped between the four intersection networks. Shared biotargets intersection has two hub genes, CASP3 and BCL2L1, that were not targeted by GEM or MO alone, while STAT3 was the only hub gene that was untargeted by any other intersections except GEM intersection.
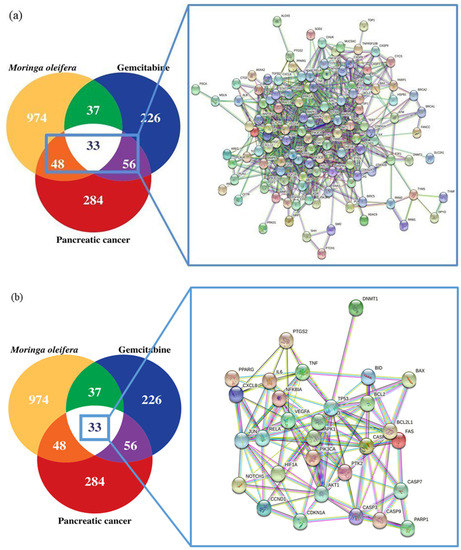
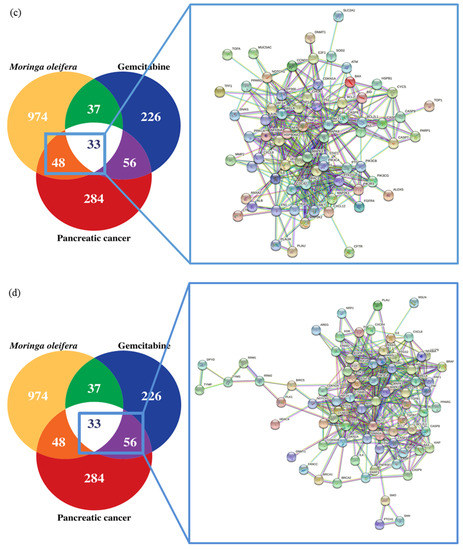
Figure 2.
Targets of MO, GEM and their combinations against pancreatic cancer using STRING database. (a) Venn diagram and PPI network of MO + GEM intersection target genes against pancreatic cancer. (b) Venn diagram and PPI network of shared biotargets intersection against pancreatic cancer. (c) Venn diagram and PPI network of MO intersection target genes against pancreatic cancer. (d) Venn diagram and PPI network of GEM intersection target genes against pancreatic cancer.
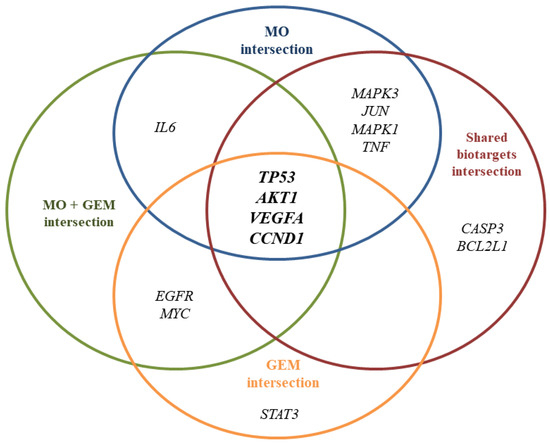
Figure 3.
Hub genes derived from the PPI network based on degree, closeness, and betweenness values. TP53, AKT1, VEGFA, and CCND1 are the hub genes that overlapped between the 4 intersection networks against pancreatic cancer.
3.5. GO and Pathway Enrichment Analysis
GO enrichment analysis was performed to further investigate the biological process, cellular components, and molecular function of the pancreatic cancer related target of MO and GEM using DAVID bioinformatics resources. The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis was also conducted to further reveal the pharmacological mechanism of the combination between MO and GEM against pancreatic cancer. Three intersection networks were investigated to determine the GO and pathway enrichment analysis, i.e., MO + GEM intersection, shared biotargets intersection, and MO intersection networks. The GO enrichment terms and KEGG pathway analysis are illustrated in Figure 4 and Figure 5. The results of GO analysis for the top 10 biological processes among the three intersection networks showed that the negative regulation of the apoptotic process was highly regulated compared to the other processes. In cellular component enrichment analysis, the top 10 significant enrichment terms were mainly involved in the cytosol and nucleus among the three intersection networks. The top 10 significant enrichment terms represented by molecular function showed that protein binding was highly regulated in MO + GEM intersection (Figure 4a), while identical protein binding was highly regulated in shared biotargets (Figure 4b) and MO intersections (Figure 4c). Besides, the KEGG pathway enrichment analysis of the three intersection networks was mainly associated with pathways in cancer and hepatitis B. Among the three networks, pancreatic cancer was one of the related pathways, where it was ranked as third and fourth in the MO + GEM intersection (Figure 5a) and MO intersection (Figure 5c), respectively. However, pancreatic cancer was found in the last rank of the shared biotargets intersection (Figure 5b).
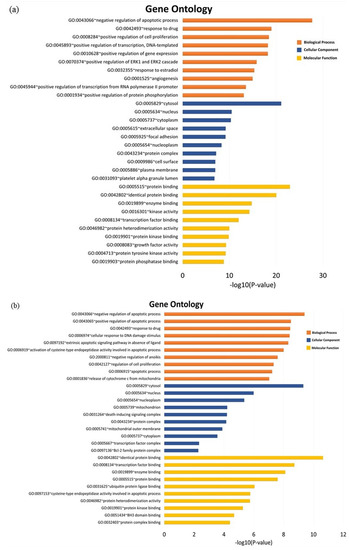
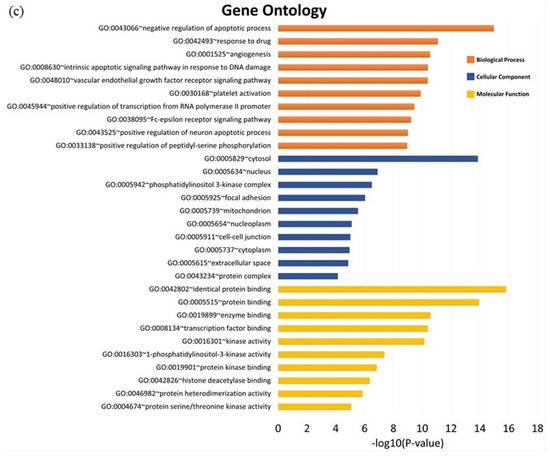
Figure 4.
GO enrichment terms analysis of MO, and its combination with GEM against pancreatic cancer. (a) The bar chart represents the top 10 significant enrichment terms represented by the biological process, cellular component, and molecular function of MO + GEM intersection target genes against pancreatic cancer. (b) The bar chart represents the top 10 significant enrichment terms represented by the biological process, cellular component, and molecular function of shared biotargets intersection against pancreatic cancer. (c) The bar chart represents the top 10 significant enrichment terms represented by the biological process, cellular component, and molecular function of MO intersection target genes against pancreatic cancer.
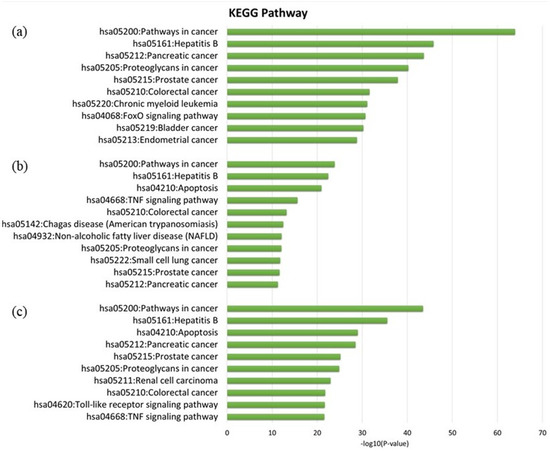
Figure 5.
The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of MO, and its combination with GEM against pancreatic cancer. (a) The bar chart represents the top 10 KEGG pathway analysis of MO + GEM intersection target genes against pancreatic cancer. (b) The bar chart represents the top 11 KEGG pathway analysis of shared biotargets intersection against pancreatic cancer. (c) The bar chart represents the top 10 KEGG pathway analysis of MO intersection target genes against pancreatic cancer.
3.6. Compound-Target-Pathway Network Constructions
The compound-target-pathway network (Figure 6) was constructed based on the significantly enriched pathways by using Cytoscape software to further understand the pharmacological mechanism of MO and GEM against pancreatic cancer. Compound-target network analysis identified catechin, kaempferol, and quercetin as hub compounds in the network of MO + GEM intersection target genes against pancreatic cancer, since the degree value of these compounds is greater than the average degree of node, 14.910. In the network of shared biotargets intersection against pancreatic cancer with the average degree of node, 22.737, only catechin was recognized as a hub compound, while two hub compounds, which are catechin and kaempferol, were identified in the network of MO intersection target genes against pancreatic cancer with a degree value greater than the average degree of node, 30.909 (Table 3).
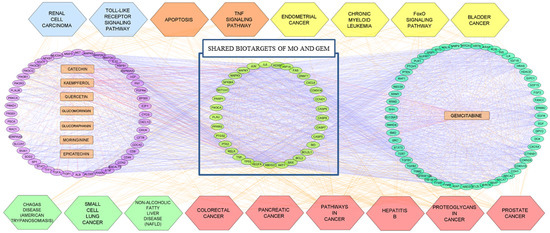
Figure 6.
The compound-target-pathway network of MO and GEM, as well as the shared biotargets intersection against pancreatic cancer, by using Cytoscape. The network contains 162 nodes (8 compounds, 17 pathways, and 137 target genes) and 3227 edges. The left and right compound-target network are the MO and GEM intersections target genes, respectively, while the center compound-target network is their combination against pancreatic cancer. The light blue hexagons represent the pathways involved in MO intersection targets, the orange hexagons represent the pathways involved in the MO and shared biotargets intersections, the yellow hexagons represent the pathways involved in the GEM intersection targets, the green hexagons represent the pathway involved in shared biotargets intersection, and the pink hexagons represent the pathway involved in three intersection networks. The oval nodes represent the target genes, the rectangle nodes represent the compounds, the hexagons represent the signaling pathway, while the edges represent the interaction between them.

Table 3.
Hub compounds in compound-target network based on the degree values.
4. Discussion
As the seventh most common cause of cancer death, most pancreatic cancer patients are less likely to survive for more than 5 years, since the relative survival rate at 5 years for pancreatic cancer is less than 10% across the globe. Traditionally, the conventional approach in drug discovery is considered as “one drug, one target, one disease” theory. In contrast, network pharmacology study has been known as “multi-drug, multi-target, multi-diseases” to explore the correlation of drugs and diseases [33]. In this present study, a network pharmacology approach was proposed to identify the potential bioactive compounds and theoretically evaluate the effect of the combination therapy of MO and GEM against pancreatic cancer.
The progression of pancreatic cancer is often associated with the mutation of oncogene and tumor suppressor genes, such as KRAS and TP53, respectively. Currently, there is no effective treatment that has been clinically approved which directly targets the mutation of KRAS oncogene in pancreatic cancer [34]. Despite the significant efforts in the research and development of the drug, gemcitabine (GEM), which is the gold-standard first-line chemotherapy against locally advanced and metastatic pancreatic cancer since 1997, has poor efficacy due to the rapid acquired resistance [35]. It has been reported in a previous study that pancreatic cancer patients with KRAS mutation had a poor response and shorter survival rates compared to those with wild-type KRAS after being treated with gemcitabine [34]. Besides, mutant TP53 has been demonstrated to induce gemcitabine resistance towards pancreatic cancer cells. Gemcitabine stabilized the mutant of p53 protein in the nucleus and caused chemoresistance, which occurred concurrently with the mutant-p53-dependent expression of CDK1 and CCNB1 genes that led to the hyperproliferation of pancreatic cancer cells [36]. Hence, this present in silico study combined GEM with MO that might help the future researchers in developing an effective treatment against pancreatic cancer.
This study revealed four potential bioactive compounds in MO, including catechin, epicatechin, kaempferol, and quercetin, that met the requirement of ADME screening. Glucomoringin, glucoraphanin, and moringinine were included in this study, as they have been found to exhibit the anticancer effect in various kinds of cancer [30,31,32]. However, only three compounds of MO, i.e., catechin, kaempferol, and quercetin, were considered as the hub compounds in MO + GEM intersection target genes against pancreatic cancer. Catechin and kaempferol were also identified to be the hub compounds in MO intersection targets against pancreatic cancer. Kaempferol was found to play a role in the regulation of various cancer cell activities, which include inflammation, apoptosis, and cell cycle [37]. Quercetin had been reported to stimulate antitumor activities through the induction of apoptosis, cell cycle, and autophagy [38,39,40,41]. Since catechin was the only compound that has been recognized as a hub compound in the shared biotargets intersection against pancreatic cancer, this compound plays a vital role against pancreatic cancer. Catechin is one of the flavonoid derivatives, which is prominently found in green tea, that possesses health-promoting properties, including anticancer and antioxidant properties [42,43,44]. It has been demonstrated that catechin, which is also known as cyclooxygenase-1 (COX-1) inhibitor, had exhibited a synergistic and additive antiproliferative effect on breast, bladder, and prostate cancer in vitro with the combination of COX-2 inhibitor [45,46]. Besides, catechin with the combination of IP6 also gives a reduction in cellular proliferation, as well as inducing the apoptosis effect in the pancreatic cancer cells, MiaPaca and Panc-1, significantly [47].
Several hub genes were identified based on the PPI network and considered to play a crucial role within the network against pancreatic cancer [8]. In this study, it is suggested that TP53, AKT1, VEGFA, and CCND1 may be the key targets for anticancer activity of MO and GEM combination, which might have major biological importance in treating pancreatic cancer. TP53 has been known to be mutated in 70% of cases in pancreatic cancer, causing the loss of DNA-binding ability, which activates the transcription of genes [48]. The progression of malignant pancreatic cancer was found to be associated with the sustained expression of mutant TP53 [49]. From the previous study, R273H mutation of P53 in MiaPaca-2 pancreatic cancer cell lines have been found to play roles in increasing colony formation and proliferation, and resistance to drug [49]. A study on MO demonstrated that the hot water leaf extract of the plant revealed the antiproliferative effect in the lung cancer cell, A549, by the activation of P53, caspases, and PARP1 cleavage, which led to the apoptosis of the cancer cell [50]. Besides, TP53 plays a role as a diagnostic marker in pancreatic cancer patients treated with GEM chemotherapy [51]. Previously, study has reported that the chemoresistance of GEM can be overcome by the combination of P53-reactivating molecules, RITA, and CP-31398, which reduced the growth rate and caused apoptosis to the pancreatic cancer cell line [36]. AKT1 is the main protein in the signaling pathway of PI3K/Akt and in the various types of cancer. The abundance of AKT1 activity has been shown through the transmission of strong antiapoptotic signals [52]. It has been demonstrated that activation of AKT was prevented by the AKT inhibitor, together with GEM, which then enhanced the cytotoxic activity of GEM [53]. In pancreatic cancer, VEGFA is the most common and prevalent angiogenic factor among other family members. Since it can promote angiogenesis and greatly increases cancer cell motility, VEGFA is responsible for inducing pancreatic cancer cell invasion and migration [54,55]. The suppression of VEGFA expression by GEM will cause the inhibition of tumor angiogenesis in the cancer cells [56]. Researchers recently are also focusing to target the pancreatic stroma, as the tumor microenvironment of pancreatic cancer plays an important role in its pathogenesis. A previous study has reported an aberrant gene expression profile in the cancer-associated stroma, which is associated with high expression of VEGFA, COX-2, and collagen I, as well as the alteration of integrin expression. These could result in the abnormal interaction of epithelial–mesenchymal that enhances the proliferation and invasiveness of cancer cells [57]. Hence, it is suggested that the MO and GEM combination may help in the inhibition of the aberrant signal generated by pancreatic cancer associated stromal cells and provides therapeutic options. Cyclin D1, encoded by CCND1 gene, is a key regulator protein in the G1–S phase transformation, which is also responsible for the regulation of cell growth and differentiation. Cyclin D1 deregulation can result in tumor formation caused by genetic instability in vivo and in vitro [58]. A study has shown that GEM causes CCND1 suppression in the human breast cancer cell line, which is due to the cell cycle arrest in S phase [59]. Moreover, cancer cells that have been treated with the leaf extract of MO were shown to have a significant reduction in cyclin D1 levels in a dose-dependent manner [60]. Therefore, it is assumed that downregulation of mutant TP53, AKT1, VEGFA, and CCND1 genes might reduce the ability of pancreatic cancer cells to survive.
Moreover, this study revealed the novel targets in the combination of MO and GEM may represent the new target proteins against pancreatic cancer, including CASP3 and BCL2L1, which were not targeted by MO or GEM alone. CASP3 is known as a proapoptotic gene that is involved in multiple apoptosis processes [61]. A previous study has shown that doxorubicin induced apoptosis in breast cancer cell lines by upregulating the expression of caspase-3, caspase-8, and BAX [62]. In contrast, BCL-xL, which is encoded by the BCL2L1 gene, is an important antiapoptotic protein that aids in the survival of pancreatic cancer cell differentiation [63]. Venetoclax, the anticancer drug against acute myeloid leukemia and chronic lymphocytic leukemia, has been reported to inhibit the binding of BCL2 to BAK or BAX, which trigger the apoptosis of the cancer cells [64]. Hence, these genes could be an ideal target of MO and GEM combination in controlling pancreatic cancer growth.
Furthermore, the possible mechanism of action and signaling pathway against pancreatic cancer was further evaluated through enrichment analysis. The result of the GO enrichment analysis demonstrated that the most enriched biological process of the target genes of MO, as well as its combination with GEM, involves the regulation of cell proliferation and apoptosis. It has been reported that the positive and negative regulation of apoptotic processes were simultaneously activated through noxious and protective signals via various pathways [65]. Thus, it is pertinent to finely balance the regulation of the two signals to increase the effectiveness of the combination therapy between MO and GEM. Besides, the positive regulation of cell proliferation might lead to the abnormal growth of the cancer cell. Therefore, targeting the genes involved in the regulation of cell proliferation might improve the treatment of pancreatic cancer. From the KEGG pathway enrichment analysis of MO, as well as its combination with GEM, the results showed that the enrichment pathways were mainly related to pancreatic cancer, prostate cancer, colorectal cancer, bladder cancer, endometrial cancer, and chronic myeloid leukemia. There were also involvement of other pathways in this study, such as hepatitis B, apoptosis, FoxO signaling pathway, Ras signaling pathway, VEGF signaling pathway, and toll-like receptor signaling pathway. On the other hand, when MO and GEM were combined, other diseases were also being targeted, such as chagas disease (American trypanosomiasis) and nonalcoholic fatty liver disease (NAFLD). These findings revealed that “multi-pathway” theory has been applied.
5. Conclusions
In conclusion, this is the first network pharmacology study reported to predict the pancreatic cancer target genes of the bioactive compounds of MO with the combination of GEM and theoretically evaluate their effect in pancreatic cancer development and progression. Multiple targets, including TP53, AKT1, VEGFA, and CCND1, were found to be functionally enhanced in multiple pathways associated with pancreatic cancer development. Furthermore, CASP3 and BCL2L1 that were not targeted by MO or GEM were found in the shared targets of MO and GEM, which may represent a new target protein against pancreatic cancer. Hence, the effect of GEM could be enhanced when it is combined with MO in treating pancreatic cancer. These findings provide new insights into the antipancreatic cancer effects of the MO and GEM combination therapy and pave the way for further in vitro and in vivo experimental validations.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/pr9101742/s1, Table S1: The bioactive compounds in Moringa oleifera, Table S2: Target genes prediction of Moringa oleifera, Table S3: Target genes prediction of gemcitabine, Table S4: Target genes prediction of pancreatic cancer.
Author Contributions
Conceptualization, K.T., S.W. and Z.S.; methodology, N.A.S., N.A.R., K.T., S.W. and Z.S.; validation, K.T., S.W. and Z.S.; formal analysis, K.T., S.W., Z.S., N.A.S. and N.A.R.; investigation, N.A.S., N.A.R. and Z.S.; resources, N.A.S., N.A.R., K.T. and Z.S.; data curation, N.A.S., N.A.R., K.T., S.W. and Z.S.; writing—original draft preparation, N.A.S.; writing—review and editing, K.T., S.W. and Z.S.; visualization, K.T., S.W. and Z.S.; supervision, K.T., S.W. and Z.S. All authors have read and agreed to the published version of the manuscript.
Funding
The publication fee was supported by Universiti Putra Malaysia.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- World Health Organization. Available online: https://www.who.int/ (accessed on 22 May 2021).
- World Health Organization; International Agency for Research on Cancer. Global Cancer Observatory—GLOBOCAN. Available online: https://gco.iarc.fr/ (accessed on 16 March 2021).
- Burris, H.; Moore, M.J.; Andersen, J.; Green, M.R.; Rothenberg, M.L.; Modiano, M.R.; Cripps, M.C.; Portenoy, R.K.; Storniolo, A.M.; Tarassoff, P.; et al. Improvements in survival and clinical benefit with gemcitabine as first-line therapy for patients with advanced pancreas cancer: A randomized trial. J. Clin. Oncol. 1997, 15, 2403–2413. [Google Scholar] [CrossRef]
- Amrutkar, M.; Gladhaug, I.P. Pancreatic cancer chemoresistance to gemcitabine. Cancers 2017, 9, 157. [Google Scholar] [CrossRef]
- Hagoel, L.; Vexler, A.; Kalich-Philosoph, L.; Earon, G.; Ron, I.; Shtabsky, A.; Marmor, S.; Lev-Ari, S. Combined effect of Moringa oleifera and ionizing radiation on survival and metastatic activity of pancreatic cancer cells. Integr. Cancer Ther. 2019, 18, 1–11. [Google Scholar] [CrossRef]
- Vergara-Jimenez, M.; Almatrafi, M.; Fernandez, M. Bioactive components in Moringa Oleifera leaves protect against chronic disease. Antioxidants 2017, 6, 91. [Google Scholar] [CrossRef]
- Alhakmani, F.; Kumar, S.; Khan, S.A. Estimation of total phenolic content, in-vitro antioxidant and anti-inflammatory activity of flowers of Moringa oleifera. Asian Pac. J. Trop. Biomed. 2013, 3, 623–627. [Google Scholar] [CrossRef]
- Wu, W.; Zhang, Z.; Li, F.; Deng, Y.; Lei, M.; Long, H.; Hou, J.; Wu, W. A network-based approach to explore the mechanisms of Uncaria Alkaloids in treating hypertension and alleviating Alzheimer’s disease. Int. J. Mol. Sci. 2020, 21, 1766. [Google Scholar] [CrossRef]
- Leung, E.L.; Cao, Z.W.; Jiang, Z.H.; Zhou, H.; Liu, L. Network-based drug discovery by integrating systems biology and computational technologies. Brief. Bioinform. 2013, 14, 491–505. [Google Scholar] [CrossRef]
- Ru, J.; Li, P.; Wang, J.; Zhou, W.; Li, B.; Huang, C.; Li, P.; Guo, Z.; Tao, W.; Yang, Y.; et al. TCMSP: A database of systems pharmacology for drug discovery from herbal medicines. J. Cheminform. 2014, 6, 13. [Google Scholar] [CrossRef]
- Xue, R.; Fang, Z.; Zhang, M.; Yi, Z.; Wen, C.; Shi, T. TCMID: Traditional Chinese medicine integrative database for herb molecular mechanism analysis. Nucleic Acids Res. 2013, 41, D1089–D1095. [Google Scholar] [CrossRef]
- White, J. PubMed 2.0. Med. Ref. Serv. Q. 2020, 39, 382–387. [Google Scholar] [CrossRef]
- Huang, Q.; Liu, R.; Liu, J.; Huang, Q.; Liu, S.; Jiang, Y. Integrated network pharmacology analysis and experimental validation to reveal the mechanism of anti-insulin resistance effects of moringa oleifera seeds. Drug Des. Dev. Ther. 2020, 14, 4069–4084. [Google Scholar] [CrossRef]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem 2019 update: Improved access to chemical data. Nucleic Acids Res. 2019, 47, D1102–D1109. [Google Scholar] [CrossRef]
- Wishart, D.S.; Feunang, Y.D.; Guo, A.C.; Lo, E.J.; Marcu, A.; Grant, J.R.; Sajed, T.; Johnson, D.; Li, C.; Sayeeda, Z.; et al. DrugBank 5.0: A major update to the DrugBank database for 2018. Nucleic Acids Res. 2018, 46, D1074–D1082. [Google Scholar] [CrossRef]
- Freshour, S.L.; Kiwala, S.; Cotto, K.C.; Coffman, A.C.; McMichael, J.F.; Song, J.J.; Griffith, M.; Griffith, O.L.; Wagner, A.H. Integration of the drug-gene interaction database (DGIdb 4.0) with open crowdsource efforts. Nucleic Acids Res. 2021, 49, D1144–D1151. [Google Scholar] [CrossRef]
- Davis, A.P.; Grondin, C.J.; Johnson, R.J.; Sciaky, D.; Wiegers, J.; Wiegers, T.C.; Mattingly, C.J. Comparative Toxicogenomics database (CTD): Update 2021. Nucleic Acids Res. 2021, 49, D1138–D1143. [Google Scholar] [CrossRef]
- Amberger, J.S.; Bocchini, C.A.; Schiettecatte, F.; Scott, A.F.; Hamosh, A. OMIM.org: Online Mendelian inheritance in man (OMIM®), an online catalog of human genes and genetic disorders. Nucleic Acids Res. 2015, 43, D789–D798. [Google Scholar] [CrossRef]
- Rappaport, N.; Nativ, N.; Stelzer, G.; Twik, M.; Guan-Golan, Y.; Stein, T.I.; Bahir, I.; Belinky, F.; Morrey, C.P.; Safran, M.; et al. MalaCards: An integrated compendium for diseases and their annotation. Database J. Biol. Databases Curation 2013, 2013, bat018. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Wan, Y.; Xu, L.; Liu, Z.; Yang, M.; Jiang, X.; Zhang, Q.; Huang, J. Utilising network pharmacology to explore the underlying mechanism of Wumei Pill in treating pancreatic neoplasms. BMC Complement. Altern. Med. 2019, 19, 158. [Google Scholar] [CrossRef]
- Dennis, G.; Sherman, B.T.; Hosack, D.A.; Yang, J.; Gao, W.; Lane, H.C.; Lempicki, R.A. DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol. 2003, 4, R60. [Google Scholar] [CrossRef]
- Fahey, J.W.; Wade, K.L.; Stephenson, K.K.; Shi, Y.; Liu, H.; Panjwani, A.A.; Warrick, C.R.; Olson, M.E. A strategy to deliver precise oral doses of the Glucosinolates or Isothiocyanates from Moringa oleifera leaves for use in clinical studies. Nutrients 2019, 11, 1547. [Google Scholar] [CrossRef]
- Nguyen, V.; Stewart, J.; Lopez, M.; Ioannou, I.; Allais, F. Glucosinolates: Natural occurrence, biosynthesis, accessibility, isolation, structures, and biological activities. Molecules 2020, 25, 4537. [Google Scholar] [CrossRef]
- Mathiron, D.; Iori, R.; Pilard, S.; Soundara Rajan, T.; Landy, D.; Mazzon, E.; Rollin, P.; Djedaïni-Pilard, F. a combined approach of NMR and mass spectrometry techniques applied to the α-Cyclodextrin/Moringin complex for a novel bioactive formulation. Molecules 2018, 23, 1714. [Google Scholar] [CrossRef]
- Michl, C.; Vivarelli, F.; Weigl, J.; De Nicola, G.R.; Canistro, D.; Paolini, M.; Iori, R.; Rascle, A. The chemopreventive phytochemical Moringin isolated from Moringa oleifera seeds inhibits JAK/STAT signaling. PLoS ONE 2016, 11, e0157430. [Google Scholar] [CrossRef]
- Parshad, H.; Frydenvang, K.; Liljefors, T.; Larsen, C.S. Correlation of aqueous solubility of salts of benzylamine with experimentally and theoretically derived parameters. A multivariate data analysis approach. Int. J. Pharm. 2002, 237, 193–207. [Google Scholar] [CrossRef]
- Parshad, H.; Frydenvang, K.; Liljefors, T.; Sorensen, H.O.; Larsen, C. Aqueous solubility study of salts of benzylamine derivatives and p-substituted benzoic acid derivatives using X-ray crystallographic analysis. Int. J. Pharm. 2004, 269, 157–168. [Google Scholar] [CrossRef]
- Fahmy, T.A.; Nahla, S.H.; Rehab, R.A. Hepatoprotective and antiproliferative activity of moringinine, chlorogenic acid and quercetin. Int. J. Res. Med. Sci. 2016, 4, 1147–1153. [Google Scholar] [CrossRef]
- Rajan, T.S.; De Nicola, G.R.; Iori, R.; Rollin, P.; Bramanti, P.; Mazzon, E. Anticancer activity of glucomoringin isothiocyanate in human malignant astrocytoma cells. Fitoterapia 2016, 110, 1–7. [Google Scholar] [CrossRef]
- Almuhayawi, M.S.; AbdElgawad, H.; Al Jaouni, S.K.; Selim, S.; Hassan, A.; Khamis, G. Elevated CO2 improves glucosinolate metabolism and stimulates anticancer and anti-inflammatory properties of broccoli sprouts. Food Chem. 2020, 328, 127102. [Google Scholar] [CrossRef]
- Sakle, N.S.; More, S.A.; Mokale, S.N. A network pharmacology-based approach to explore potential targets of Caesalpinia pulcherima: An updated prototype in drug discovery. Sci. Rep. 2020, 10, 17217. [Google Scholar] [CrossRef]
- Ryu, W.J.; Han, G.; Lee, S.H.; Choi, K.Y. Suppression of Wnt/β-catenin and RAS/ERK pathways provides a therapeutic strategy for gemcitabine-resistant pancreatic cancer. Biochem. Biophys. Res. Commun. 2021, 549, 40–46. [Google Scholar] [CrossRef]
- Kang, Y.W.; Lee, J.E.; Jung, K.H.; Son, M.K.; Shin, S.M.; Kim, S.J.; Fang, Z.; Yan, H.H.; Park, J.H.; Han, B.; et al. KRAS targeting antibody synergizes anti-cancer activity of gemcitabine against pancreatic cancer. Cancer Lett. 2018, 438, 174–186. [Google Scholar] [CrossRef]
- Fiorini, C.; Cordani, M.; Padroni, C.; Blandino, G.; Di Agostino, S.; Donadelli, M. Mutant p53 stimulates chemoresistance of pancreatic adenocarcinoma cells to gemcitabine. Biochim. Biophys. Acta 2015, 1853, 89–100. [Google Scholar] [CrossRef]
- Lee, J.; Kim, J.H. Kaempferol inhibits pancreatic cancer cell growth and migration through the blockade of EGFR-related pathway in vitro. PLoS ONE 2016, 11, e0155264. [Google Scholar] [CrossRef]
- Vidya, P.R.; Senthil, M.R.; Maitreyi, S.; Ramalingam, K.; Karunagaran, D.; Nagini, S. The flavonoid quercetin induces cell cycle arrest and mitochondria-mediated apoptosis in human cervical cancer (HeLa) cells through p53 induction and NF-κB inhibition. Eur. J. Pharmacol. 2010, 649, 84–91. [Google Scholar] [CrossRef]
- Psahoulia, F.H.; Moumtzi, S.; Roberts, M.L.; Sasazuki, T.; Shirasawa, S.; Pintzas, A. Quercetin mediates preferential degradation of oncogenic Ras and causes autophagy in Ha-RAS-transformed human colon cells. Carcinogenesis 2007, 28, 1021–1031. [Google Scholar] [CrossRef]
- Mouria, M.; Gukovskaya, A.S.; Jung, Y.; Buechler, P.; Hines, O.J.; Reber, H.A.; Pandol, S.J. Food-derived polyphenols inhibit pancreatic cancer growth through mitochondrial cytochrome C release and apoptosis. Int. J. Cancer 2002, 98, 761–769. [Google Scholar] [CrossRef]
- Senthilkumar, K.; Arunkumar, R.; Elumalai, P.; Sharmila, G.; Gunadharini, D.N.; Banudevi, S.; Krishnamoorthy, G.; Benson, C.S.; Arunakaran, J. Quercetin inhibits invasion, migration and signalling molecules involved in cell survival and proliferation of prostate cancer cell line (PC-3). Cell Biochem. Funct. 2011, 29, 87–95. [Google Scholar] [CrossRef]
- Musial, C.; Kuban-Jankowska, A.; Gorska-Ponikowska, M. Beneficial properties of green tea catechins. Int. J. Mol. Sci. 2020, 21, 1744. [Google Scholar] [CrossRef]
- Bae, J.; Kim, N.; Shin, Y.; Kim, S.; Kim, Y. Activity of catechins and their applications. Biomed. Dermatol. 2020, 4, 8. [Google Scholar] [CrossRef]
- Sun, H.; Yin, M.; Hao, D.; Shen, Y. Anti-cancer activity of catechin against A549 lung carcinoma cells by induction of cyclin kinase inhibitor p21 and suppression of Cyclin E1 and P–AKT. Appl. Sci. 2020, 10, 2065. [Google Scholar] [CrossRef]
- Farivar-Mohseni, H.; Kandzari, S.J.; Zaslau, S.; Riggs, D.R.; Jackson, B.J.; McFadden, D.W. Synergistic effects of Cox-1 and -2 inhibition on bladder and prostate cancer in vitro. Am. J. Surg. 2004, 188, 505–510. [Google Scholar] [CrossRef]
- McFadden, D.; Riggs, D.; Jackson, B.; Cunningham, C. Additive effects of Cox-1 and Cox-2 inhibition on breast cancer in vitro. Int. J. Oncol. 2006, 29, 1019–1023. [Google Scholar] [CrossRef] [PubMed]
- McMillan, B.; Riggs, D.R.; Jackson, B.J.; Cunningham, C.; McFadden, D.W. Dietary influence on pancreatic cancer growth by Catechin and Inositol Hexaphosphate. J. Surg. Res. 2007, 141, 115–119. [Google Scholar] [CrossRef]
- Cicenas, J.; Kvederaviciute, K.; Meskinyte, I.; Meskinyte-Kausiliene, E.; Skeberdyte, A.; Cicenas, J. KRAS, TP53, CDKN2A, SMAD4, BRCA1 and BRCA2 mutations in pancreatic cancer. Cancers 2017, 9, 42. [Google Scholar] [CrossRef] [PubMed]
- Jahedi, H.; Fahud, A.L.; Lim, C.L. Role of p53 family isoforms in enhancing aggressiveness and chemoresistance in pancreatic cancer—Review. World Acad. Sci. J. 2019, 1, 236–246. [Google Scholar] [CrossRef]
- Tiloke, C.; Anand, K.; Gengan, R.M.; Chuturgoon, A.A. Moringa oleifera and their phytonanoparticles: Potential antiproliferative agents against cancer. Biomed. Pharmacother. 2018, 108, 457–466. [Google Scholar] [CrossRef]
- Sinn, M.; Sinn, B.V.; Treue, D.; Keilholz, U.; Damm, F.; Schmuck, R.B.; Lohneis, P.; Klauschen, F.; Striefler, J.K.; Bahra, M.; et al. TP53 mutations predict sensitivity to adjuvant gemcitabine in patients with pancreatic ductal adenocarcinoma: Next-generation sequencing results from the CONKO-001 trial. Clin. Cancer Res. 2020, 26, 3732–3739. [Google Scholar] [CrossRef]
- Parsons, C.M.; Muilenburg, D.; Bowles, T.L.; Virudachalam, S.; Bold, R.J. The role of Akt activation in the response to chemotherapy in pancreatic cancer. Anticancer. Res. 2010, 30, 3279–3289. [Google Scholar] [PubMed]
- Wang, Z.; Luo, G.; Qiu, Z. Akt inhibitor MK-2206 reduces pancreatic cancer cell viability and increases the efficacy of gemcitabine. Spandidos Publ. 2020, 19, 1999–2004. [Google Scholar] [CrossRef] [PubMed]
- Costache, M.I.; Ioana, M.; Iordache, S.; Ene, D.; Costache, C.A.; Săftoiu, A. VEGF expression in pancreatic cancer and other malignancies: A review of the literature. Rom. J. Intern. Med. 2015, 53, 199–208. [Google Scholar] [CrossRef] [PubMed]
- Doi, Y.; Yashiro, M.; Yamada, N.; Amano, R.; Noda, S.; Hirakawa, K. VEGF-A/VEGFR-2 signaling plays an important role for the motility of pancreas cancer cells. Ann. Surg. Oncol. 2012, 19, 2733–2743. [Google Scholar] [CrossRef]
- Ikeda, R.; Vermeulen, L.C.; Jiang, Z.; Lau, E.; Kolesar, J.M. Gemcitabine and paclitaxel suppress the production of vascular endothelial growth factor induced by deferoxamine in human non-small cell lung cancer A549 cells. Exp. Ther. Med. 2010, 1, 853–857. [Google Scholar] [CrossRef]
- Korc, M. Pancreatic cancer associated stroma production. Am. J. Surg. 2007, 194, S84–S86. [Google Scholar] [CrossRef]
- Bachmann, K.; Neumann, A.; Hinsch, A.; Nentwich, M.F.; El Gammal, A.T.; Vashist, Y.; Perez, D.; Bockhorn, M.; Izbicki, J.R.; Mann, O. Cyclin D1 is a strong prognostic factor for survival in pancreatic cancer: Analysis of CD G870A polymorphism, FISH and immunohistochemistry. J. Surg. Oncol. 2015, 111, 316–323. [Google Scholar] [CrossRef]
- Hernández-Vargas, H.; Rodríguez-Pinilla, S.M.; Julián-Tendero, M.; Sánchez-Rovira, P.; Cuevas, C.; Antón, A.; Rios, M.J.; Palavios, J.; Moreno-Bueno, G. Gene expression profiling of breast cancer cells in response to gemcitabine: NF-κB pathway activation as a potential mechanism of resistance. Breast Cancer Res. Treat. 2007, 102, 157–172. [Google Scholar] [CrossRef]
- Kou, X.; Li, B.; Olayanju, J.; Drake, J.; Chen, N. Nutraceutical or pharmacological potential of Moringa oleifera Lam. Nutrients 2018, 10, 343. [Google Scholar] [CrossRef]
- Radziszewska, A.; Schroer, S.A.; Choi, D.; Tajmir, P.; Radulovich, N.; Ho, J.C.; Wang, L.; Liadis, N.; Hakem, R.; Tsao, M.S.; et al. Absence of caspase-3 protects pancreatic {beta}-cells from c-Myc-induced apoptosis without leading to tumor formation. J. Biol. Chem. 2009, 284, 10947–10956. [Google Scholar] [CrossRef]
- Pilco-Ferreto, N.; Calaf, G.M. Influence of doxorubicin on apoptosis and oxidative stress in breast cancer cell lines. Int. J. Oncol. 2016, 49, 753–762. [Google Scholar] [CrossRef] [PubMed]
- Loo, L.; Soetedjo, A.; Lau, H.H.; Ng, N.; Ghosh, S.; Nguyen, L.; Krishnan, V.G.; Choi, H.; Roca, X.; Hoon, S.; et al. BCL-xL/BCL2L1 is a critical anti-apoptotic protein that promotes the survival of differentiating pancreatic cells from human pluripotent stem cells. Cell Death Dis. 2020, 11, 378. [Google Scholar] [CrossRef]
- Roberts, A.W. Therapeutic development and current uses of BCL-2 inhibition. Hematology 2020, 2020, 1–9. [Google Scholar] [CrossRef]
- Solary, E.; Droin, N.; Bettaieb, A.; Corcos, L.; Dimanche-Boitrel, M.T.; Garrido, C. Positive and negative regulation of apoptotic pathways by cytotoxic agents in hematological malignancies. Leukemia 2000, 14, 1833–1849. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).