Abstract
According to reports published, the aberrant expression of microRNAs (miRNAs), a class of 19–25 nucleotide-long small non-coding RNAs, is responsible for human cancers, including nasopharyngeal cancer (NPC). The dysregulation of miRNAs that act either as a tumor suppressor or oncogene, leading to a wide range of NPC pathogenesis pathways, includes the proliferation, invasion, migration as well as the metastasis of NPC cells. This article reviews and highlights recent advances in the studies of miRNAs in NPC, with a specific demonstration of the functions of miRNA, especially circulating miRNAs, in the pathway of NPC pathogenesis. Additionally, the possible use of miRNAs as early screening and prognostic biomarkers and for therapeutic molecular monitoring has been extensively studied.
1. Introduction
In 1993, Victor Ambros and colleagues discovered the first microRNA (miRNA), namely lin-4, in Caenorhabditis elegans. Up to date, with in-depth research on microRNA (miRNA), a new era of early diagnosis, prognosis of human cancers as well as cancer treatment has commenced [1,2]. miRNAs are the family of naturally occurring small non-coding RNAs within 19–25 nucleotides (nts) in length. They play multiple key roles in regulating a wide array of biological processes, including cell proliferation, differentiation, metabolism, stress response and apoptosis through the regulation of numerous target genes [3,4,5]. miRNAs regulate biological processes by binding to sequences in the 3′-untranslated region (3′-UTR) of their target mRNAs, eventually resulting in the degradation and/or repression of mRNA [3,4,5]. Additionally, miRNAs have been observed to interact with other regions of target genes, including 5′-UTR, as well as promoter and coding regions. Each miRNA is reported to be the mediator of multiple genes by acting at the post-transcriptional levels. Consequently, both the mutation of miRNAs and dysfunction of miRNA biogenesis may lead to various human diseases, including cancers [6]. Growing evidences have indicated that miRNAs are frequently dysregulated in various cancers. Therefore, understanding their profile in cancer may identify them as powerful post-transcriptional biomarkers as well as highlight their great biomarker potential for the early screening, prognosis and therapeutic targets of human cancers.
Cancers are a serious disease to human life and health and have become the second leading cause of human death globally, accounting for an estimated 9.6 million deaths in 2018. It was emphasized that new cases of cancer could reach 23.6 million in 2030 [7]. Among them, nasopharyngeal carcinoma (NPC), a prevalent nasopharyngeal malignant tumor with remarkable differences in distribution and which gravitates toward Southern Asia, is considered to be the most common head and neck cancer. Even though improvements in nasopharyngeal cancer treatment have been achieved, diagnosis at an advanced stage reduces the success rate of treatment as well as the survival of patients. Thus, early screening and diagnosis represent beneficial opportunities to increase the survival of patients as well as the effects of nasopharyngeal cancer treatment. However, the non-specific symptoms related to the early stage of NPC, as well as the deeply seated location of the nasopharynx, are major obstacles to an early screening of NPC [8]. Therefore, effective biomarkers are truly needed [9]. At present, much effort has been made to identify early biomarkers by focusing on the etiological factors that lead to nasopharyngeal tumorigenesis. In addition to the etiology of Epstein–Barr virus infection, recently, the aberrant expression of miRNAs has been demonstrated to play key roles in the pathogenesis of NPC. Hence, an adequate understanding of the roles of miRNAs in NPC pathogenesis will provide more opportunities as well as better strategies to identify them as effective biomarkers of NPC. Additionally, it will pave the way for a better application of their functions in clinics, i.e., developing them into a promising approach in NPC therapies. In this article, we will summarize the key roles of miRNAs in the pathogenesis of NPC. Additionally, updated miRNA-related research data will be presented. Finally, we will demonstrate the application of miRNAs in NPC early screening, prognosis as well as cancer treatment.
2. Brief Understanding of miRNAs Biogenesis
Mature miRNAs are generated through a two-step processing of primary miRNAs in both the nuclear and cytoplasmic regions. miRNAs can be divided into intronic or intergenic miRNAs (Figure 1) [5,10,11]. Almost half of miRNA genes are located in the intergenic region (between two genes) and may exist either as a cluster of miRNA genes or a single miRNA gene under the control of its own promoter [5,11].
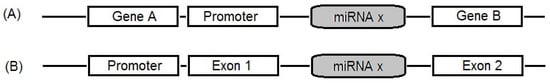
Figure 1.
Location of the microRNA (miRNAs) gene in the genome: (A) intergenic miRNAs: miRNAs located between two genes; (B) intronic miRNA: miRNAs located in the region of intron in a gene.
The biogenesis of miRNAs is classified into two categories: the canonical pathway and non-canonical pathway. In the canonical pathway, the primary miRNAs (pri-miRNAs) are transcribed from their genes and cleaved into pre-miRNAs by two main enzymes, Drosha and Dicer [5]. In this dominant pathway, the biogenesis of miRNAs, as well as the mechanism of regulation of their target genes’ expression, is summarized in the steps shown in Figure 2. The mature miRNA originates from a long primary miRNA (pri-miRNA) with the structure of 5′-7 methyl-guanosine capped and 3′ polyadenylated, and it is transcribed by RNA-polymerase II (RNA-polyII) from its own promoter or the promoter of the host genes in which it is contained; it is controlled by RNA-polyII-associated transcription factors and epigenetic regulators [12,13,14]. Pri-miRNA contains a hair-spin structure which varies from hundreds to thousands of base pairs in length [10,15]. Pri-miRNA is cleaved at the stem of the hair-spin structure, leading to the release of a ~60–70 nucleotide hair-spin structure, which is termed as the precursor miRNA (pre-miRNA). This process is catalyzed by the microprocessor, which contains the nuclear RNAse III-type protein Drosha and its cofactor, the DiGeorge syndrome critical region eight (DGCR8) protein found in humans, and takes place in the nucleus [16]. The pre-miRNA with a stem-loop structure is exported from the nucleus into the cytoplasm for further processing by exportin-5 (EXP 5), originally known as a minor export factor for tRNAs, and the Ras-related nuclear protein guanosine triphosphate (RAN-GTP) [11,15,17]. Then, the terminal loop of the pre-miRNA is removed by RNA III Dicer and its cofactor transactivation-responsive RNA-binding protein (TRBP), thereby releasing a ~20–22 nucleotide miRNA-duplex, which contains two 5′ phosphorylated sequence strands with 3′ overhangs, named the mature miRNA guide strand (miRNA) and complementary passenger strand (miRNA*) [17,18]. The miRNA-duplex is loaded into an Argonaute protein (Ago protein) to generate a RNA-induced silencing complex (RISC), and the miRNA passenger is degraded. The mature miRNA is incorporated into the miRNA-induced silencing complex (miRISC), which participates in regulating the gene expression via the interaction between a region called the “seed” (2–8 nt), a site located at the 5′ part of miRNA (positions 2–7) and the target mRNAs 3′-UTR by a Watson–Crick complementary sequence, leading to the degradation of the mRNA (in case of perfect pairing between the miRNAs/mRNA target) or the blocking of mRNA translation (in case of imperfect pairing (Figure 3)) [3,11,19]. Up to date, some miRNAs are generated by different non-canonical pathways, including: (1) miTrons (pri-miRNAs are encoded in the introns of coding genes); (2) Dicer-independent miRNAs; (3) tRNA-derived miRNAs [5]. These non-canonical pathways use many different combinations of proteins which are involved in another pathway—the canonical pathway, including Dicer, exportin-5 and AGO2. It is noted that the Dicer protein is necessary in both canonical and non-canonical pathways, while Drosha and DGCR8 are only functioned to canonical miRNAs. In other words, the non-canonical miRNAs could be generated in the case of a Drosha and DGCR8 absence [5].
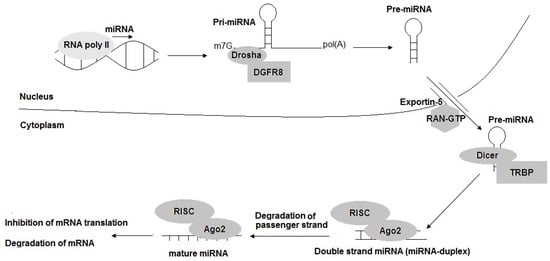
Figure 2.
The biogenesis of miRNAs: the canonical pathway. pri-miRNA: primary miRNA; pre-miRNA: precursor miRNA; DGCR8: DiGeorge syndrome critical region 8; RAN-GTP: Ras-related nuclear protein guanosine triphosphate; TRBP: transactivation-responsive RNA-binding protein; Ago: Argonaute protein; RISC: RNA-induced silencing complex; miRISC: miRNA-induced silencing complex.

Figure 3.
The interaction types of miRNA–mRNA (target mRNA). Site matching between the seed region of miRNA (at positions 2–8) and the 3′-end of the target mRNA. The character of N represents the single Watson–Crick paring nucleotide. Pol(A): Poly-A tail; A: Adenine.
3. The Roles of miRNAs in NPC Tumorigenesis: Potential Biomarkers for NPC Early Screening, Prognosis, and Therapy
The first study to reveal the roles of miRNA in nasopharyngeal tumorigenesis was published in 2008 by Sengupta and colleges [20]. In their report, by using microarray-based approaches, they identified eight miRNAs with significantly different expressions in laser-capture micro-dissected NPC samples compared with normal nasopharyngeal epithelial samples. Among them, six miRNAs, including hsa-miR-29c, hsa-miR-34b/c, hsa-miR-212, hsa-miR-216, and hsa-miR-217, exhibited a lower expression in tumor cells, and two miRNAs, hsa-miR-151 and hsa-miR-192, showed a higher expression in NPC tumor cells. Particularly, the target genes were reported to be associated with the lower expression of hsa-miR-29c, and its 15 target genes’ levels, including FLJ12505, COL4A1, COL4A2, COL5A2, COL3A1, COL1A2, FBN1, SPARC, COL15A1, FUSIP1, COL1A1, TFEC, IFNG, LAMC1, LAMC1, showed a two- and six-fold increase in NPC tumors compared with normal nasopharyngeal epithelium. Their abnormal expression was reported to be associated with tumor cell invasiveness, metastasis and metastatic potential, which are prominent characteristics of NPC. Hence, since the first report, growing evidences on miRNA regulatory networks in NPC progression have emerged. Over the years, numerous studies have indicated that a high proportion of miRNAs play a significant role in NPC pathogenesis. The dysregulation of miRNAs that act either as tumor suppressors (down-regulation) or oncogenes (up-regulation) leads to the abnormal regulation of a range of NPC-associated biological pathways, such as cell proliferation, apoptosis, survival and metastasis, which play a crucial role in nasopharyngeal carcinogenesis (Figure 4) [21,22].
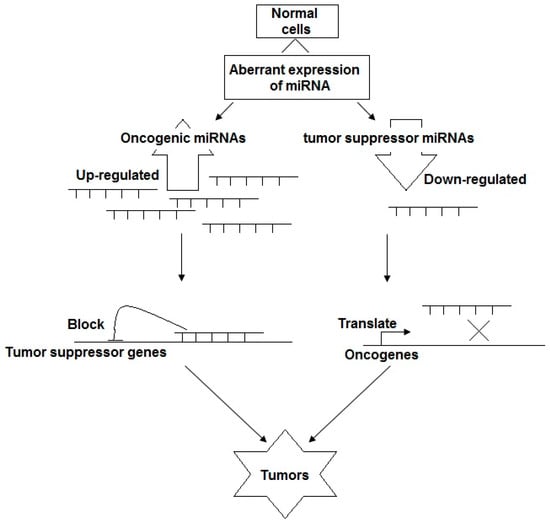
Figure 4.
The biological function of miRNAs: tumor suppressors or oncogenic miRNAs.
In terms of the mechanism, miRNAs act as oncogenic and tumor suppressor miRNAs in tumorigenic events. In the function of tumor suppressor miRNAs, miRNAs are down-regulated, resulting in a potentially enhanced expression of oncogenes, promoting cell proliferation, migration, angiogenesis, invasion, and metastasis, whereas oncogenic miRNAs are over-expressed in nasopharyngeal tumorigenesis, leading to the inhibition of tumor suppressor genes, and, as a result, they increase the proliferation, migration and invasion of NPC cells as well as the loss of cell cycle regulation and they inhibit cell apoptosis.
3.1. miRNAs as Tumor Suppressor Genes
Generally, tumor suppressor miRNAs can down-regulate different oncogenes, which are revealed to promote cell proliferation, migration, angiogenesis, invasion, and metastasis [23]. miRNAs that act as tumor suppressors are reported to be often down-regulated in NPC pathogenesis [22,24,25]. The down-regulation of tumor repressor miRNAs, which are negative regulators of protein-coding genes, leads to an up-regulation of their target genes and the subsequent alterations of the associated cellular pathways in NPC [24]. Many miRNAs (including hsa-Let-7, hsa-miR-9, hsa-miR-26, hsa-miR-29, hsa-miR-30, hsa-miR-34, hsa-miR-101, hsa-miR-200, hsa-miR-218, hsa-miR-223, hsa-miR-331, hsa-miR-429, etc.) are reported to be tumor suppressor genes that play vital roles in nasopharyngeal carcinoma [26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55]. Bioinformatic prediction tools in combination with experimental analysis, including NPC culture cell analysis, case-control studies, etc., have demonstrated that miRNAs could influence multiple steps of tumorigenesis by interacting with target genes (Table 1). Understanding the role of tumor suppressor genes in NPC has the potential to establish the prognostic biomarkers and therapeutic targets of NPC.

Table 1.
List of miRNAs that act as tumor suppressor genes in nasopharyngeal carcinoma (NPC).
3.2. miRNAs as Oncogenes
Several miRNAs are reported as oncogenes, known as oncogenic miRNAs (onco-miRNAs), which are over-expressed in nasopharyngeal tumorigenesis. Their activities have been found to increase the proliferation, migration and invasion of NPC cells, the loss of cell cycle regulation and inhibition the cell apoptosis [22,24,56]. Many onco-miRNAs (including hsa-miR-10a/b, hsa-miR-17, hsa-miR-18, hsa-miR-21, hsa-miR-93, hsa-miR-141, hsa-miR-144, hsa-miR-155, hsa-miR-205, hsa-miR-214, hsa-miR-378, hsa-miR-421, hsa-miR-663, hsa-miR-3135, etc.) are reported to be associated with nasopharyngeal tumorigenesis (Table 2) [57,58,59,60,61,62,63,64,65,66,67,68,69,70,71,72,73,74,75,76,77,78,79,80,81,82,83,84,85,86,87,88,89,90,91,92].

Table 2.
List of miRNAs as oncogenes in NPC.
4. Circulating miRNA: Potential Values in Nasopharyngeal Carcinoma Early Screening, Prognosis and Cancer Treatment
As described above, the over-expression of oncogenic miRNAs or down-expression of tumor suppressor miRNAs are reported to be strongly associated with nasopharyngeal tumorigenesis, progression, tumor colony formation, invasion, metastasis as well as radio/chemical resistance. Notably, recent growing evidence indicates that several miRNAs could be detected in biological fluids, including blood serum/plasma, tears, urine, etc., which reflect the pathophysiological condition of cancer [93,94,95,96,97]. These miRNAs are termed circulating miRNAs (c-miRNAs) or extracellular miRNA (ECmiRNA) [94]. Two major populations of c-miRNAs (including non-vesicle-associated c-miRNAs and vesicle-associated c-miRNAs, which reflect the mechanism of c-miRNAs release) have been identified. However, the mechanism of c-miRNAs release is still unclear [98]. The first profile of c-miRNAs was identified by Chim et al. in 2008 [99]. They discovered the existence of placental miRNAs, including miR-141, miR-149, miR-299-5p, and miR-135b, in maternal plasma using quantitative reverse-transcription real-time PCR [99]. Two months later, Lawrie et al. in 2008 reported that the levels of tumor-related miRNAs, including miR-155, miR-210, and miR-21, were significantly higher in the serum from diffuse large B-cell lymphoma compared with the control sera. Notably, the high expression of miR-21 was associated with relapse-free survival [100]. Hence, the study highlighted c-miRNAs as non-invasive powerful diagnostic markers for human cancers. Since then, the new class of miRNA, c-miRNAs, has opened up new molecular markers for human cancer monitoring.
Due to their many essential characteristics, c-miRNAs have been considered as valuable biomarkers for the early screening and diagnosis of human cancers, including NPC. c-miRNAs are stable in circulation and resistant to RNAse activities in the extracellular environment [93,94]. Additionally, c-miRNAs remain stable with a considerable level of expression even when they are subjected to harsh conditions, including an extreme pH level (high or low pH), extended storage, boiling, and multiple freeze-thaw cycles [93,94,101]. However, the mechanisms by which c-miRNAs maintain their remarkable stability in a RNAse-rich extracellular environment as well as in harsh conditions are still not well understood. The levels of c-miRNAs could easily be evaluated by various methods, such as qRT-PCR and oligonucleotide microarray [102,103]. Therefore, c-miRNAs are identified as suitable molecular targets for human cancer early screening, prognosis as well as treatment.
In several reports, interestingly, the potential uses of the c-miRNA expression profile are reported as biomarkers for early screening, and the prognosis and diagnosis of NPC [96,104]. In the study conducted by Zeng et al. in 2012, they performed the evaluation of serum miRNA profiling on the sera of twenty NPC patients and twenty non-cancerous controls. Four serum c-miRNAs, c-miR-223, c-miR-29c, c-miR-20a and c-miR-17, were significantly expressed in NPC patients’ sera compared with the expression in the non-cancerous controls. Thus, serum-based biomarkers, especially the four serum c-miRNAs, are potential biomarkers for NPC diagnosis [105]. Yi et al. in 2019 reported that the low expression of c-miR-31-5p was significantly associated with the NPC tumor-node-metastasis (TNM) stage: Stage I + II vs. III + IV, T1 vs. T2 + T3 + T4 and N1 + N3 vs. N0. Moreover, c-miR-31-5p showed a moderate diagnostic performance. Therefore, c-miR-31-5p was concluded as the non-invasive valuable, and as a novel biomarker for the early diagnosis of NPC. Especially, it was reported to be an attractive therapeutic molecular target for NPC treatment [106]. Exosome-associated miRNAs, a type of vesicle-associated c-miRNA, are derived from cancer and could interact with endothelial cells, thereby enhancing proliferation, migration as well as angiogenesis [107]. Exosomal miR-24-3p was reported to be involved in nasopharyngeal pathogenesis by mediating T-cell suppression via the repression of fibroblast growth factor 11 (FGF11), and it may serve as a potential biomarker in NPC [108]. Up to date, exosome-associated miRNAs are still of interest in finding potential biomarkers for NPC. Other exosome-associated miRNAs, such as metastasis-associated miR-23a [109], exosomal miR-9 [107], exosomal miR-34c [110], etc., have been reported. Their data suggested that exosomal miRNAs reflect the pathogenesis of NPC and that there is the possibility of exosome-based therapies for NPC in the future.
5. miRNAs as Novel Molecular Targets for NPC Therapies
As described, strikingly different miRNA expressions have been observed in NPC; therefore, the activation of tumor suppressor miRNAs and/or the inhibition of oncogenic miRNAs may open up a novel approach for the development of therapeutics/treatments for NPC. Up to date, an increasing number of studies have focused on the possibility of miRNAs serving as molecular targets for NPC therapies. In the study conducted by Kang et al. in 2016, they investigated the effects of miR-24 on the radio-sensitivity of NPC cells. They found that the miR-24/SP1 pathway contributed to the reduction in radio-resistance in human NPC [111]. Therefore, the finding of the miR-24/SP1 pathway may help our understanding of radio-sensitivity in NPC and represents a promising therapeutic molecular target. Thus, based on the roles of miRNAs, more and more studies are applying miRNAs to NPC therapy.
6. Conclusions
Since the first discovery of miRNAs, numerous studies have focused on their roles in human tumorigenesis, including NPC. As described in this article, the abnormal expression of miRNAs, especially c-miRNAs, have been shown to play important roles in the hallmarks of NPC by targeting many pathways, including cell proliferation, invasion and migration. Especially, such detectable changes in non/less invasive samples (plasma, serum, etc.) make them promising non-invasive biomarkers for the early screening, prognosis, and therapeutic molecular target monitoring of NPC.
Author Contributions
All authors contributed to the design and conception of the study. T.D.L.: writing original draft preparation; T.A.H.L.: review and editing the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research received funding sponsored by Ho Chi Minh City Open University, Ho Chi Minh City, Vietnam.
Acknowledgments
We wish to express our thanks to the research project sponsored by Ho Chi Minh City Open University, Ho Chi Minh City, Vietnam.
Conflicts of Interest
The authors declare that there is no conflict of interest regarding the publication of this paper.
References
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Li, K.; Zhu, X.; Li, L.; Ning, R.; Liang, Z.; Zeng, F.; Su, F.; Huang, S.; Yang, X.; Qu, S. Identification of non-invasive biomarkers for predicting the radiosensitivity of nasopharyngeal carcinoma from serum microRNAs. Sci. Rep. 2020, 10, 5161. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Bushati, N.; Cohen, S.M. microRNA functions. Annu. Rev. Cell Dev. Biol. 2007, 23, 175–205. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef]
- Stahlhut Espinosa, C.E.; Slack, F.J. The role of microRNAs in cancer. Yale J. Biol. Med. 2006, 79, 131–140. [Google Scholar]
- Si, W.; Shen, J.; Zheng, H.; Fan, W. The role and mechanisms of action of microRNAs in cancer drug resistance. Clin. Epigenet. 2019, 11, 25. [Google Scholar] [CrossRef]
- Li, M.; Wang, C.; Yu, B.; Zhang, X.; Shi, F.; Liu, X. Diagnostic value of RASSF1A methylation for breast cancer: A meta-analysis. Biosci. Rep. 2019, 39, BSR20190923. [Google Scholar] [CrossRef]
- Tabuchi, K.; Nakayama, M.; Nishimura, B.; Hayashi, K.; Hara, A. Early detection of nasopharyngeal carcinoma. Int. J. Otolaryngol. 2011, 2011, 638058. [Google Scholar] [CrossRef]
- Felekkis, K.; Touvana, E.; Stefanou, C.; Deltas, C. microRNAs: A newly described class of encoded molecules that play a role in health and disease. Hippokratia 2010, 14, 236–240. [Google Scholar]
- Wong, L.L.; Wang, J.; Liew, O.W.; Richards, A.M.; Chen, Y.T. MicroRNA and Heart Failure. Int. J. Mol. Sci. 2016, 17, 502. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Hagedorn, C.H.; Cullen, B.R. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA 2004, 10, 1957–1966. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Kim, M.; Han, J.; Yeom, K.H.; Lee, S.; Baek, S.H.; Kim, V.N. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004, 23, 4051–4060. [Google Scholar] [CrossRef] [PubMed]
- Lao, D.T.; Truong, K.P.; Le, H.A.T. miRNA-141 as the Biomarker for Human Cancers. AJPRHC 2018, 10, 42–49. [Google Scholar] [CrossRef]
- Peng, Y.; Croce, C.M. The role of MicroRNAs in human cancer. Signal Transduct. Target. Ther. 2016, 1, 15004. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Ahn, C.; Han, J.; Choi, H.; Kim, J.; Yim, J.; Lee, J.; Provost, P.; Rådmark, O.; Kim, S.; et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 2003, 425, 415–419. [Google Scholar] [CrossRef]
- Kim, V.N.; Han, J.; Siomi, M.C. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 2009, 10, 126–139. [Google Scholar] [CrossRef]
- Inui, M.; Martello, G.; Piccolo, S. MicroRNA control of signal transduction. Nat. Rev. Mol. Cell Biol. 2010, 11, 252–263. [Google Scholar] [CrossRef]
- Witkos, T.M.; Koscianska, E.; Krzyzosiak, W.J. Practical Aspects of microRNA Target Prediction. Curr. Mol. Med. 2011, 11, 93–109. [Google Scholar] [CrossRef]
- Sengupta, S.; den Boon, J.A.; Chen, I.H.; Newton, M.A.; Stanhope, S.A.; Cheng, Y.J.; Chen, C.J.; Hildesheim, A.; Sugden, B.; Ahlquist, P. MicroRNA 29c is down-regulated in nasopharyngeal carcinomas, up-regulating mRNAs encoding extracellular matrix proteins. Proc. Natl. Acad. Sci. USA 2008, 105, 5874–5878. [Google Scholar] [CrossRef]
- Bruce, J.P.; Liu, F.F. MicroRNAs in nasopharyngeal carcinoma. Chin. J. Cancer 2014, 33, 539–544. [Google Scholar] [CrossRef] [PubMed]
- Spence, T.; Bruce, J.; Yip, K.W.; Liu, F.F. MicroRNAs in nasopharyngeal carcinoma. Chin. Clin. Oncol. 2016, 5, 17. [Google Scholar] [CrossRef] [PubMed]
- Shenouda, S.K.; Alahari, S.K. MicroRNA function in cancer: Oncogene or a tumor suppressor? Cancer Metastasis Rev. 2009, 28, 369–378. [Google Scholar] [CrossRef] [PubMed]
- Tan, G.; Tang, X.; Tang, F. The role of microRNAs in nasopharyngeal carcinoma. Tumour Biol. 2015, 36, 69–79. [Google Scholar] [CrossRef] [PubMed]
- Wong, T.S.; Man, O.Y.; Tsang, C.M.; Tsao, S.W.; Tsang, R.K.; Chan, J.Y.; Ho, W.K.; Wei, W.I.; To, V.S. MicroRNA let-7 suppresses nasopharyngeal carcinoma cells proliferation through downregulating c-Myc expression. J. Cancer Res. Clin. Oncol. 2011, 137, 415–422. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Wu, K.; Yang, Z.; Wu, A. High-mobility group A2 overexpression is an unfavorable prognostic biomarker for nasopharyngeal carcinoma patients. Mol. Cell Biochem. 2015, 409, 155–162. [Google Scholar] [CrossRef]
- Wu, A.; Wu, K.; Li, J.; Mo, Y.; Lin, Y.; Wang, Y.; Shen, X.; Li, S.; Li, L.; Yang, Z. Let-7a inhibits migration, invasion and epithelial-mesenchymal transition by targeting HMGA2 in nasopharyngeal carcinoma. J. Transl. Med. 2015, 13, 105. [Google Scholar] [CrossRef]
- Cai, K.; Wan, Y.; Sun, G.; Shi, L.; Bao, X.; Wang, Z. Let-7a inhibits proliferation and induces apoptosis by targeting EZH2 in nasopharyngeal carcinoma cells. Oncol. Rep. 2012, 28, 2101–2106. [Google Scholar] [CrossRef]
- Lu, J.; Luo, H.; Liu, X.; Peng, Y.; Zhang, B.; Wang, L.; Xu, X.; Peng, X.; Li, G.; Tian, W.; et al. miR-9 targets CXCR4 and functions as a potential tumor suppressor in nasopharyngeal carcinoma. Carcinogenesis 2014, 35, 554–563. [Google Scholar] [CrossRef]
- Lu, J.; Xu, X.; Liu, X.; Peng, Y.; Zhang, B.; Wang, L.; Luo, H.; Peng, X.; Li, G.; Tian, W.; et al. Predictive value of miR-9 as a potential biomarker for nasopharyngeal carcinoma metastasis. Br. J. Cancer 2014, 110, 392–398. [Google Scholar] [CrossRef]
- Gao, F.; Zhao, Z.L.; Zhao, W.T.; Fan, Q.R.; Wang, S.C.; Li, J.; Zhang, Y.Q.; Shi, J.W.; Lin, X.L.; Yang, S.; et al. miR-9 modulates the expression of interferon-regulated genes and MHC class I molecules in human nasopharyngeal carcinoma cells. Biochem. Biophys. Res. Commun. 2013, 431, 610–616. [Google Scholar] [CrossRef]
- Sam, C.K.; Brooks, L.A.; Niedobitek, G.; Young, L.S.; Prasad, U.; Rickinson, A.B. Analysis of Epstein-Barr virus infection in nasopharyngeal biopsies from a group at high risk of nasopharyngeal carcinoma. Int. J. Cancer 1993, 53, 957–962. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Tang, L.L.; Sun, Y.; Cui, R.X.; Wang, H.Y.; Huang, B.J.; He, Q.M.; Jiang, W.; Ma, J. MiR-29c suppresses invasion and metastasis by targeting TIAM1 in nasopharyngeal carcinoma. Cancer Lett. 2013, 329, 181–188. [Google Scholar] [CrossRef]
- He, Q.; Ren, X.; Chen, J.; Li, Y.; Tang, X.; Wen, X.; Yang, X.; Zhang, J.; Wang, Y.; Ma, J.; et al. miR-16 targets fibroblast growth factor 2 to inhibit NPC cell proliferation and invasion via PI3K/AKT and MAPK signaling pathways. Oncotarget 2016, 7, 3047–3058. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Q.; Zhang, Y.; Zhao, M.; Li, Q.; Chen, R.; Long, X.; Fang, W.; Liu, Z. miR-16 induction after CDK4 knockdown is mediated by c-Myc suppression and inhibits cell growth as well as sensitizes nasopharyngeal carcinoma cells to chemotherapy. Tumour Biol. 2016, 37, 2425–2433. [Google Scholar] [CrossRef] [PubMed]
- Alajez, N.M.; Shi, W.; Hui, A.B.; Bruce, J.; Lenarduzzi, M.; Ito, E.; Yue, S.; O’Sullivan, B.; Liu, F.F. Enhancer of Zeste homolog 2 (EZH2) is overexpressed in recurrent nasopharyngeal carcinoma and is regulated by miR-26a, miR-101, and miR-98. Cell Death Dis. 2010, 1, e85. [Google Scholar] [CrossRef]
- Yu, L.; Lu, J.; Zhang, B.; Liu, X.; Wang, L.; Li, S.Y.; Peng, X.H.; Xu, X.; Tian, W.D.; Li, X.P. miR-26a inhibits invasion and metastasis of nasopharyngeal cancer by targeting EZH2. Oncol. Lett. 2013, 5, 1223–1228. [Google Scholar] [CrossRef]
- Wang, H.Y.; Li, Y.Y.; Fu, S.; Wang, X.P.; Huang, M.Y.; Zhang, X.; Shao, Q.; Deng, L.; Zeng, M.S.; Zeng, Y.X.; et al. MicroRNA-30a promotes invasiveness and metastasis in vitro and in vivo through epithelial-mesenchymal transition and results in poor survival of nasopharyngeal carcinoma patients. Exp. Biol. Med. 2014, 239, 891–898. [Google Scholar] [CrossRef]
- Li, H.; Li, X.; Ge, X.; Jia, L.; Zhang, Z.; Fang, R.; Yang, J.; Liu, J.; Peng, S.; Zhou, M.; et al. MiR-34b-3 and miR-449a inhibit malignant progression of nasopharyngeal carcinoma by targeting lactate dehydrogenase A. Oncotarget 2016, 23, 54838–54851. [Google Scholar] [CrossRef]
- Li, Y.Q.; Ren, X.Y.; He, Q.M.; Xu, Y.F.; Tang, X.R.; Sun, Y.; Zeng, M.S.; Kang, T.B.; Liu, N.; Ma, J. MiR-34c suppresses tumor growth and metastasis in nasopharyngeal carcinoma by targeting MET. Cell Death Dis. 2015, 22, e1618. [Google Scholar] [CrossRef][Green Version]
- Peng, X.H.; Huang, H.R.; Lu, J.; Liu, X.; Zhao, F.P.; Zhang, B.; Lin, S.X.; Wang, L.; Chen, H.H.; Xu, X.; et al. MiR-124 suppresses tumor growth and metastasis by targeting Foxq1 in nasopharyngeal carcinoma. Mol. Cancer 2014, 13, 186. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Lv, X.B.; Wang, X.P.; Sang, Y.; Xu, S.; Hu, K.; Wu, M.; Liang, Y.; Liu, P.; Tang, J.; et al. MiR-138 suppressed nasopharyngeal carcinoma growth and tumorigenesis by targeting the CCND1 oncogene. Cell Cycle 2012, 11, 2495–2506. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Qi, X.; Li, J.; Zhou, C.; Lv, C.; Tian, M. MiR-142-3p Suppresses SOCS6 Expression and Promotes Cell Proliferation in Nasopharyngeal Carcinoma. Cell Physiol. Biochem. 2015, 36, 1743–1752. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; He, Q.; Wen, X.; Hong, X.; Yang, X.; Tang, X.; Zhang, P.; Lei, Y.; Sun, Y.; Zhang, J.; et al. EZH2-DNMT1-mediated epigenetic silencing of miR-142-3p promotes metastasis through targeting ZEB2 in nasopharyngeal carcinoma. Cell Death Differ. 2019, 26, 1089–1106. [Google Scholar] [CrossRef]
- Wang, L.; Tian, W.D.; Xu, X.; Nie, B.; Lu, J.; Liu, X.; Zhang, B.; Dong, Q.; Sunwoo, J.B.; Li, G.; et al. Epstein-Barr virus nuclear antigen 1 (EBNA1) protein induction of epithelial-mesenchymal transition in nasopharyngeal carcinoma cells. Cancer 2014, 120, 363–372. [Google Scholar] [CrossRef]
- Yang, X.; Ni, W.; Lei, K. miR-200b suppresses cell growth, migration and invasion by targeting Notch1 in nasopharyngeal carcinoma. Cell Physiol. Biochem. 2013, 32, 1288–1298. [Google Scholar] [CrossRef]
- Ma, L.; Deng, X.; Wu, M.; Zhang, G.; Huang, J. Down-regulation of miRNA-204 by LMP-1 enhances CDC42 activity and facilitates invasion of EBV-associated nasopharyngeal carcinoma cells. FEBS Lett. 2014, 588, 1562–1570. [Google Scholar] [CrossRef]
- Deng, M.; Tang, H.; Zhou, Y.; Zhou, M.; Xiong, W.; Zheng, Y.; Ye, Q.; Zeng, X.; Liao, Q.; Guo, X.; et al. miR-216b suppresses tumor growth and invasion by targeting KRAS in nasopharyngeal carcinoma. J. Cell Sci. 2011, 124 Pt 17, 2997–3005. [Google Scholar] [CrossRef]
- Deng, M.; Liu, J.F.; Gu, Y.X.; Zheng, G.P.; He, Z.M. miR-216b suppresses cell proliferation and invasion by targeting PKCα in nasopharyngeal carcinoma cells. Zhonghua Zhong Liu Za Zhi 2013, 35, 645–650. [Google Scholar]
- Alajez, N.M.; Lenarduzzi, M.; Ito, E.; Hui, A.B.; Shi, W.; Bruce, J.; Yue, S.; Huang, S.H.; Xu, W.; Waldron, J.; et al. MiR-218 suppresses nasopharyngeal cancer progression through downregulation of survivin and the SLIT2-ROBO1 pathway. Cancer Res. 2011, 71, 2381–2391. [Google Scholar] [CrossRef]
- Gao, L.; Xiong, X. MiR-223 inhibits the proliferation, invasion and EMT of nasopharyngeal carcinoma cells by targeting SSRP1. Int. J. Clin. Exp. Pathol. 2018, 11, 4374–4384. [Google Scholar]
- Yang, W.; Lan, X.; Li, D.; Li, T.; Lu, S. MiR-223 targeting MAFB suppresses proliferation and migration of nasopharyngeal carcinoma cells. BMC Cancer 2015, 15, 461. [Google Scholar] [CrossRef]
- Hui, A.B.; Bruce, J.P.; Alajez, N.M.; Shi, W.; Yue, S.; Perez-Ordonez, B.; Xu, W.; O’Sullivan, B.; Waldron, J.; Cummings, B.; et al. Significance of dysregulated metadherin and microRNA-375 in head and neck cancer. Clin Cancer Res. 2011, 17, 7539–7550. [Google Scholar] [CrossRef]
- Hou, J.; Yan, J.; Ren, X.Y.; Zhu, K.; Du, X.Y.; Li, J.J.; Xu, M. Long noncoding RNA ROR1-AS1 induces tumor metastasis and epithelial-mesenchymal transition by sponging miR-375 in nasopharyngeal carcinoma. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 174–180. [Google Scholar] [CrossRef]
- Liu, N.; Jiang, N.; Guo, R.; Jiang, W.; He, Q.M.; Xu, Y.F.; Li, Y.Q.; Tang, L.L.; Mao, Y.P.; Sun, Y.; et al. MiR-451 inhibits cell growth and invasion by targeting MIF and is associated with survival in nasopharyngeal carcinoma. Mol. Cancer 2013, 12, 123. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Gao, R.; Yu, Y.; Kaul, Z.; Wang, J.; Kalra, R.S.; Zhang, Z.; Kaul, S.C.; Wadhwa, R. Tumor suppressor activity of miR-451: Identification of CARF as a new target. Sci. Rep. 2018, 8, 375. [Google Scholar] [CrossRef] [PubMed]
- Kavitha, N.; Vijayarathna, S.; Jothy, S.L.; Oon, C.E.; Chen, Y.; Kanwar, J.R.; Sasidharan, S. MicroRNAs: Biogenesis, roles for carcinogenesis and as potential biomarkers for cancer diagnosis and prognosis. Asian Pac. J. Cancer Prev. 2014, 15, 7489–7497. [Google Scholar] [CrossRef] [PubMed]
- Ørom, U.A.; Nielsen, F.C.; Lund, A.H. MicroRNA-10a binds the 5’UTR of ribosomal protein mRNAs and enhances their translation. Mol. Cell 2008, 30, 460–471. [Google Scholar] [CrossRef]
- Sun, X.J.; Liu, H.; Zhang, P.; Zhang, X.D.; Jiang, Z.W.; Jiang, C.C. miR-10b promotes migration and invasion in nasopharyngeal carcinoma cells. Asian Pac. J. Cancer Prev. 2013, 14, 5533–5537. [Google Scholar] [CrossRef]
- Allaya, N.; Khabir, A.; Sallemi-Boudawara, T.; Sellami, N.; Daoud, J.; Ghorbel, A.; Frikha, M.; Gargouri, A.; Mokdad-Gargouri, R.; Ayadi, W. Over-expression of miR-10b in NPC patients: Correlation with LMP1 and Twist1. Tumour Biol. 2015, 36, 3807–3814. [Google Scholar] [CrossRef]
- Chen, C.; Lu, Z.; Yang, J.; Hao, W.; Qin, Y.; Wang, H.; Xie, C.; Xie, R. MiR-17-5p promotes cancer cell proliferation and tumorigenesis in nasopharyngeal carcinoma by targeting p21. Cancer Med. 2016, 5, 3489–3499. [Google Scholar] [CrossRef]
- Hu, Z.; Zhou, S.; Luo, H.; Ji, M.; Zheng, J.; Huang, F.; Wang, F. miRNA-17 promotes nasopharyngeal carcinoma radioresistance by targeting PTEN/AKT. Int. J. Clin. Exp. Pathol. 2019, 12, 229–240. [Google Scholar]
- Luo, Z.; Dai, Y.; Zhang, L.; Jiang, C.; Li, Z.; Yang, J.; McCarthy, J.B.; She, X.; Zhang, W.; Ma, J.; et al. miR-18a promotes malignant progression by impairing microRNA biogenesis in nasopharyngeal carcinoma. Carcinogenesis 2013, 34, 415–425. [Google Scholar] [CrossRef]
- Mai, S.; Xiao, R.; Shi, L.; Zhou, X.; Yang, T.; Zhang, M.; Weng, N.; Zhao, X.; Wang, R.; Liu, J.; et al. MicroRNA-18a promotes cancer progression through SMG1 suppression and mTOR pathway activation in nasopharyngeal carcinoma. Cell Death Dis. 2019, 10, 819. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.; Bian, G.; Pan, Y.; Han, X.; Sun, Y.; Wang, Y.; Shen, G.; Cheng, M.; Fang, X.; Hu, S. MiR-20a-5p promotes radio-resistance by targeting Rab27B in nasopharyngeal cancer cells. Cancer Cell Int. 2017, 17, 32. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Yan, L.; Zhang, W.; Wang, H.; Chen, W.; Hu, N.; Ou, H. miR-21 inhibitor suppresses proliferation and migration of nasopharyngeal carcinoma cells through down-regulation of BCL2 expression. Int. J. Clin. Exp. Pathol. 2014, 7, 3478–3487. [Google Scholar]
- Ou, H.; Li, Y.; Kang, M. Activation of miR-21 by STAT3 induces proliferation and suppresses apoptosis in nasopharyngeal carcinoma by targeting PTEN gene. PLoS ONE 2014, 9, e109929. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Li, H.; Guo, X.; Wang, Z.; Liang, S.; Dang, C. IGF-I Induces Epithelial-to-Mesenchymal Transition via the IGF-IR-Src-MicroRNA-30a-E-Cadherin Pathway in Nasopharyngeal Carcinoma Cells. Oncol. Res. 2016, 24, 225–231. [Google Scholar] [CrossRef]
- Lyu, X.; Fang, W.; Cai, L.; Zheng, H.; Ye, Y.; Zhang, L.; Li, J.; Peng, H.; Cho, W.C.; Wang, E.; et al. TGFβR2 is a major target of miR-93 in nasopharyngeal carcinoma aggressiveness. Mol. Cancer 2014, 13, 51. [Google Scholar] [CrossRef]
- Xu, Y.F.; Mao, Y.P.; Li, Y.Q.; Ren, X.Y.; He, Q.M.; Tang, X.R.; Sun, Y.; Liu, N.; Ma, J. MicroRNA-93 promotes cell growth and invasion in nasopharyngeal carcinoma by targeting disabled homolog-2. Cancer Lett. 2015, 363, 146–155. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, Z. miR-93 enhances cell proliferation by directly targeting CDKN1A in nasopharyngeal carcinoma. Oncol. Lett. 2018, 15, 1723–1727. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Wang, W.; Li, S.; Yang, Q.; Hu, J.; Zeng, N.; Gao, C. MicroRNA 141 represses nasopharyngeal carcinoma growth through inhibiting BMI1. Oncol Lett. 2018, 16, 6479–6487. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhao, R.; Wang, H.; Luo, Y.; Wang, X.; Niu, W.; Zhou, Y.; Wen, Q.; Fan, S.; Li, X.; et al. miR-141 is involved in BRD7-mediated cell proliferation and tumor formation through suppression of the PTEN/AKT pathway in nasopharyngeal carcinoma. Cell Death Dis. 2016, 7, e2156. [Google Scholar] [CrossRef] [PubMed]
- Lao, T.D.; Nguyen, T.V.; Nguyen, D.H.; Nguyen, M.T.; Nguyen, C.H.; Le, T.H.A. miR-141 is up-regulated in biopsies from Vietnamese patients with nasopharyngeal carcinoma. Braz. Oral Res. 2018, 32, e126. [Google Scholar] [CrossRef]
- Zhang, L.Y.; Ho-Fun Lee, V.; Wong, A.M.; Kwong, D.L.; Zhu, Y.H.; Dong, S.S.; Kong, K.L.; Chen, J.; Tsao, S.W.; Guan, X.Y.; et al. MicroRNA-144 promotes cell proliferation, migration and invasion in nasopharyngeal carcinoma through repression of PTEN. Carcinogenesis 2013, 34, 454–463. [Google Scholar] [CrossRef]
- Song, L.; Chen, L.; Luan, Q.; Kong, Q. miR-144-3p facilitates nasopharyngeal carcinoma via crosstalk with PTEN. J. Cell Physiol. 2019, 234, 17912–17924. [Google Scholar] [CrossRef]
- Wu, C.W.; Wang, S.G.; Lin, M.L.; Chen, S.S. Downregulation of miR-144 by triptolide enhanced p85α-PTEN complex formation causing S phase arrest of human nasopharyngeal carcinoma cells. Eur. J. Pharmacol. 2019, 855, 137–148. [Google Scholar] [CrossRef]
- Kong, Y.G.; Cui, M.; Chen, S.M.; Xu, Y.; Xu, Y.; Tao, Z.Z. LncRNA-LINC00460 facilitates nasopharyngeal carcinoma tumorigenesis through sponging miR-149-5p to up-regulate IL6. Gene 2018, 639, 77–84. [Google Scholar] [CrossRef]
- Luo, Z.; Zhang, L.; Li, Z.; Jiang, C.; Dai, Y.; Liu, X.; Zheng, Y.; Yu, H.; Xiang, J.; Li, G. miR-149 promotes epithelial-mesenchymal transition and invasion in nasopharyngeal carcinoma cells. Zhong Nan Da Xue Xue Bao Yi Xue Ban 2011, 36, 604–609. [Google Scholar] [CrossRef]
- Zhu, X.; Wang, Y.; Sun, Y.; Zheng, J.; Zhu, D. MiR-155 up-regulation by LMP1 DNA contributes to increased nasopharyngeal carcinoma cell proliferation and migration. Eur. Arch. Otorhinolaryngol. 2014, 271, 1939–1945. [Google Scholar] [CrossRef]
- Zuo, W.N.; Zhu, H.; Li, L.P.; Jin, A.Y.; Wang, H.Q. MiR-155 promotes proliferation and inhibits apoptosis of nasopharyngeal carcinoma cells through targeting PTEN-PI3K/AKT pathway. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 7935–7942. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Xie, D.L.; Dai, X.Y. Down-regulation of miR-155 promotes apoptosis of nasopharyngeal carcinoma CNE-1 cells by targeting PI3K/AKT-FOXO3a signaling. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 7391–7398. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y.; Wu, S.; Zhao, R.; Deng, Q. MiR-205 promotes proliferation, migration and invasion of nasopharyngeal carcinoma cells by activation of AKT signalling. J. Int. Med. Res. 2016, 44, 231–240. [Google Scholar] [CrossRef] [PubMed]
- Nie, G.; Duan, H.; Li, X.; Yu, Z.; Luo, L.; Lu, R.; Ji, Z.; Zhang, W. MicroRNA 205 promotes the tumorigenesis of nasopharyngeal carcinoma through targeting tumor protein p53-inducible nuclear protein 1. Mol. Med. Rep. 2015, 12, 5715–5722. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhang, Z.C.; Li, Y.Y.; Wang, H.Y.; Fu, S.; Wang, X.P.; Zeng, M.S.; Zeng, Y.X.; Shao, J.Y. Knockdown of miR-214 promotes apoptosis and inhibits cell proliferation in nasopharyngeal carcinoma. PLoS ONE 2014, 9, e86149. [Google Scholar] [CrossRef]
- Deng, M.; Ye, Q.; Qin, Z.; Zheng, Y.; He, W.; Tang, H.; Zhou, Y.; Xiong, W.; Zhou, M.; Li, X.; et al. miR-214 promotes tumorigenesis by targeting lactotransferrin in nasopharyngeal carcinoma. Tumour Biol. 2013, 34, 1793–1800. [Google Scholar] [CrossRef]
- Yu, B.L.; Peng, X.H.; Zhao, F.P.; Liu, X.; Lu, J.; Wang, L.; Li, G.; Chen, H.H.; Li, X.P. MicroRNA-378 functions as an onco-miR in nasopharyngeal carcinoma by repressing TOB2 expression. Int. J. Oncol. 2014, 44, 1215–1222. [Google Scholar] [CrossRef]
- Chen, L.; Tang, Y.; Wang, J.; Yan, Z.; Xu, R. miR-421 induces cell proliferation and apoptosis resistance in human nasopharyngeal carcinoma via downregulation of FOXO4. Biochem. Biophys. Res. Commun. 2013, 435, 745–750. [Google Scholar] [CrossRef]
- Zhao, L.; Tang, M.; Hu, Z.; Yan, B.; Pi, W.; Li, Z.; Zhang, J.; Zhang, L.; Jiang, W.; Li, G.; et al. miR-504 mediated down-regulation of nuclear respiratory factor 1 leads to radio-resistance in nasopharyngeal carcinoma. Oncotarget 2015, 6, 15995–16018. [Google Scholar] [CrossRef]
- Liang, S.; Zhang, N.; Deng, Y.; Chen, L.; Zhang, Y.; Zheng, Z.; Luo, W.; Lv, Z.; Li, S.; Xun, T. Increased Serum Level of MicroRNA-663 Is Correlated with Poor Prognosis of Patients with Nasopharyngeal Carcinoma. Dis. Markers 2016, 2016, 7648215. [Google Scholar] [CrossRef]
- Yu, Q.; Zhang, F.; Du, Z.; Xiang, Y. Up-regulation of serum miR-744 predicts poor prognosis in patients with nasopharyngeal carcinoma. Int. J. Clin. Exp. Med. 2015, 8, 13296–13302. [Google Scholar] [PubMed]
- Mo, X.; Yin, W.; Huang, Y.; Guo, W.; Zhou, M.; Ye, H. Expression of miR-3182 and EBV-miR-BART8-3p in nasopharyngeal carcinoma is correlated with distant metastasis. Int. J. Clin. Exp. Pathol. 2018, 11, 3134–3140. [Google Scholar] [PubMed]
- Ma, R.; Jiang, T.; Kang, X. Circulating microRNAs in cancer: Origin, function and application. J. Exp. Clin. Cancer Res. 2012, 31, 38. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, K.P.; Ramphul, E.; Howard, L.; Gallagher, W.M.; Malone, C.; Kerin, M.J.; Dwyer, R.M. Circulating MicroRNAs in Cancer. Methods Mol. Biol. 2017, 1509, 123–139. [Google Scholar]
- Cufaro, M.C.; Pieragostino, D.; Lanuti, P.; Rossi, C.; Cicalini, I.; Federici, L.; De Laurenzi, V.; Del Boccio, P. Extracellular Vesicles and Their Potential Use in Monitoring Cancer Progression and Therapy: The Contribution of Proteomics. J. Oncol. 2019, 2019, 1639854. [Google Scholar] [CrossRef]
- Sohel, M.H. Extracellular/Circulating MicroRNAs: Release Mechanisms, Functions and Challenges. Achiev. Life Sci. 2016, 10, 175–186. [Google Scholar] [CrossRef]
- Xu, X.; Lu, J.; Wang, F.; Liu, X.; Peng, X.; Yu, B.; Zhao, F.; Li, X. Dynamic Changes in Plasma MicroRNAs Have Potential Predictive Values in Monitoring Recurrence and Metastasis of Nasopharyngeal Carcinoma. Biomed. Res. Int. 2018, 7329195. [Google Scholar] [CrossRef]
- Wen, W.; Mai, S.J.; Lin, H.X.; Zhang, M.Y.; Huang, J.L.; Hua, X.; Lin, C.; Long, Z.Q.; Lu, Z.J.; Sun, X.Q.; et al. Identification of two microRNA signatures in whole blood as novel biomarkers for diagnosis of nasopharyngeal carcinoma. J. Transl. Med. 2019, 17, 186. [Google Scholar] [CrossRef]
- Cui, M.; Wang, H.; Yao, X.; Zhang, D.; Xie, Y.; Cui, R.; Zhang, X. Circulating MicroRNAs in Cancer: Potential and Challenge. Front. Genet. 2019, 10, 626. [Google Scholar] [CrossRef]
- Chim, S.S.; Shing, T.K.; Hung, E.C.; Leung, T.Y.; Lau, T.K.; Chiu, R.W.; Lo, Y.M. Detection and characterization of placental microRNAs in maternal plasma. Clin. Chem. 2008, 54, 482–490. [Google Scholar] [CrossRef]
- Lawrie, C.H.; Gal, S.; Dunlop, H.M.; Pushkaran, B.; Liggins, A.P.; Pulford, K.; Banham, A.H.; Pezzella, F.; Boultwood, J.; Wainscoat, J.S.; et al. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br. J. Haematol. 2008, 141, 672–675. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A.; et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar] [CrossRef] [PubMed]
- Lodes, M.J.; Caraballo, M.; Suciu, D.; Munro, S.; Kumar, A.; Anderson, B. Detection of cancer with serum miRNAs on an oligonucleotide microarray. PLoS ONE 2009, 4, e6229. [Google Scholar] [CrossRef] [PubMed]
- Resnick, K.E.; Alder, H.; Hagan, J.P.; Richardson, D.L.; Croce, C.M.; Cohn, D.E. The detection of differentially expressed microRNAs from the serum of ovarian cancer patients using a novel real-time PCR platform. Gynecol. Oncol. 2009, 112, 55–59. [Google Scholar] [CrossRef]
- Zeng, X.; Xiang, J.; Wu, M.; Xiong, W.; Tang, H.; Deng, M.; Li, X.; Liao, Q.; Su, B.; Luo, Z.; et al. Circulating miR-17, miR-20a, miR-29c, and miR-223 combined as non-invasive biomarkers in nasopharyngeal carcinoma. PLoS ONE 2012, 7, e46367. [Google Scholar] [CrossRef]
- Yi, S.J.; Liu, P.; Chen, B.L.; Ou-Yang, L.; Xiong, W.M.; Su, J.P. Circulating miR-31-5p may be a potential diagnostic biomarker in nasopharyngeal carcinoma. Neoplasma 2019, 66, 825–829. [Google Scholar] [CrossRef]
- Lu, J.; Liu, Q.H.; Wang, F.; Tan, J.J.; Deng, Y.Q.; Peng, X.H.; Liu, X.; Zhang, B.; Xu, X.; Li, X.P. Exosomal miR-9 inhibits angiogenesis by targeting MDK and regulating PDK/AKT pathway in nasopharyngeal carcinoma. J. Exp. Clin. Cancer Res. 2018, 13, 147. [Google Scholar] [CrossRef]
- Ye, S.B.; Zhang, H.; Cai, T.T.; Liu, Y.N.; Ni, J.J.; He, J.; Peng, J.Y.; Chen, Q.Y.; Mo, H.Y.; Jun-Cui Zhang, X.S.; et al. Exosomal miR-24-3p impedes T-cell function by targeting FGF11 and serves as a potential prognostic biomarker for nasopharyngeal carcinoma. J. Pathol. 2016, 240, 329–340. [Google Scholar] [CrossRef]
- Bao, L.; You, B.; Shi, S.; Shan, Y.; Zhang, Q.; Yue, H.; Zhang, J.; Zhang, W.; Shi, Y.; Liu, Y.; et al. Metastasis-associated miR-23a from nasopharyngeal carcinoma-derived exosomes mediates angiogenesis by repressing a novel target gene TSGA10. Oncogene 2018, 37, 2873–2889. [Google Scholar] [CrossRef]
- Wan, F.Z.; Chen, K.H.; Sun, Y.C.; Chen, X.C.; Liang, R.B.; Chen, L.; Zhu, X.D. Exosomes overexpressing miR-34c inhibit malignant behavior and reverse the radioresistance of nasopharyngeal carcinoma. J. Transl. Med. 2020, 18, 12. [Google Scholar] [CrossRef]
- Kang, M.; Xiao, J.; Wang, J.; Zhou, P.; Wei, T.; Zhao, T.; Wang, R. MiR-24 enhances radiosensitivity in nasopharyngeal carcinoma by targeting SP1. Cancer Med. 2016, 5, 1163–1173. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).