1. Introduction
The intensifying crisis of multi-drug resistance is increasing concerns about untreatable infections being a significant threat to public health [
1,
2]. Hence, the necessity for alternative treatments that are efficient and have little possibility to affect resistance has arisen. For this reason, the area of antimicrobial compounds has, since its inception, gone through in-depth research displaying their apparent suitability as alternatives to antibiotics [
3]. A group of antimicrobials, named bacteriocins, have been studied for their great potential in controlling antibiotic-resistant pathogens. Antimicrobial peptides (AMPs) exist as a favorable choice as medications to prevent infections by bacteria that are not efficiently curable with traditional antibiotics [
4,
5,
6].
Antimicrobial peptides are elements of the natural immune response of surviving organisms and have a broad spectrum of action ranging from parasites to viruses at a minimal concentration [
7,
8]. AMPs are mostly smaller in size (˂10 kDa), being cation-rich peptides that demonstrate a wide array of immunological and antimicrobial activities. Moreover, different
Bacillus bacteria can produce various peptides, including bacteriocins, glycopeptides, lipopeptides, and cyclic peptides [
9,
10,
11]. This provides a diverse collection of AMPs with numerous essential chemical compositions that gain their microbicidal action through the interference of the microbial membrane in combination with the cellular mechanisms of action. A few of them show properties that are concentration-dependent [
12,
13]. AMPs have good characteristics, and the production of antimicrobial peptides by
Bacillus genera is currently being portrayed. Numerous peptides generated by these bacteria have been extensively utilized in the fermentation process to manufacture antibiotics [
14,
15].
The production of an antimicrobial peptide by microbes is vastly reliant on the strains along with the nutritive and bacterial culture requirements [
16]. Culture media optimization is crucial since every substance that maintains bacterial evolution can be a prospective substrate [
17]. Thus, devising a suitable culture medium in addition to deciding the requirements for cultures are vital to increase the antibiotic yield [
18]. The production of antimicrobial peptides has been optimized by employing the response surface methodology (RSM), which belongs to a compilation of statistical procedures used for model development to define a combination of factor levels. It generates an optimum response under the influence of one or more independent factors such as media composition, pH, and incubation time [
19]. It applies a polynomial equation to discover the sort of connection between the elements and their significant and collective effects on the required response. Synergy has a valuable impact on the aspect of antimicrobial activity. We state that the synergy of a peptide enhances the activity, which demonstrates low potency and killing kinetics.
The capability of some microbes to develop a biofilm, which is an accumulation of microorganisms implanted within a self-formed base of components, complicates treatment, resulting in biofilm-covered bacteria being 10–1000 times more resistant to traditional antibiotics compared to their planktonic equivalents [
20,
21]. In the previous report, we observed short peptides generated by bacteria that reduced or destroyed other microbes by disrupting the preliminary phases of biofilm development [
22]. Biofilms have been recognized to contribute to physicochemical protection from antimicrobials. Besides that, the strength of antibiotics is dependent on eradicating the biofilm, which is required to destroy planktonic units [
23,
24]. Apart from antimicrobial resistance, bacteria in a biofilm similarly appear resistant to several physicochemical pressures, facilitating the preservation of bacteria in even the toughest of circumstances [
25]. Owing to the elevated resistance of biofilms to regular antibacterial agents, the first unconventional way to address this challenge is to use bacteriocins as antibacterials, individually or in combination [
26,
27]. Because their efficacy, as well as potency against bacterial biofilms are equivalent to traditional antibiotics, AMPs might be used as therapeutic options to tackle biofilms [
28].
In the current analysis, we intend to purify and characterize a novel antimicrobial peptide from Bacillus genera, which was isolated from the Korean fermented food kimchi, by optimizing the culture conditions with the response surface methodology. We tried to establish its antimicrobial, as well as antibiofilm ability, which may contribute to this area. The leading light peptide of the set, MS07, was assessed against the biofilm of Gram-positive and Gram-negative pathogens. Our discoveries implied that the cyclic lipopeptide MS07 could be established as a novel synthetic peptide under physiological conditions.
2. Materials and Methods
2.1. Reagents and Materials
The chemicals and solvents used for our study were extra pure grades. The Sephadex G-50 and DEAE-Sephadex A-50 columns were procured from Pharmacia (Uppsala, Sweden). Bacterial media Muller–Hinton (MH) and Man–Rogosa–Sharpe (MRS) were procured from Dickinson & co., Becton, sparks, MD, USA. Bacterial strain CBSMS07 was obtained from the Korean fermented food kimchi. All reagents used for experimental purposes were of analytical quality. Lipopeptide MS07 was dissolved in 5% DMSO (dimethyl sulfoxide) for in vitro experiments.
2.2. Bacterial Isolates and Nutrition Media
The bacterial strain CBSMS07, proficient in making AMP, was obtained from the Korean fermented food kimchi. The method of isolation for the bacterial strain was done following our earlier statement [
29]. About 1 g of kimchi was incubated at 37 °C for 24 h by mixing with 0.85% NaCl. Serial dilutions were executed, and steaking was performed to check the suitable CFU (colony-forming unit)/mL. The stock was mixed with glycerol (20%) and kept at -75 °C. The identification of CBSMS07 was performed based on the consistent morphology with Bergey’s Manual of Systematic Bacteriology. For further identification, the sequencing study of the 16S rRNA gene was executed [
30]. The optimization of culture media with different percentages of carbon and nitrogen sources (0.25%, 0.5%, 0.75%, 1%, 1.25%, 1.5%, 1.75%, and 2%) and different percentages of metal ion sources (0.005%, 0.0075%, 0.01%, 0.0125%, and 0.015%) for the bacterial strain was performed with various carbon sources (maltose, glucose, sucrose, lactose, fructose, starch, mannitol, sorbitol), nitrogen sources (peptone, tryptone, beef extract, yeast extract, malt, soymeal, oatmeal), and metal ion sources (MgCl
2, KCl, NaCl, CaCl
2, KH
2PO
4, Na
2HPO
4, ZnSO
4, FeSO
4) in different proportions. In contrast, MRS and MH were used as controls.
2.3. Bioprocess Design and Optimization by the Response Surface Methodology
The peptide MS07 obtained from bacterial strain CBSMS07 showed more significant activity with mannitol as the carbon source, peptone as the nitrogen source, and NaCl as the metal ion source. The Box–Behnken design was utilized to verify the highly efficient media arrangement amount by employing Design-Expert software (Version 12.0, Stat-Ease Inc., Minneapolis, MN, USA) [
31,
32]. By using this model, the most important independent variables, named A, B, and C, were incorporated, and each factor could be assessed at three different levels, low (−), the center point (0), and high (+), whereas bacteriocin activity (AU/mL) was the dependent variable (
Table 1).
In total, seventeen investigational runs were performed in a partial factorial model (
Table 2). The software provided the predicted bacteriocin activity (AU/mL). The impact of each significant variable on bacteriocin activity was determined statistically by higher than 95% (
p ˂ 0.05) confidence level.
The bacterial strain CBSMS07 was cultured in fully optimized media at 37 °C containing 1.773% mannitol, 1.249% peptone, and 0.012% NaCl (MPN media (mannitol, peptone, NaCl media)). Production was regulated in 2L Erlenmeyer flasks, including 400 mL culture media with constant rocking at 155 rpm. The antimicrobial compound was designated as MS07, produced by the strain CBSMS07.
2.4. Purification of Peptide MS07
Mass culture was carried out in MPN media with continuous shaking for 30 h at 37 °C. The supernatant was taken out by centrifugation (Supra 22k, Hanil Science industrial, Gimpo, Korea) (10,000× g) at 4 °C for 30 min. The salting out process was conducted by mixing diammonium sulfate (20–60%) with harvested culture supernatant with overnight stirring at 4 °C. The active precipitate was collected employing centrifugation at 10,000× g for 50 min at 4 °C, resuspended with buffer 10mM Tris-HCl (pH 7.4). Hereafter, ultrafiltration (Millipore co., Billerica, MA, USA) through the molecular cut-off (MWCO) membrane (10 and 1 kDa) was carried out for further column chromatography. The active fraction was loaded on a column Sephadex G-50 (2.5 × 85 cm) (Pharmacia, Uppsala, Sweden) next to the column DEAE-Sephadex A-50 (1.5 × 37 cm) using equal buffer. Further, active fractions were collected and concentrated, employing chloroform precipitation. Furthermore, the precipitate was subjected to the Pierce peptide desalting spin column (Thermo Scientific, Rockford, IL, USA) and analyzed for purity.
2.5. Tricine-SDS Polyacrylamide Gel Electrophoresis and Bioassay
Estimation of peptide MS07 concentration was done by the Bradford method [
33] by applying bovine serum albumin (BSA) as the standard. Hereafter, tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis (tricine-SDS-PAGE) [
34] was performed to evaluate the purity and molecular weight of peptide MS07. The gel was stained with Coomassie Brilliant Blue R-250 and destained with a mixture of distilled water, methanol, and glacial acetic acid (8:1:1). According to our previous report [
35], the in situ inhibitory activity was assessed against the indicator organisms (1.5 × 10
7 CFU/mL). The gel was rinsed with Tris-HCl buffer (50 mM/L; pH 7.4) containing Triton X-100 (2.5%) several times. Later, the gel was composed of agar (0.6%) on MH broth (Difco, Sparks, MD, USA) and incubated for 10 h at 37 °C.
2.6. Intact Molecular Weight Determination by MALDI-TOF
Molecular weight determination of MS07 was assessed through the MALDI-TOF MS (Matrix-Assisted Laser Desorption/Ionisation-Time of Flight Mass Spectrometry) method using Ultraflex III (Burker, Bremen, Germany). MALDI-TOF/MS assessments were done in the range of m/z 500–6000 and 2000–40,000 for both mass spectra.
2.7. Stability and Solubility of Peptide MS07
The thermostability of MS07 was analyzed by subjecting it to various treatments of temperature at 0, 10, 20, 40, 50, 60, 80, and 100 °C for 60 min and 121 °C/105 kPa for 15 min prior to evaluating the remaining activity. The samples were tested against the indicator microbial strain Micrococcus luteus ATCC 9341. Likewise, pH stability was checked against a wide array of buffers, for instance citric acid-sodium phosphate (pH 4.0–6.0), Tris-HCl (pH 7.0–9.5), and KCl-NaOH (pH 11.0–12.5). The solubility of MS07 was checked with different organic and inorganic compounds for which bacitracin was the control.
2.8. Antimicrobial Inhibitory Spectrum
To determine the antimicrobial inhibitory spectrum, the agar dilution method [
36] was applied to check the MIC (minimum inhibiting concentration) of peptide MS07. Bacitracin and vancomycin were taken as reference antibiotics to compare the MIC value of MS07. Twofold serial dilution was performed using 5% DMSO to check the MIC and minimum bactericidal concentration (MBC) of lipopeptide MS07. MIC was distinct as the reciprocal of the dilution that could reduce the noticeable growth of selected microorganisms (1.5 × 10
8 CFU/mL). MIC was carried out against both Gram-negative and Gram-positive microorganisms. The tested microorganisms were applied to the MH-agar medium containing different concentrations of peptide MS07 and reference antibiotics, further incubated at 37 °C. Gram-negative bacteria:
A. faecalis ATCC 1004,
E. coli KCTC 1923,
S. typhimurium KCTC 1925, Extended-spectrum beta-lactamase V4
(Escherichia coli), Extended-spectrum beta-lactamase 31
, Extended-spectrum beta-lactamase W
1, Pseudomonas aeruginosa KCTC 1637; Gram-positive bacteria:
B. subtilis ATCC 6633,
E. faecalis ATCC 29212, MRSA B15,
M. luteus ATCC 9341,
M. smegmatis ATCC 9341,
S. aureus KCTC 1928, VRE 4. MBC was obtained by using the broth dilution method. MBC was assessed as the minimum concentration inhibiting the bacterial growth entirely (≥99.9%) [
37].
2.9. Time-Kill Kinetics Assay
The activities of peptide MS07 were investigated in time-kill studies tested with
Escherichia coli cells in the log (Log
10) growing phase [
38]. Concentrations half the MIC, equivalent to MIC, two times the MIC, four times the MIC, and eight times the MIC of the peptide were made. Furthermore, an inoculum volume of 5.0 × 10
5 CFU/mL was selected and incubated at 37 °C. One milliliter aliquots of the medium were prepared with different time intervals of initial, two, four, six, eight, and twenty-four hours for bacteria and inoculated into 10 mL of nutrient medium. Incubation was performed at 37 °C for 24 h. The colony forming unit (CFU) of the bacteria was determined. The growth control (GC) test was performed for the organisms without a reference antibiotic or peptide. The graph of the log
10 (CFU/mL) was plotted against the growth time.
2.10. Synergism of Peptide MS07 with Melittin
The synergistic effect between the peptide MS07 and melittin was evaluated by the combination assay [
39]. It is noteworthy that for the synergism assay, twofold dilutions of MIC concentration samples were prepared against the pathogenic microorganism
Escherichia coli. The synergistic antibacterial activity against
E. coli was tested at half of the MIC and the MIC concentration of the peptide MS07 with half of the MIC and the MIC concentration of melittin. The study was also conducted in the absence of melittin as the control.
2.11. Antibiofilm Properties of Peptide MS07
The antibiofilm activity of Gram-positive bacteria
Staphylococcus aureus KCTC 1928 and Gram-negative bacteria
Escherichia coli KCTC 1923 and
Pseudomonas aeruginosa KCTC 1637 was assessed with ampicillin and oxacillin in combination with MS07. Bacteria cells (100 µL) were positioned in 96 well TCPs (tissue culture plates) in MH broth accompanied with glucose (0.2%). The plate was incubated at 37 °C for 24 h. The solution, not including cells, was treated as the control. The culture was removed, cleaned with PBS, and resuspended in 100 µL methanol (100%). Further, the culture media was incubated for 15 min to calculate the biofilm formed. After the elimination of methanol, biofilm development was measured by crystal violet (0.4%) staining for 30 min. It was washed with distilled water (DW), and the well plate was thoroughly air-dried. Absorbance was measured at 620 nm by adding 100% ethanol to each well [
35]. The experiment was carried out in triplicate to validate the process.
2.12. Viability Assessment of Biofilm Cells with MS07
The viability test of the preformed biofilm cells with peptide MS07 was investigated on
P. aeruginosa KCTC 1637 and
E. coli KCTC 1923 at sub-inhibitory concentrations [
40]. In brief, into a 96 well plate was poured 600 µL of 1.0 × 10
6 CFU/mL in tryptic soy broth (TSB), and incubation was performed at 37 °C for 24 h. The medium was separated, and well plates were soaked two times with PBS (pH 7.5). Peptide MS07 (200µL of various MIC) was dissolved in TSB. After inoculation at 37 °C for 12 h, the supernatant was removed, and two-hundred microliters of PBS were mixed with 50 µg of methyl-thiazol-tetrazolium (MTT). Afterward, the viability assay was established upon the development of the insoluble purple formazan from a decrease of MTT by the respiratory reduction of the bacterial cells. The crystal form of formazan was suspended in DMSO. The absorptions were taken at 570 nm with a microplate reader. Three separate tests were conducted for each concentration.
To determine the minimum biofilm eradication concentration (MBEC), Gram-positive bacteria
M. luteus ATCC 9341 and Gram-negative bacteria
P. aeruginosa KCTC 1637 and
E. coli KCTC 1923 were used. A 96 well revival microtiter plate was used, like every well, including 200 µL TSB medium [
41]. The microplate was closed and incubated for 4 hours at 37 °C. The bacteria properly showed regrowth of planktonic cells from the persisting biofilm. Afterward, the minimum biofilm eradication concentration (MBEC) was described as the lowest peptide intensity stopping the regrowth of bacteria from the biofilm. The minimum bactericidal concentration on the biofilm (MBCb) was described as the minimum strength of peptide that was essential to reduce viable biofilm cell numbers by 99.9% (≥3 log
10).
2.13. Inhibition of Bacterial Biofilm Imaging by Confocal Microscopy
Confocal laser scanning microscopy was used for imagining the formation of bacterial biofilm (
Pseudomonas aeruginosa) in the absence and presence of peptide MS07.
Pseudomonas aeruginosa (1.5 × 10
8 CFU/mL) was incubated in a 96 well microplate or three well chamber slides per well under microaerobic conditions for three days to grow the biofilm. Then, the supernatants were aspirated and treated with different MIC concentrations of MS07 for 48 h. After the incubation period, the resident biofilm was measured under flood cells and stained employing the Live/Dead BacLight viability stain (Life Technologies Corporation), visualized using a confocal microscopy process [
42].
3. Results
3.1. Strain Identification and Culture Media
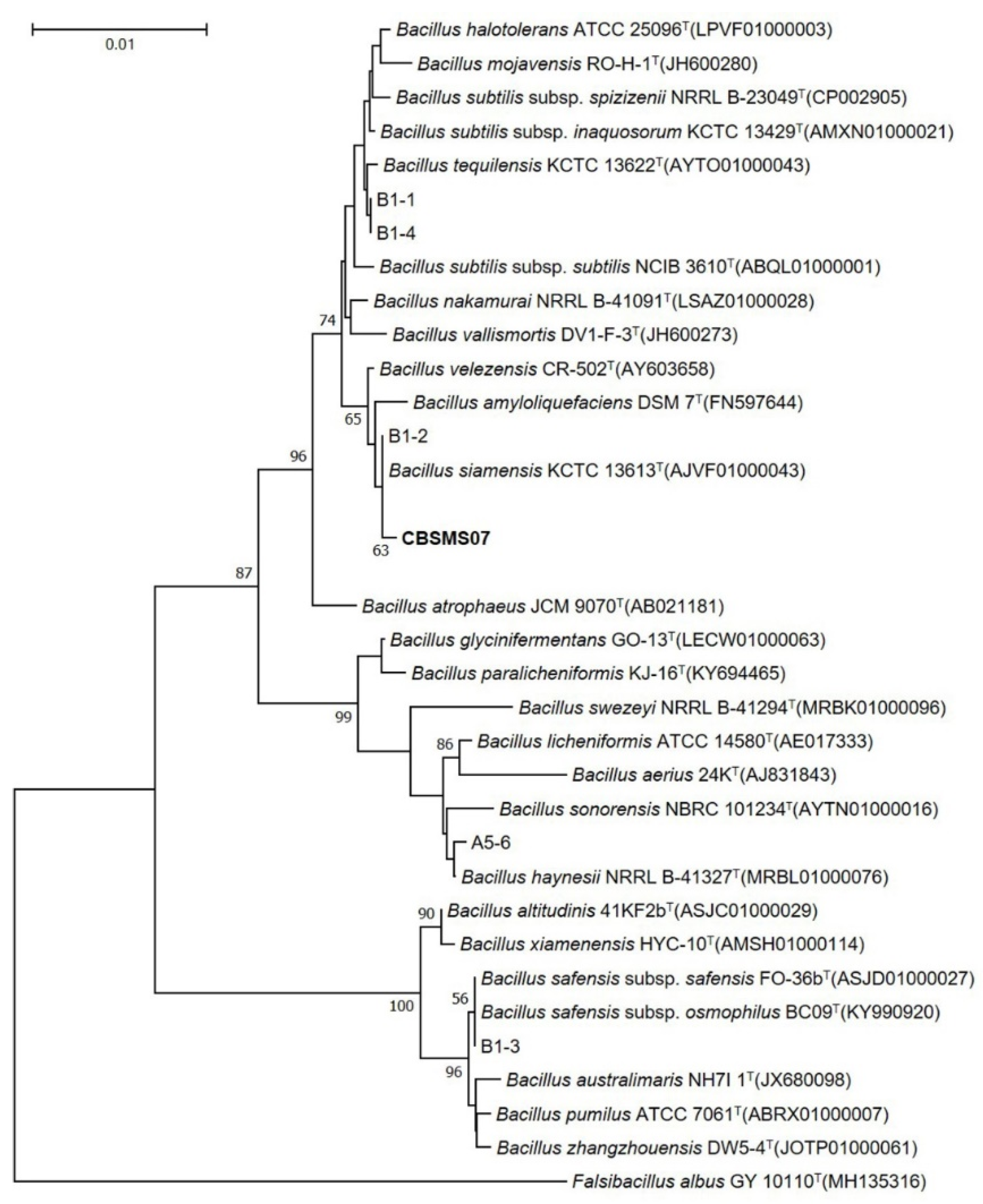
Bacterial strains proficient in producing AMPs were collected and isolated from the Korean fermented food kimchi. The strain CBSMS07 presented a high degree morphological resemblance to the
Bacillus strain. The 16S rRNA sequence of the isolate showed the closest character to
Bacillus siamensis KCTC 13613
T (Accession No. AJVF01000043) with a pairwise similarity of 99.9%. A phylogenetic tree constructed from the 16S rRNA sequence analysis is shown in
Figure 1. The optimization of nutritional media was carried out with numerous carbon sources (0.25%–2%), nitrogen sources (0.25%–2%), and metal ion sources (0.005%–0.015%) for antimicrobial peptide (AMP) MS07, which was produced by bacterial strain
B. siamensis CBSMS07. Among all the nutritional sources, one-point-seven-seven-three percent mannitol, 1.249% peptone, and 0.012% NaCl were the most significant carbon source, nitrogen source, and metal ion source, respectively. Antimicrobial peptide MS07 was grown on optimized media at 37 °C for 30 h at 160 rpm. The culture conditions showed a significant effect on producing bacteriocin.
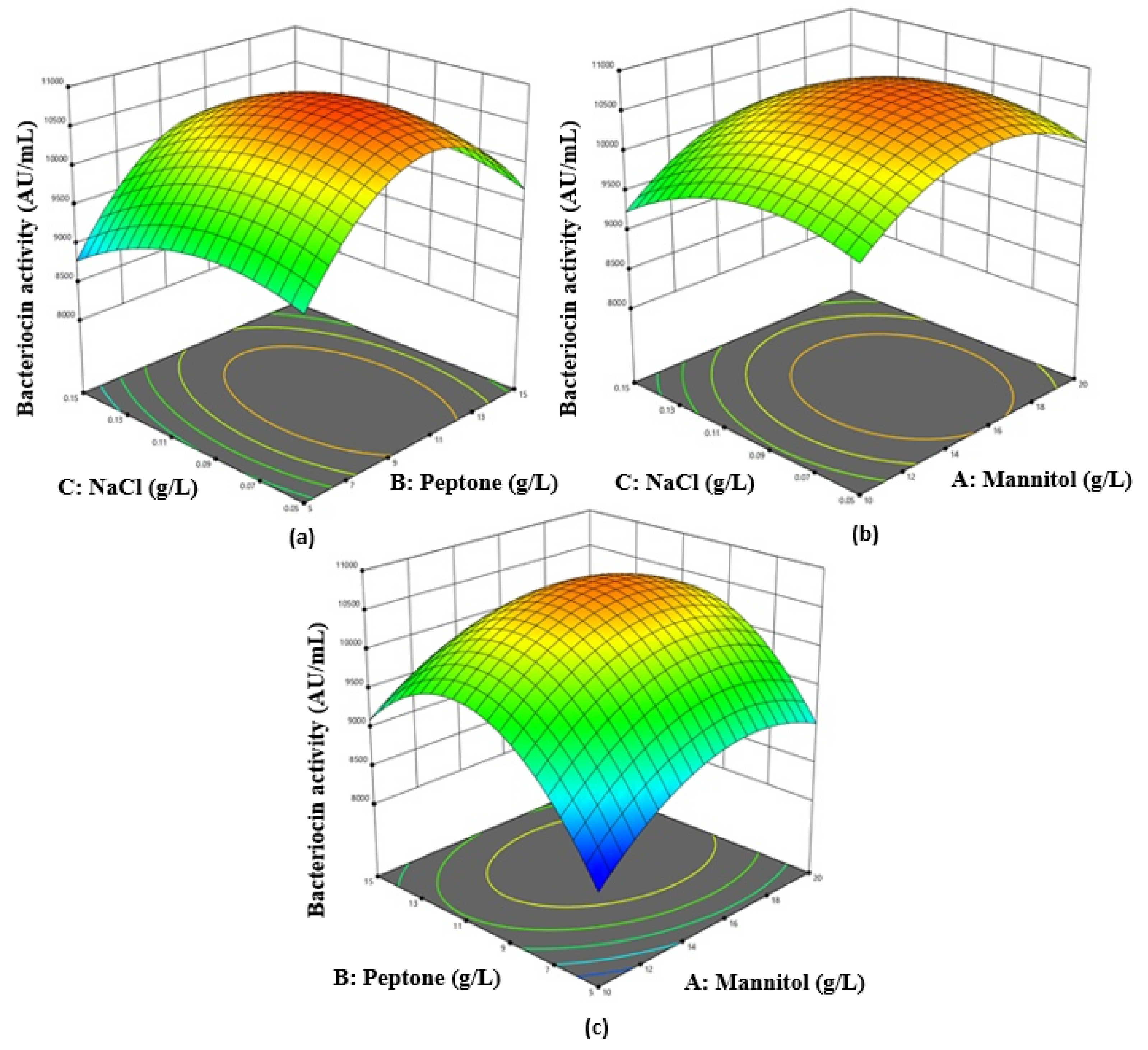
3.2. Box–Behnken Design and Response Surface Analysis
Corresponding to the theory of RSM, the higher amounts obtained from the levels of the three independent variables mannitol (A), peptone (B), and NaCl (C) were further examined each at three distinct levels. The bacteriocin activity differed significantly from 8370 to 10,990 AU/mL at various concentrations of several elements in the culture medium. The fitness of bacteriocin activity was calculated by the ANOVA test. From the ANOVA test, the straight terms A, B, and C and interface terms AC, AB, and BC were observed to be statistically significant (
p ˂ 0.05) by the analysis of bacteriocin action. After evaluation of the data, we found that the interactions of AB, AC, and BC matched the maximum peak for bacteriocin activity appearing at a NaCl concentration of 0.0123 g/L, a mannitol concentration of 1.773 g/L, and a peptone concentration of 1.249 g/L, where the gradual increase in their level resulted in higher bacteriocin activity being achieved (
Figure 2). By employing multiple regressing evaluations to the investigational data, the ultimate equation to describe bacteriocin activity is:
Bacteriocin activity = −1857.50 + 795.80A + 1002.70B + 23620.00C − 5.20AB + 160.00AC + 200.00BC − 23.46A2 − 44.06B2 − 1.626 × 105C2
Accordingly, the match of the model was conveyed by coefficient determination (R2) as exponential, which was determined to be 0.92, which was closer to 1.00 to predict a better response.
3.3. Purification and Molecular Weight Determination of MS07
MS07 was cultured in improved media at a temperature of 37 °C with constant shaking. The purification of MS07 from the supernatant of CBSMS07 (ammonium sulfate; 20–60% saturation) is reviewed in
Table 3. Peptide MS07 was purified to uniformity by two phases of chromatographic partition on the Sephadex G-50 (
Figure 3a) and the DEAE-Sephadex A-50 column (
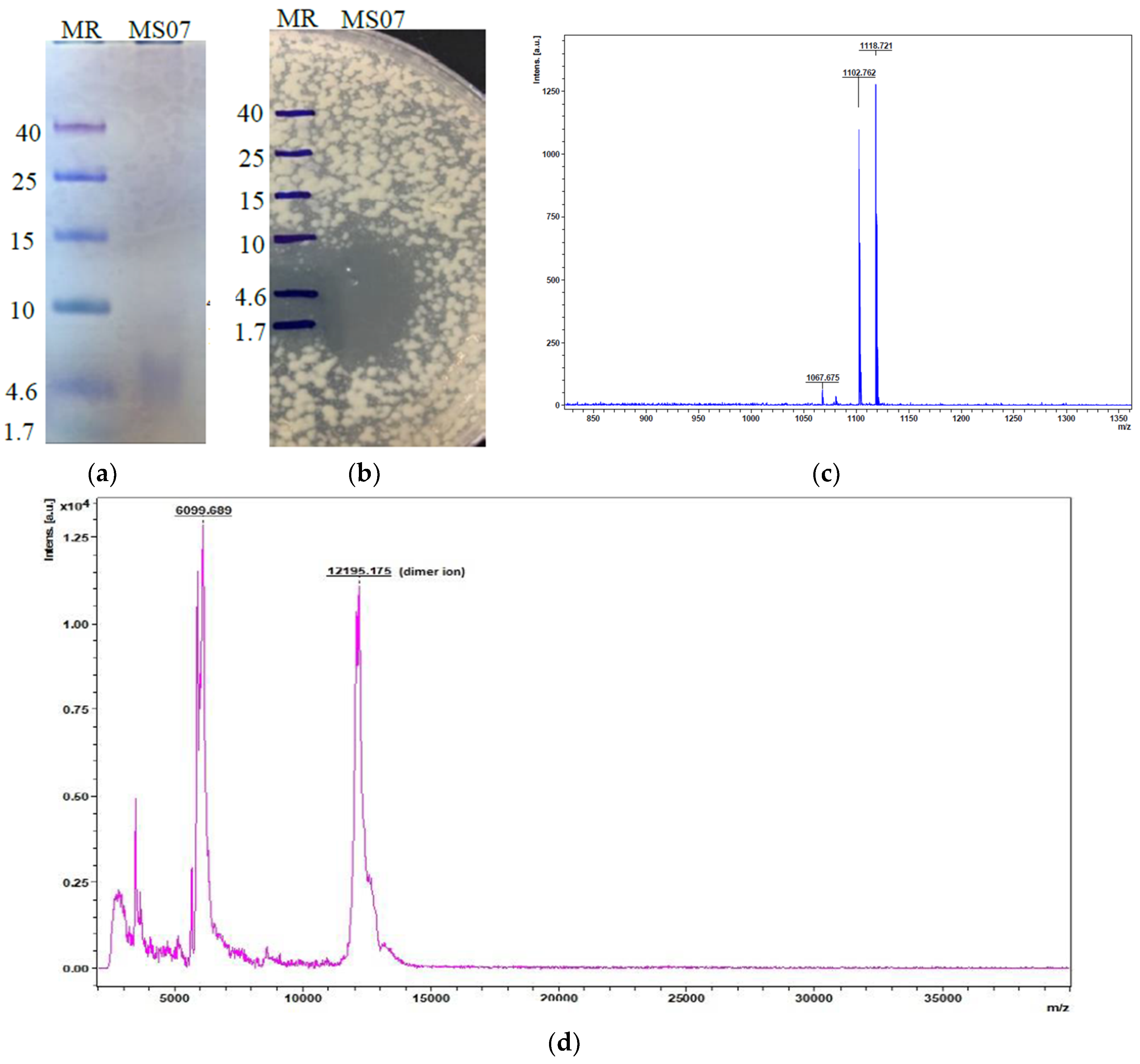
Figure 3b). The active fractions were collected based on protein concentrations and antimicrobial activities. Peptide MS07 was purified with a total activity of 27,000 AU with 27.77-fold and 7.71% activity revival. Tricine-SDS-PAGE gel electrophoresis assessment showed a single band protein of MS07 relating to a molecular weight of 6.099 kDa (
Figure 4a). The protein band was proven to be an AMP by the bioassay activity of the SDS-PAGE gel, showing a clear lysis zone in the MH agar plate in the same region against
Mycobacterium smegmatis (
Figure 4b).
3.4. Molecular Weight Determination by MALDI-TOF
MALDI-TOF assessed the molecular weight of MS07 in the acquisition mass range of 500 Da~40 kDa. The molecular weight of peptide MS07 was = 6099.689 Da (
Figure 4d), which showed a single major peak (n + 1). Rather than a significant peak, it showed some minor detected peaks also, which had similar masses to lipopeptide (fatty acid) compounds. It had low-intensity peaks at m/z = 1067.675, 1102.762, and 1118.721, corresponding to the C13, C14, and C15 molecules (
Figure 4c), which were observed in the spectra of the sample by alignment with standard units. The peak at m/z = 1067.675 that related to potassium adduct iturin A C13 was highly plentiful as a fatty acid chain in the spectrum. The peak at m/z = 1102.762 corresponded to the sodium adduct fengycin A C14, which also resembled a fatty acid chain. We found a peak at m/z = 1118.721, corresponding to puwainaphycin F analogs.
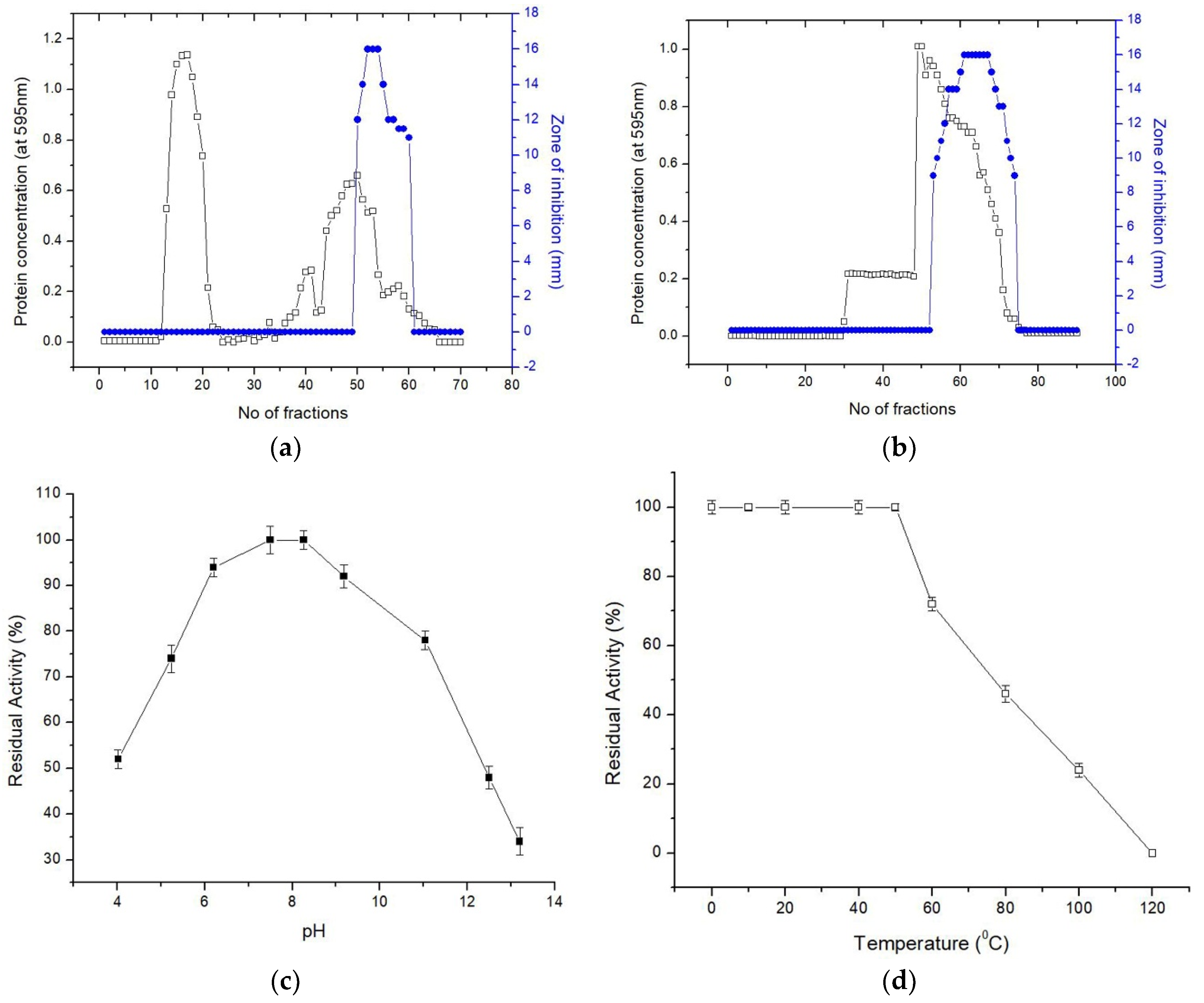
3.5. Stability and Solubility Study of MS07
To verify the thermal stability of MS07, the remaining activity was checked according to the retained antimicrobial activity. MS07 remained stable up to 50 °C (
Figure 3d). Although its activity was gradually decreased after incubation for 60 min, it retained about 72% of the activity at 60 °C. Complete loss of activity occurred in the autoclave environment at 121 °C and 105 kPa for 15 min. The ability to retain the activity of MS07 across a broad array of pH values could be the consequence of the mutable protein denaturation. To analyze the stability of pH, peptide MS07 was incubated in different pH buffers (
Figure 3c). It was observed that MS07 endured a broad array of pH values (4.02 to 11.04). MS07 showed a broad range of pH and temperature stability.
Peptide MS07 was discovered to be soluble in distilled water, methanol, ethanol, acetone, chloroform, and butanol, whereas it was not soluble in ethyl acetate, acetonitrile, and n-hexene.
3.6. Antimicrobial Spectrum, Time-Kill Kinetics Assay, and Synergistic Effect of MS07
The antimicrobial spectrum of peptide MS07, established in terms of MIC, was strongly effective against Gram-negative bacteria (MIC of 16–32 µg/mL). It was relatively effective against Gram-positive bacteria (MIC of 16–128 µg/mL). We compared the MIC of our recently purified peptide MS07 to that of bacitracin and vancomycin. Peptide MS07 had a stronger effect than commercial bacitracin and vancomycin on various Gram-negative pathogens such as Alcaligenes faecalis, Escherichia coli, Salmonella typhimurium, Extended-spectrum beta-lactamase (V4, 31, W1) (E. coli), and Pseudomonas aeruginosa, as well as some Gram-positive pathogens such as MRSA B15, Micrococcus luteus, Bacillus subtilis, and Mycobacterium smegmatis. The minimum bactericidal activity (MBC) of MS07 against various pathogens was 320–1280 µg/mL, showing high MBC activity.
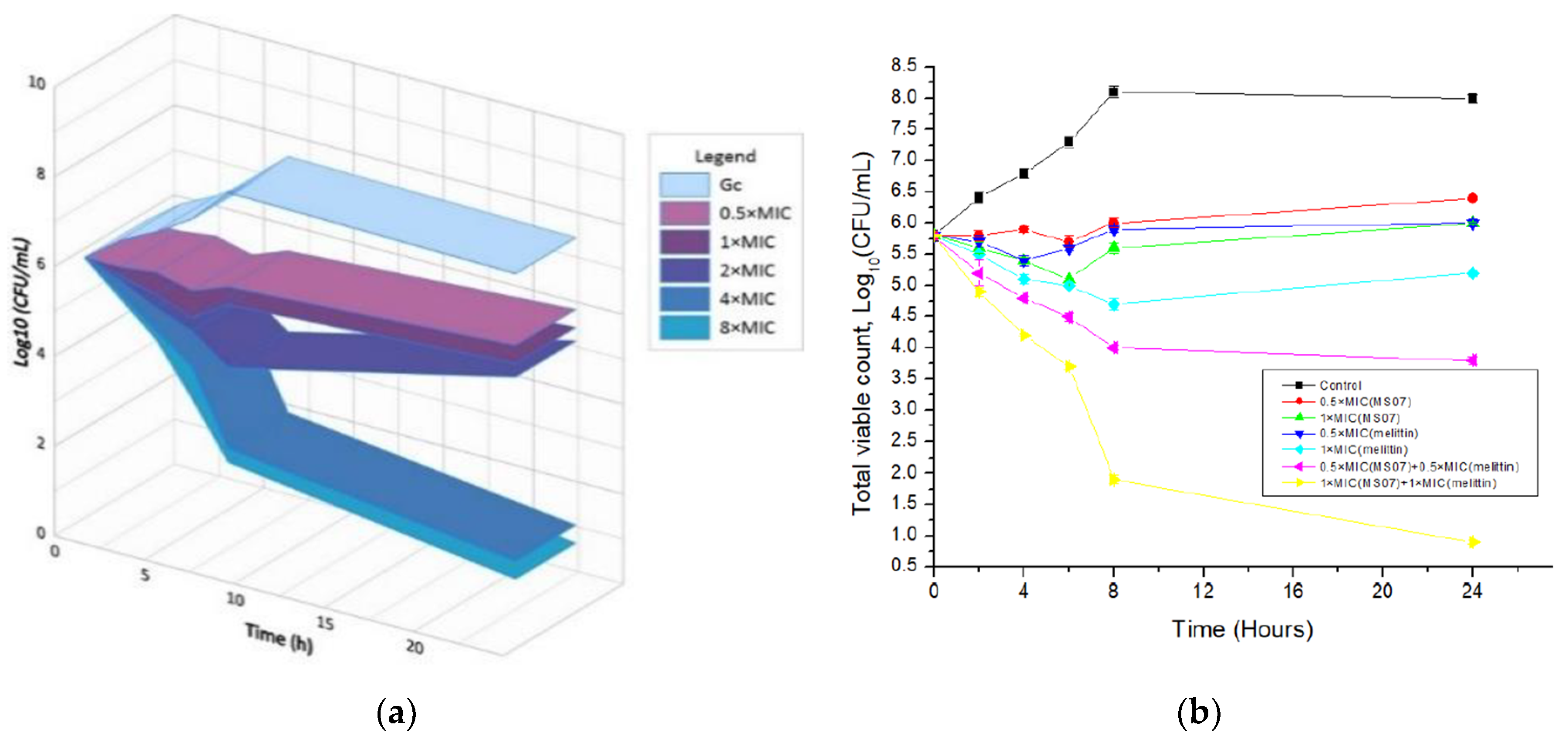
The time-kill kinetics report of peptide MS07 against the test organism
E. coli at the tested concentrations demonstrated a decline in the count of viable cells around the initial 2, 4, 6, 8, and 24 hours, correspondingly afterward and up to the 24 h for
E. coli (
Figure 5a). MS07 was tested for its antimicrobial activity using colony count assays. At the MIC level (32 µg/mL), MS07 killed bacteria up to 6 h. At low peptide concentrations, an initial decline in the count of surviving
E. coli cells was noticed, whereas, after 6–8 h,
E. coli cells surviving the peptide action started growing again. Regrowth was observed up to 2 × MIC, whereas 4 × MIC and 8 × MIC showed a complete reduction of the bacterial count.
To visualize the synergistic effect of MS07 and melittin,
E. coli cell suspensions were spread on the MH agar plate. The result showed that the combination of peptide MS07 and melittin had intense synergistic antibacterial activity against
E. coli (
Figure 5b). This indicated a potentially valuable synergistic effect compared to the peptide MS07 applied alone.
3.7. Antibiofilm Activity of Peptide MS07
For biofilms, bacteria develop as multicellular accumulates inside an extracellular substance, which shields against the host’s defense cell. MS07 was assessed for its antibiofilm activity against Gram-positive and Gram-negative bacteria. Our result demonstrated that biofilm development was significantly diminished when treated in combination with the referenced antibiotics. Therefore, the ability of MS07 to inhibit biofilm development might be more efficient more in combination with ampicillin or oxacillin. So a combination of peptide MS07 and other antibiotics, could represent an applicable therapeutic. The ANOVA test was conducted to compare the mean, p ˂ 0.05, with the result.
3.8. Viability Assays of Biofilm Cells Treated with MS07
We studied the anti-biofilm action of peptide MS07 on
Pseudomonas aeruginosa KCTC 1637 and
Escherichia coli KCTC 1923 at sub-inhibitory strengths. As shown in
Figure 6b, after employing peptide MS07 against the biofilms, the biomass was diminished with rising strengths (1 × MIC to 8 × MIC). The MIC concentration was decreased by about 15% and 11% for the
P. aeruginosa and
E. coli biofilms. The viability of
Pseudomonas aeruginosa and
Escherichia coli biofilms was decreased after 24 h at an elevated concentration of peptide MS07. A comparison of the means to the untreated control was done using the ANOVA test, ***
p ˂ 0.001, **
p ˂ 0.01, *
p ˂ 0.05. Experiments were done in triplicate.
MS07 was checked for anti-biofilm activity against Gram-positive strains (
M. luteus ATCC 9341) as well as Gram-negative strains (
E. coli KCTC 1923 and
Pseudomonas aeruginosa KCTC 1637). The MBECs of the peptide MS07 showed higher anti-biofilm activity against Gram-positive strain
Micrococcus luteus (MBEC, 1.5 µM), which was consistent with its MIC concentrations (
Table 4). MS07 exhibited MBCb on biofilms of 6 µM and 8 µM, respectively, against the Gram-negative
P. aeruginosa and E. coli biofilms, respectively.
3.9. Anti-Biofilm Action of Peptide MS07 Determined by Confocal Microscopy
Consistent with the antimicrobial activity, peptide MS07 reduced and interrupted the biofilm formation in a concentration-dependent manner, as established by a BacLight live/dead stain (
Figure 6c). Confocal microscopy images explained the formation of the biofilm on the slide after shaking for three days anaerobically. The plane surface of the image of
P. aeruginosa biofilm exhibited a distinct structure with green fluorescence. Treatment with MS07 significantly reduced biofilm formation in a concentration-dependent manner. This showed that peptide MS07 not only had bactericidal activity, but also could disrupt the biofilm.
4. Discussion
Our study aimed to purify and characterize a novel peptide MS07 and study the prospect of its potential application for treatment. We performed an initial investigation of different purified AMP [
20,
21,
22,
35] for antimicrobial and antibiofilm activity.
Bacillus strain CBSMS07 was obtained from a traditional Korean food (kimchi). Based on morphological factors and 16S rRNA gene sequences (
Figure 1), the bacterial strain was detected. Purification of peptide MS07 was performed using the process of sequential gel filtration, causing the elution of the peptide, which was free of undesirable proteins and retaining vigorous antimicrobial activity. In our present study, we checked the culture medium composition on AMP production compared with available media in the market, MH, and MRS broth. Here, we determined the optimized media by the response surface methodology with three different independent variables’ results and fixed the growth media (1.773% mannitol, 1.249% peptone, and 0.012% NaCl) for large-scale production.
Peptide MS07 was in the category of antimicrobial peptides that are quite small in size (˂10 kDa) and demonstrated a wide range of activities against several microorganisms. MS07 was lipophilic, and it moved towards the lipophilic solutions during the solubility check. The peptide was purified 27.77-fold, with a recovery of 7.71%. The purified MS07 had a lower molecular weight (6.099 kDa), which was detected by MALDI-TOF, compared to other bacteriocin purified from Bacillus strains [
43,
44]. The Edman degradation method was not effective at determining the N-terminus because it was blocked in the Edman reaction by the modification of its cyclic structure. MS07 was displayed to be active against both Gram-negative and Gram-positive bacteria, as shown in
Table 5. The salinity of the buffer was crucial because it was a measured factor when the peptide was used to make a pharmaceutical drug for oral and systemic use. The peptide was very stable at pH 4–11 and up to 50 °C, which was similar to the result of other peptides from different
Bacillus strains [
35,
45]. The antimicrobial effect of MS07 against both Gram-positive and Gram-negative bacteria was comparable with the action of bacitracin and vancomycin, as presented in
Table 5. Although the lytic activity of peptide MS07 was seemingly more active against Gram-negative bacteria than Gram-positive bacteria, the result is more effective than other peptides produced from
Bacillus sp. isolated from kimchi [
43,
46]. In vitro, the microdilution method showed comparable results providing real-time data on bacterial growth by continuous recording of bacterial production. We observed the regrowth of
E. coli after 6 h of incubation until the concentration was 2 × MIC, but did not see regrowth at a 4 × MIC and an 8 × MIC concentration. The result of the time-kill kinetics was compared to another antimicrobial peptide [
47]. Investigation of the mode of action of peptide MS07 showed its activity by causing the development of transmembrane openings dependent on both AMP and melittin concentration, as both peptides indicated a relationship between their mechanisms of action [
48]. Melittin alone showed low activity compared to a combination of MS07 with melittin, which showed increased activity against bacteria by disrupting the function of the lipid membrane throughout pore formation. Due to the probable similarity of the mechanisms, a synergy of action was found in combination with melittin.
Biofilms are significantly resistant to clinical treatment by conventional antibiotics [
49,
50] and are a vital aspect of the pathogenicity of bacteria associated with chronic diseases like respiratory infections. Besides, their susceptibility concerning antibiotics decreases when they are present in biofilms. The search for new AMP that eliminate bacterial biofilms has, thus, turned out to be exceedingly pressing. In this study, the peptide MS07 was efficient at preventing the development of
P. aeruginosa and
Escherichia coli biofilms and diminishing the viability of already developed biofilms. It was more effective when it was applied in combination with oxacillin and ampicillin. The study exhibited that 256 µg/mL AMP MS07 resulted in a 58% reduction in
Pseudomonas aeruginosa biofilm formation and a 64% decrease in
Escherichia coli biofilm formation. This process was consistent with the earlier findings that the reduction of biofilm formation by the peptide increased with its concentration [
51]. The capacity of peptide MS07 to reduce biofilm development was related to a reduced count of microbial cells on the surface. Peptide MS07 could eradicate the biofilm cells at a high concentration with potent antibacterial activity. The concentration of MS07 that destroyed 99.9% of the biofilm cells was tested against both Gram-positive and Gram-negative strains. Moreover, MS07 was shown to inhibit biofilm growth in vitro with its mechanism of action. In our study, we identified that the application of MS07 prompted a deficit of green fluorescence that was highly significant, accompanied by the cell cluster edge, which was consistent with other AMP [
51]. In preventing the development of
P. aeruginosa and
Escherichia coli biofilms by 58% and 64%, respectively, with the concentration of 8× MIC for, it was found that the cyclic lipopeptide MS07 was more than 50% effective at disrupting biofilm formation.