Phenol Degradation Kinetics by Free and Immobilized Pseudomonas putida BCRC 14365 in Batch and Continuous-Flow Bioreactors
Abstract
1. Introduction
2. Model Description
2.1. Model Assumptions
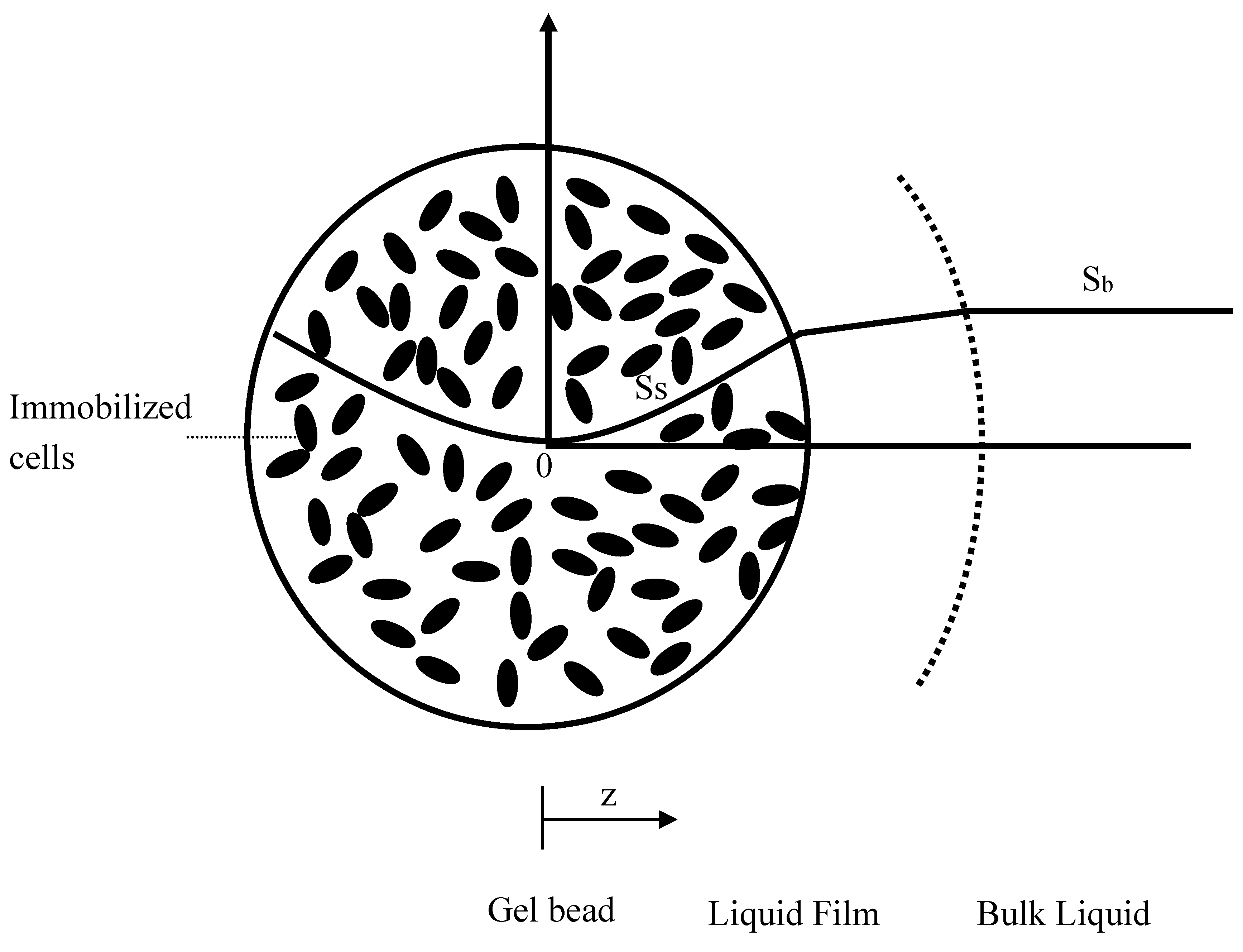
2.2. Kinetic Model in the Immobilized Cells
2.3. Model Numerical Solution
2.4. Evaluation of Biokinetic Parameters
2.5. Mass Transfer Coefficients
3. Materials and Methods
3.1. Chemicals
3.2. Cultivation of Pseudomonas putida Cells
3.3. Batch Experiments
3.4. Immobilization Protocol
3.5. Analysis of P. putida Cells and Phenol
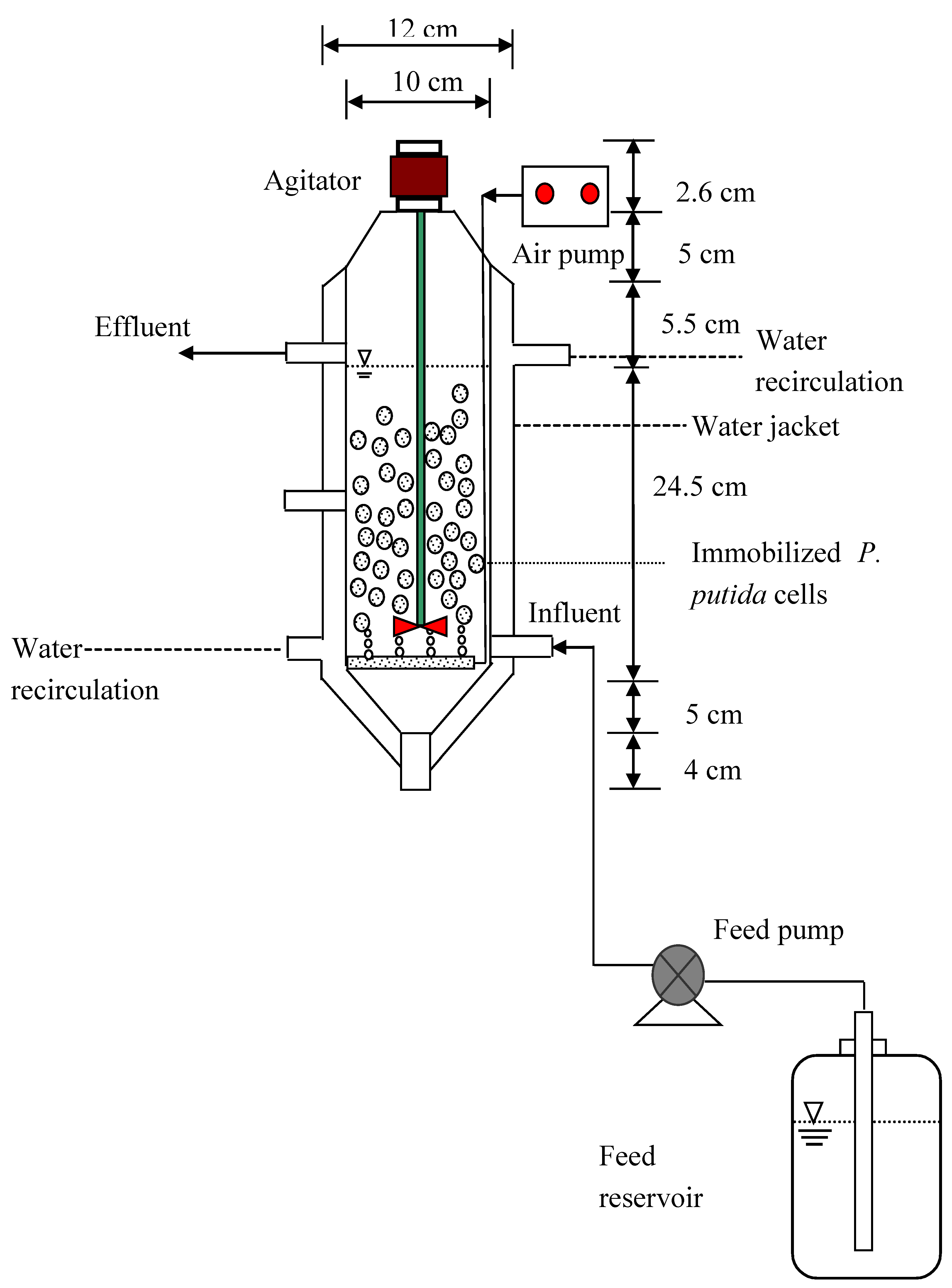
3.6. Bioreactor Setup
4. Results and Discussion
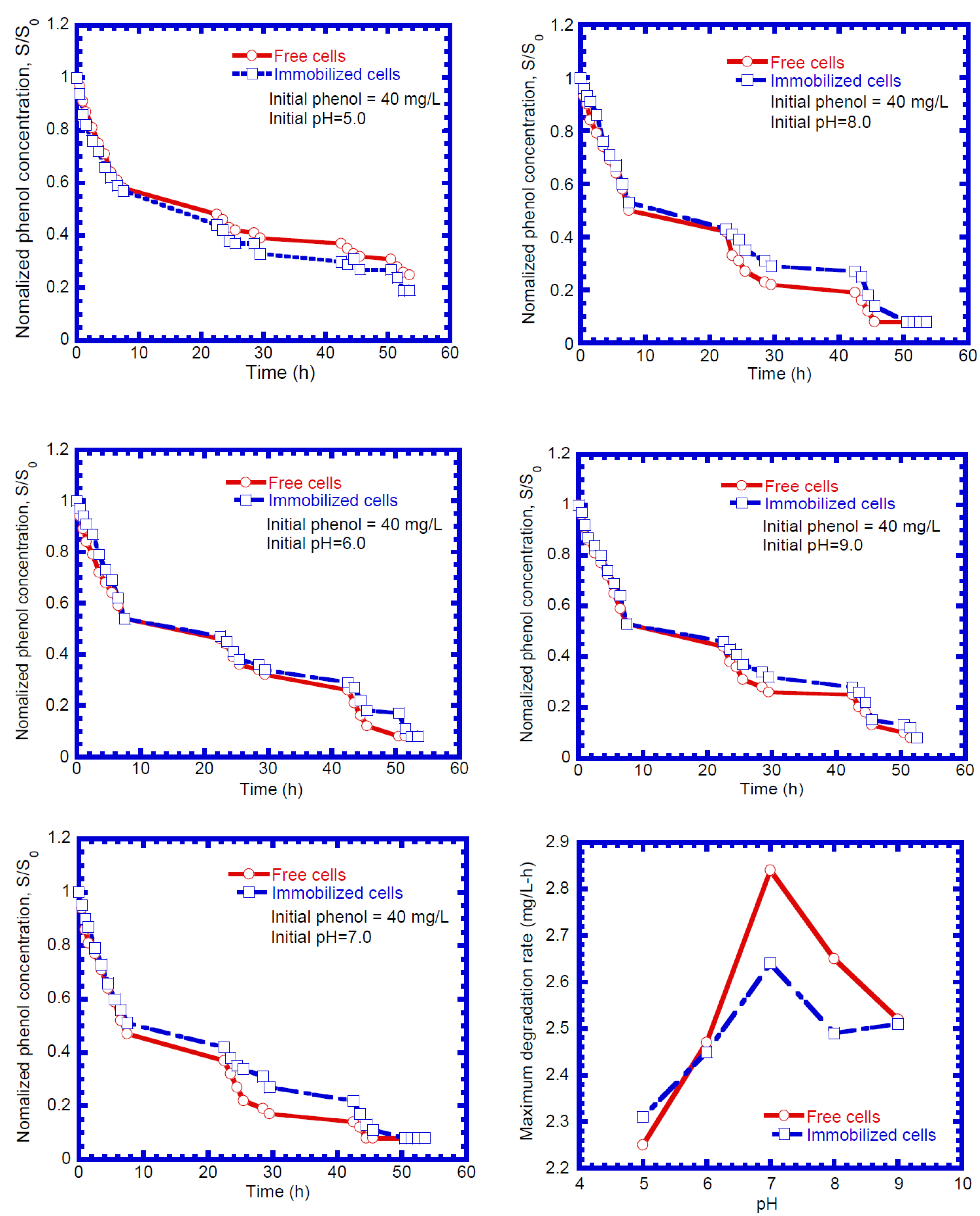
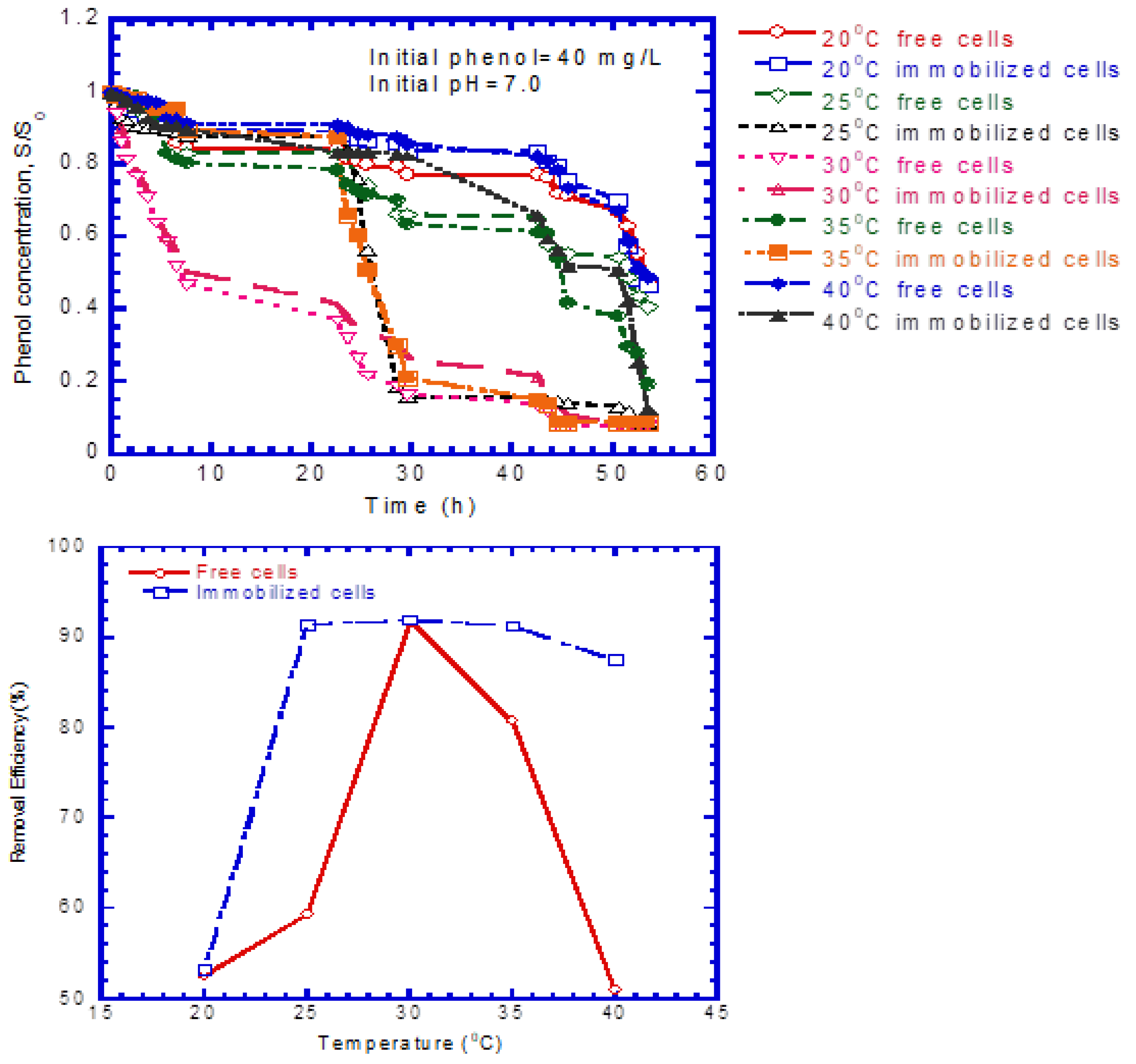
4.1. Phenol Degradation by Free and Immobilized Cells
4.2. Batch Experiments for Free Cells
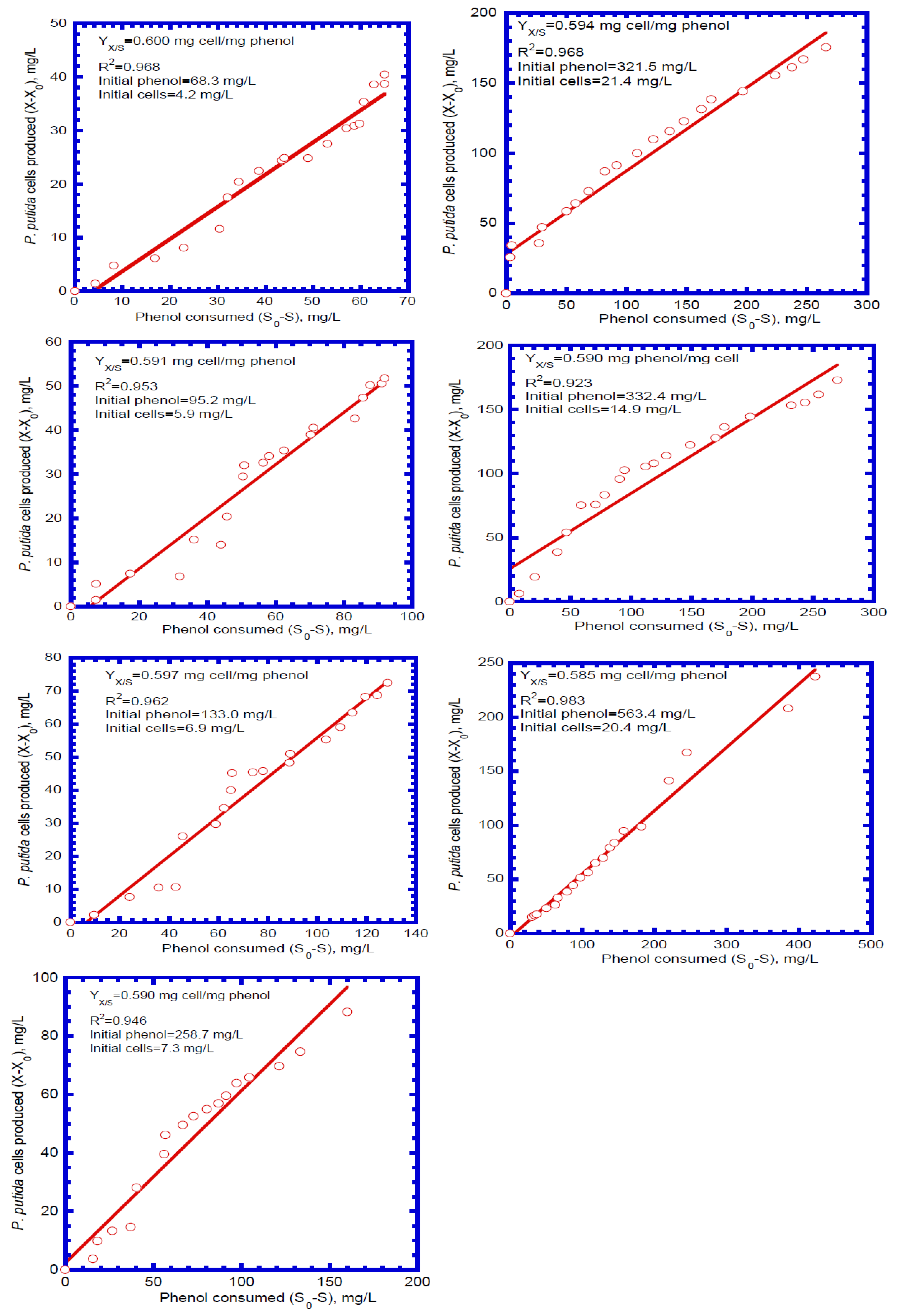
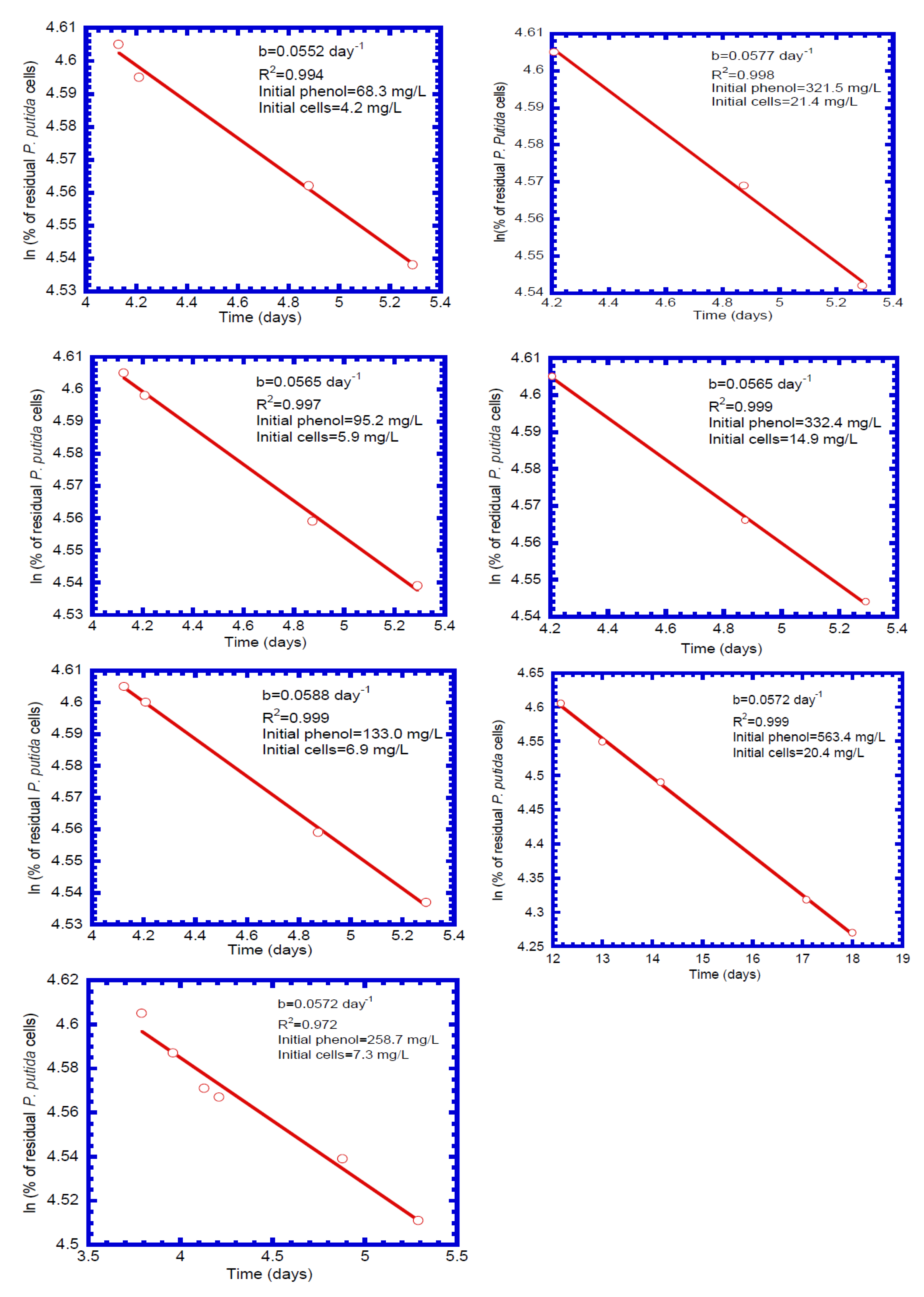
4.3. Determination of Biokinetic Parameters
4.4. Determination of Mass Transfer Coefficients
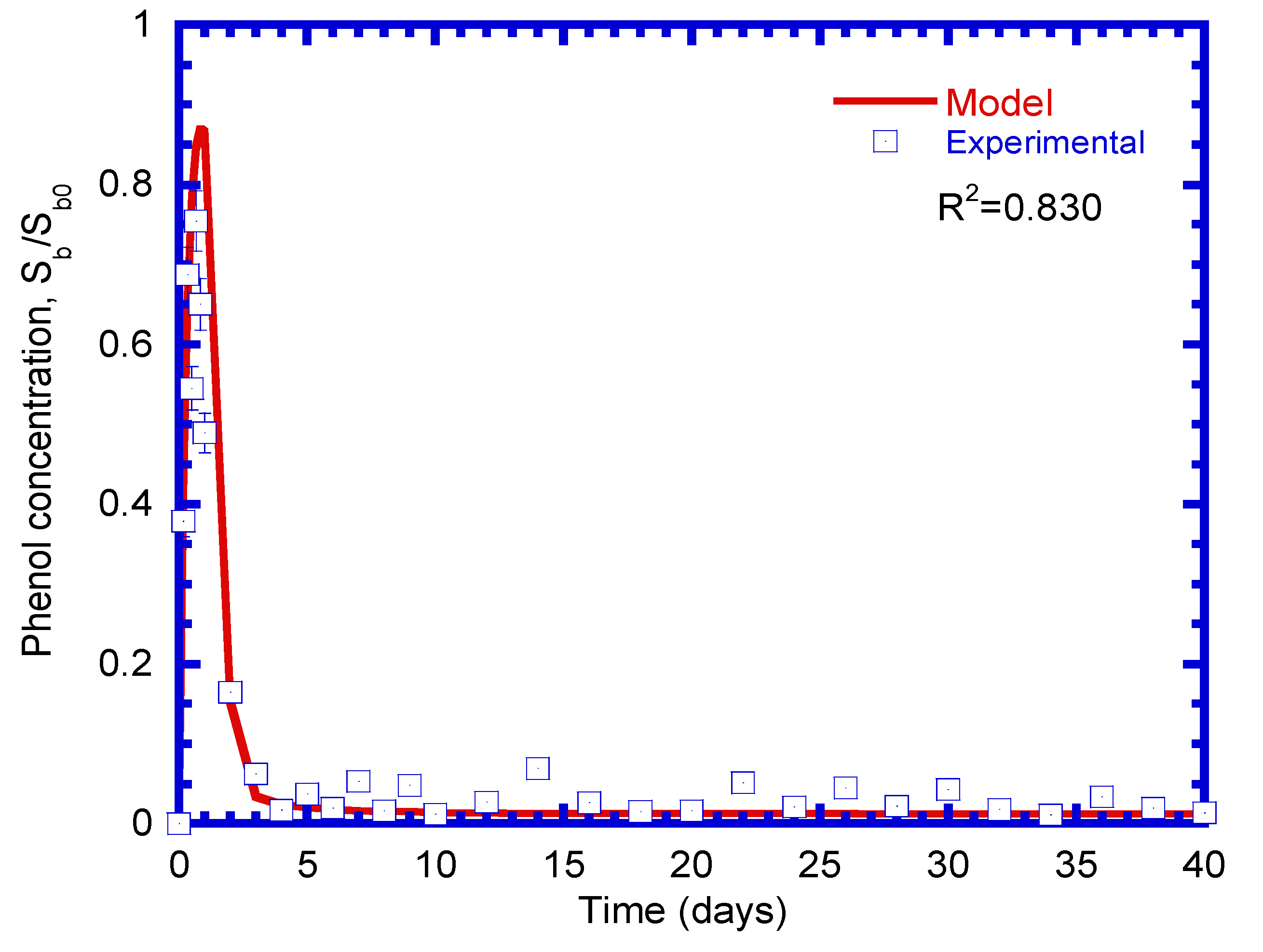
4.5. Phenol Biodegradation
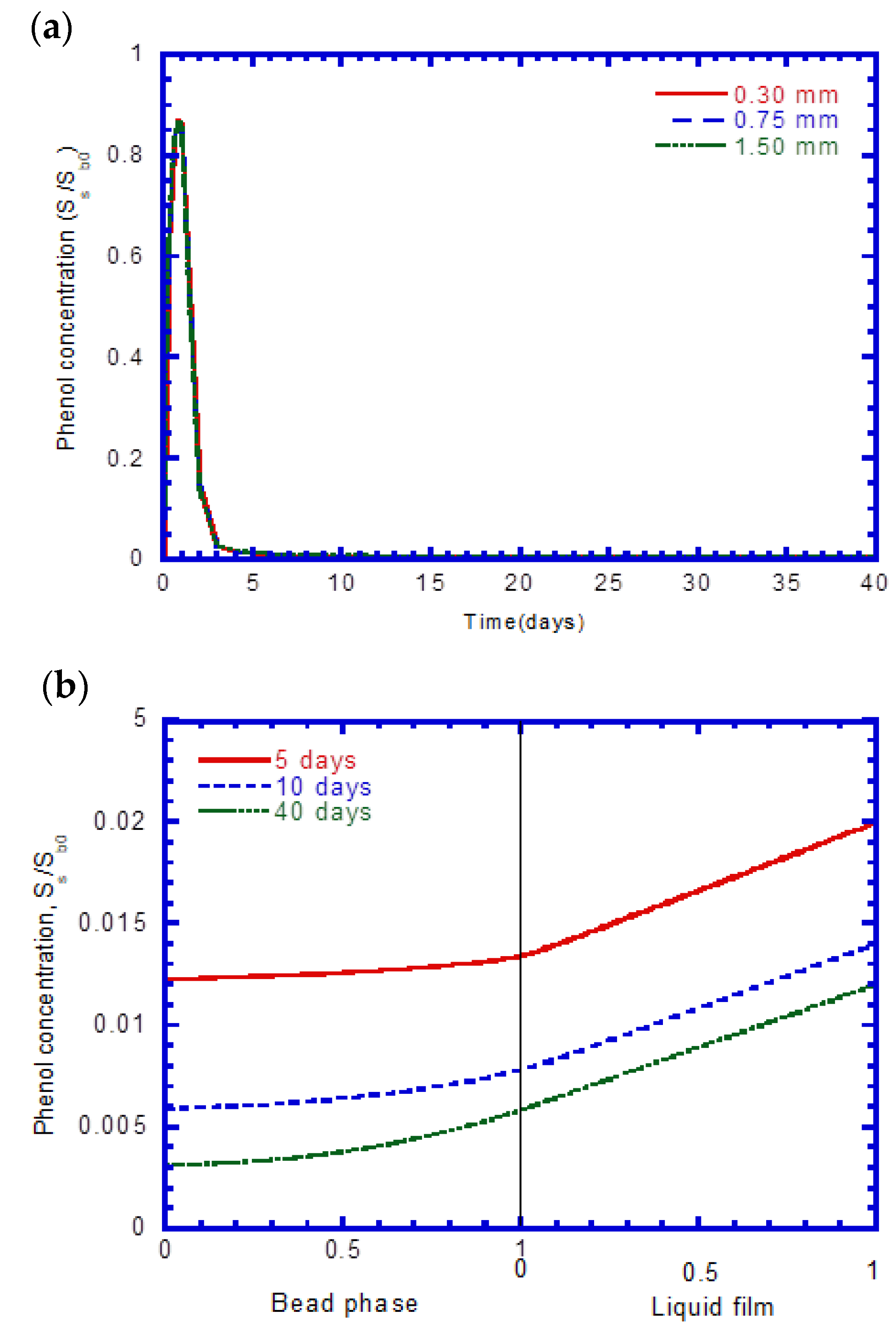
4.6. Flux into Beads
4.7. Immobilized Cells Growth
4.8. Phenol Concentration Profiles
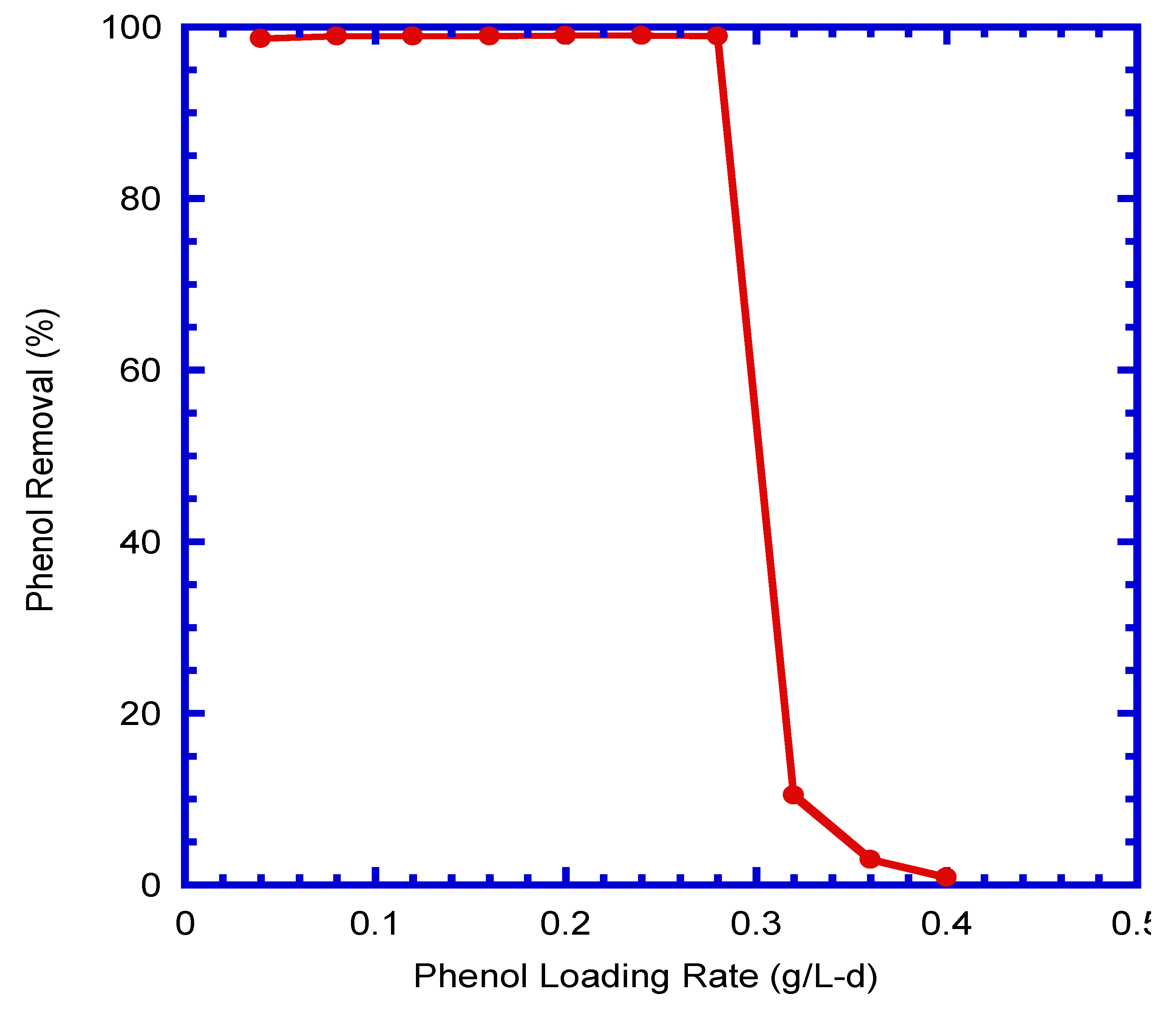
4.9. Phenol Removal at Various Loading Rates
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Dionisi, D.; Etteh, C.C. Effect of process conditions on the aerobic biodegradation of phenol and paracetamol by open mixed microbial cultures. J. Environ. Chem. Eng. 2019, 7, 1–8. [Google Scholar] [CrossRef]
- Papazi, A.; Pappas, I.; Kotzabasis, K. Combinational system for biodegradation of olive oil mill wastewater phenolics and high yield of bio-hydrogen production. J. Biotechnol. 2019, 306, 47–53. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Bennett, G.N. Biodegradation of xenobiotic by anaerobic bacteria. Appl. Microbiol. Biotechnol. 2005, 67, 600–618. [Google Scholar] [CrossRef]
- Geng, A.; En-WeiSoh, A.; Lim, C.J.; Loke, C.T.L. Isolation and characterization of a phenol-degrading bacterium from an industrial activated sludge. Appl. Microbiol. Biotechnol. 2006, 71, 728–735. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Deng, T.; Shang, Y.; Yang, K.; Wang, H. Biodegradation of phenol by entrapped cell of Debaryomyces sp. with nano-Fe3O4 under hypersaline conditions. Int. Biodeterior. Biodegrad. 2017, 123, 37–45. [Google Scholar] [CrossRef]
- Kamali, M.; Gameiro, T.; Costa, M.E.; Capela, I.; Aminabhavi, T.M. Enhanced biodegradation of phenolic wastewaters with acclimated activated sludge—A kinetic study. Chem. Eng. J. 2019, 378, 1–7. [Google Scholar] [CrossRef]
- Dong, R.; Chen, D.; Li, N.; Xu, Q.; Li, H.; He, J.; Lu, J. Removal of phenol from aqueous solution using acid-modified Pseudomonas putida-sepiolite/ZIF-8 bio-nanocomposites. Chemosphere 2020, 239, 1–11. [Google Scholar] [CrossRef]
- Li, H.; Meng, F.; Duan, W.; Lin, Y.; Zhang, Y. Biodegradation of phenol in saline or hypersaline environments by bacteria: A review. Ecotoxicol. Environ. Saf. 2019, 184, 1–9. [Google Scholar] [CrossRef]
- Chae, Y.; Kim, D.; Cui, R.; Lee, J.; An, Y.J. Deriving hazardous concentrations of phenol in soil ecosystems using a species sensitivity distribution approach. J. Hazard. Mater. 2020. [Google Scholar] [CrossRef]
- Afsharnia, M.; Saeidi, M.; Zarei, A.; Narooie, M.R.; Biglari, H. Phenol removal from aqueous environment by adsorption onto pomegranate peel carbon. Electron. Phys. 2016, 8, 3248–3256. [Google Scholar] [CrossRef]
- Quan, X.; Shi, H.; Liu, H.; Wang, J.; Qian, Y. Removal of 2,4-dichlorophenol in a conventional activated sludge system through bioaugmentation. Process Biochem. 2004, 39, 1701–1707. [Google Scholar] [CrossRef]
- Atlow, S.C.; Bonadonna-Aparo, L.; Klibanov, A.M. Dephenolization of industrial wastewaters catalyzed by polyphenol oxidase. Biotechnol. Bioeng. 1984, 6, 599–603. [Google Scholar] [CrossRef] [PubMed]
- Verma, S.R.; Tonk, I.P.; Gupta, A.K.; Saxena, M. Evaluation of an application factor for determining the safe concentration of agricultural and industrial chemicals. Water Res. 1984, 18, 111–115. [Google Scholar] [CrossRef]
- Kim, J.S.; Chin, P. Acute and chronic toxicity of phenol to mysid, Archaeomysis kokuboi. Korean J. Fish. Aquat. Sci. 1995, 28, 87–97. [Google Scholar]
- Banerjee, A.; Ghoshal, A.K. Phenol degradation performance by isolated Bacillus cereus immobilized in alginate. Int. Biodeterior. Biodegrad. 2011, 65, 1052–1060. [Google Scholar] [CrossRef]
- Parvanova-Mancheva, T.; Vasileva, E.; Beschkov, V.; Gerginova, M.; Stoilova-Disheva, M.; Alexieva, Z. Biodegradation potential of Pseudomonas putida to phenol compared to xanthobacter autotrophicus GJ10 and Pseudomonas denitrificans strains. J. Chem. Technol. Metall. 2020, 55, 23–27. [Google Scholar]
- Kurzbaum, E.; Raizner, Y.; Cohen, O.; Suckeveriene, R.Y.; Kulikov, A.; Hakimi, B.; Kruh, L.I.; Armon, R.; Farber, Y.; Menashe, O. Encapsulated Pseudomonas putida for phenol biodegradation: Use of a structural membrane for construction of a well-organized confined particle. Water Res. 2017, 121, 37–45. [Google Scholar] [CrossRef]
- Al-Zuhair, S.; El-Naas, M. Immobilization of Pseudomonas putida in PVA gel particles for biodegradation of phenol at high concentrations. Biochem. Eng. J. 2011, 56, 46–50. [Google Scholar] [CrossRef]
- Tanake, H.; Matsumura, M.; Veliky, I.A. Diffusion characteristics of substrates in Ca-alginate gel beads. Biotechnol. Bioeng. 1984, 26, 53–58. [Google Scholar] [CrossRef]
- Aksu, Z.; Bülbül, G. Determination of the effective diffusion coefficient of phenol in Ca-alginate-immobilized P. putida beads. Enzyme Microb. Technol. 1999, 25, 344–348. [Google Scholar] [CrossRef]
- Zulfadhly, Z.; Mashitah, M.; Bhatia, S. Heavy metals removal in fixed-bed column by the macro fungus Pycnoporus sanguineus. Environ. Pollut. 2001, 112, 463–470. [Google Scholar] [CrossRef]
- Wu, J.; Yu, H. Biosorption of 2,4-dichlorophenol from aqueous solutions by immobilized Phanerochaete chrysosporium biomass in a fixed-bed column. Chem. Eng. J. 2008, 138, 128–135. [Google Scholar] [CrossRef]
- Febrianto, J.; Kosasih, J.; Sunarso, Y.; Ju, N. Equilibrium and kinetic studies in adsorption of heavy metals using biosorbent: A summary of recent studies. J. Hazard. Mater. 2009, 162, 616–645. [Google Scholar] [CrossRef] [PubMed]
- Xiao, M.T.; Huang, Y.Y.; Ye, J.; Guo, Y.H. Study on the kinetic characteristics of the asymmetric production of R-(−)-mandelic acid with immobilized Saccharomyces cerevisiae FD11b. Biochem. Eng. J. 2008, 39, 311–318. [Google Scholar] [CrossRef]
- Wang, S.J.; Loh, K.C. Modeling the role of metabolic intermediates in kinetics of phenol biodegradation. Enzyme Microb. Technol. 1999, 25, 177–184. [Google Scholar] [CrossRef]
- Onysko, K.A.; Budman, H.M.; Robinson, C.W. Effect of temperature on the inhibition kinetics of phenol biodegradation by Pseudomonas putida Q5. Biotechnol. Bioeng. 2000, 70, 291–299. [Google Scholar] [CrossRef]
- Vinod, A.V.; Reddy, G.V. Simulation of biodegradation process of phenolic wastewater at higher concentrations in the fluidized-bed bioreactor. Biochem. Eng. J. 2005, 24, 1–10. [Google Scholar] [CrossRef]
- Reardon, K.F.; Mosteller, D.C.; Rogers, J.D.B. Biodegradation kinetics of benzene, toluene, and phenol as single and mixed substrates for Pseudomonas putida F1. Biotechnol. Bioeng. 2000, 69, 386–400. [Google Scholar] [CrossRef]
- Chung, T.P.; Wu, C.Y.; Juang, R.S. Improved dynamic analysis on cell growth with substrate inhibition using two-phase models. Biochem. Eng. J. 2005, 25, 209–217. [Google Scholar] [CrossRef]
- Kumar, A.; Kumar, S.; Kumar, S. Biodegradation kinetics of phenol and catechol using Pseudomonas putida MTCC 1194. Biochem. Eng. J. 2005, 22, 151–159. [Google Scholar] [CrossRef]
- Wakao, N.; Smith, J.M. Diffusion and reaction in porous catalysts. Ind. Eng. Chem. Fund. 1964, 2, 123–127. [Google Scholar] [CrossRef]
- Korgel, B.A.; Rotem, A.; Monbonquett, H.G. Effective diffusivity of galactose in calcium alginate gels containing immobilized Zymonmonas mobilis. Biotechnol. Prog. 1992, 8, 111–117. [Google Scholar] [CrossRef] [PubMed]
- Brian, P.L.T.; Hales, H.B. Effect of transpiration and changing diameter on heat and mass transfer to sphere. AIChE J. 1969, 15, 419–425. [Google Scholar] [CrossRef]
- Juang, R.S.; Tsai, S.Y. Growth kinetics of Pseudomonas putida in the biodegradation of single and mixed phenol and sodium salicylate. Biochem. Eng. J. 2006, 31, 133–140. [Google Scholar] [CrossRef]
- Tsai, S.Y.; Juang, R.S. Biodegradation of phenol and sodium salicylate mixtures by suspended Pseudomonas putida CCRC 14365. J. Hazard. Mater. 2006, 138, 125–132. [Google Scholar] [CrossRef]
- Chung, T.P.; Tseng, H.Y.; Juang, R.S. Mass transfer effect and intermediate detection for phenol degradation in immobilized Pseudomonas putida systems. Process Biochem. 2003, 38, 1497–1507. [Google Scholar] [CrossRef]
- Faridnasr, M.; Ghanbari, B.; Sassani, A. Optimization of the moving-bed biofilm sequencing batch reactor (MBSBR) to control aeration time by kinetic computational modeling: Simulated sugar-industry wastewater treatment. Bioresour. Technol. 2016, 208, 149–160. [Google Scholar] [CrossRef]
- Rosenkranz, F.; Cabrol, L.; Carballa, M.; Donoso-Bravo, A.; Cruz, L.; Ruiz-Filippi, G.; Chamy, R.; Lema, J.M. Relationship between phenol degradation efficiency and microbial community structure in an anaerobic SBR. Water Res. 2013, 47, 6739–6749. [Google Scholar] [CrossRef]
- Vital-Jacome, M.; Buitrón, G.; Moreno-Andrade, I.; Garcia-Rea, V.; Thalasso, F. Microrespirometric determination of the effectiveness factor and biodegradation kinetics of aerobic granules degrading 4-chlorophenol as the sole carbon source. J. Hazard. Mater. 2016, 313, 112–121. [Google Scholar] [CrossRef]
- Angelucci, D.M.; Clagnan, E.; Brusetti, L.; Tomei, M.C. Anaerobic phenol biodegradation: Kinetic study and microbial community shifts under high-concentration dynamic loading. Appl. Microbiol. Biotechnol. 2020. [Google Scholar] [CrossRef]
- Sahinkaya, E.; Dilek, F.B. Effects of 2,4-dichlorophenol on activated sludge. Appl. Microbiol. Biotechnol. 2002, 59, 361–367. [Google Scholar] [PubMed]
- Pirbazari, M.; Ravindran, V.; Badriyha, B.N.; Kim, S.H. Hybrid membrane filtration process for leachate treatment. Water Res. 1996, 30, 2691–2706. [Google Scholar] [CrossRef]
- Bajaj, M.; Gallert, C.; Winter, J. Phenol degradation kinetics of an aerobic mixed culture. Biochem. Eng. J. 2009, 46, 205–209. [Google Scholar] [CrossRef]
- Monteiro, Á.A.; Boaventura, R.A.; Rodrigues, A.E. Phenol biodegradation by Pseudomonas putida DSM 548 in a batch reactor. Biochem. Eng. J. 2000, 6, 45–49. [Google Scholar] [CrossRef]
- Abuhamed, T.; Bayraktar, E.; Mehmetoğlu, T.; Mehmetoğlu, Ü. Kinetics model for growth of Pseudomonas putida F1 during benzene, toluene and phenol biodegradation. Process Biochem. 2004, 39, 983–988. [Google Scholar] [CrossRef]
- Banerjee, I.; Modak, J.M.; Bandopadhyay, K.; Das, D.; Maiti, B.R. Mathematical model for evaluation of mass transfer limitations in phenol biodegradation by immobilized Pseudomonas putida. J. Biotechnol. 2001, 87, 211–223. [Google Scholar] [CrossRef]
- Ju, L.K.; Ho, C.S. Correlation of cell volume fractions with cell concentrations in fermentation media. Biotechnol. Bioeng. 1988, 32, 95–99. [Google Scholar] [CrossRef]
- Wilke, C.E.; Chang, P. Correlation of diffusion coefficients in dilute solutions. AIChE J. 1955, 1, 264–270. [Google Scholar] [CrossRef]
- El-Naas, M.H.; Al-Muhtaseb, S.A.; Makhlouf, S. Biodegradation of phenol by Pseudomonas putida immobilized in polyvinyl alcohol (PVA) gel. J. Hazard. Mater. 2009, 164, 720–725. [Google Scholar] [CrossRef]
- González, G.; Herrera, G.; García, M.T.; Peña, M. Biodegradation of phenolic industrial wastewater in a fluidized bed bioreactor with immobilized cells of Pseudomonas putida. Bioresour. Technol. 2001, 80, 137–142. [Google Scholar] [CrossRef]
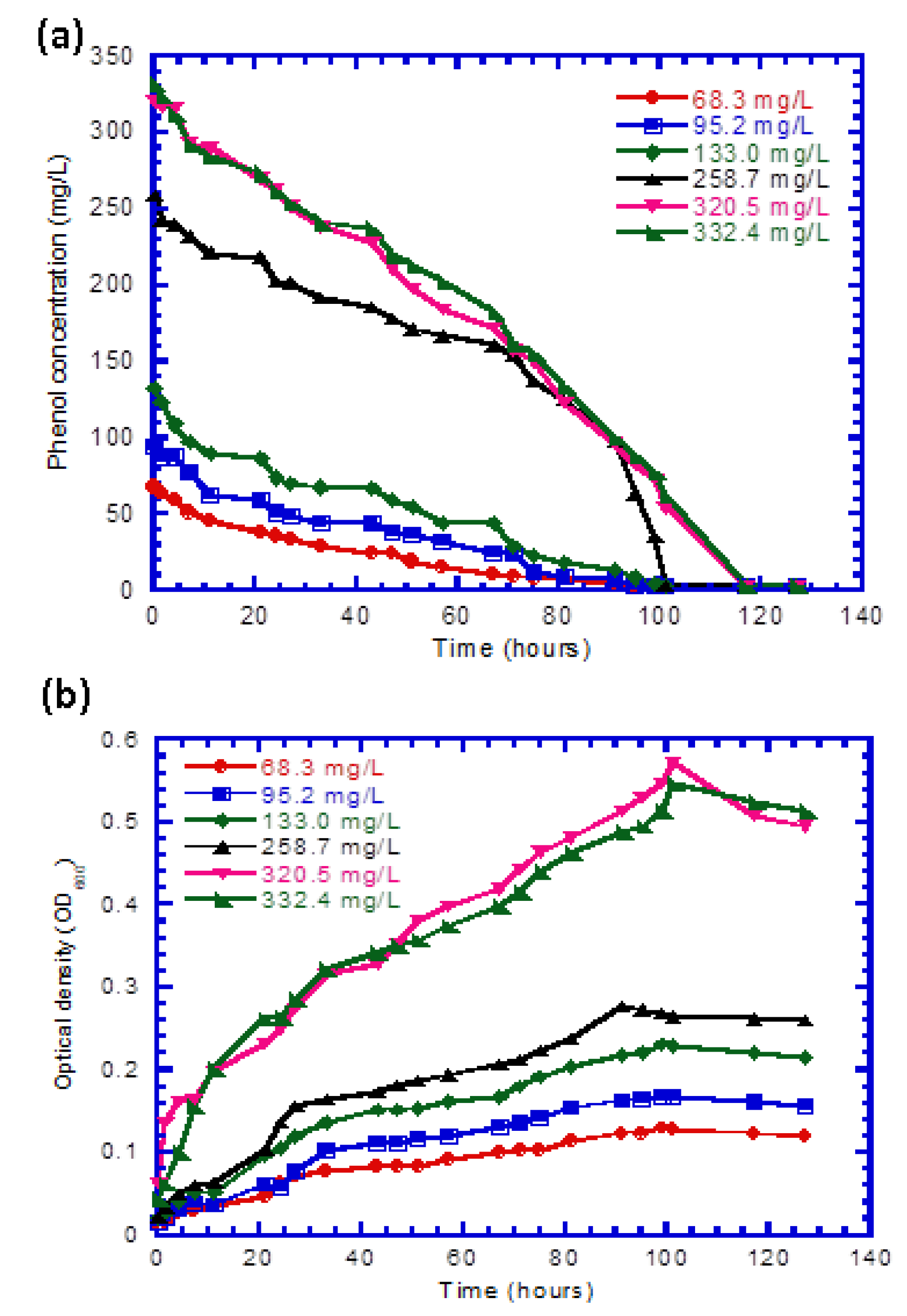
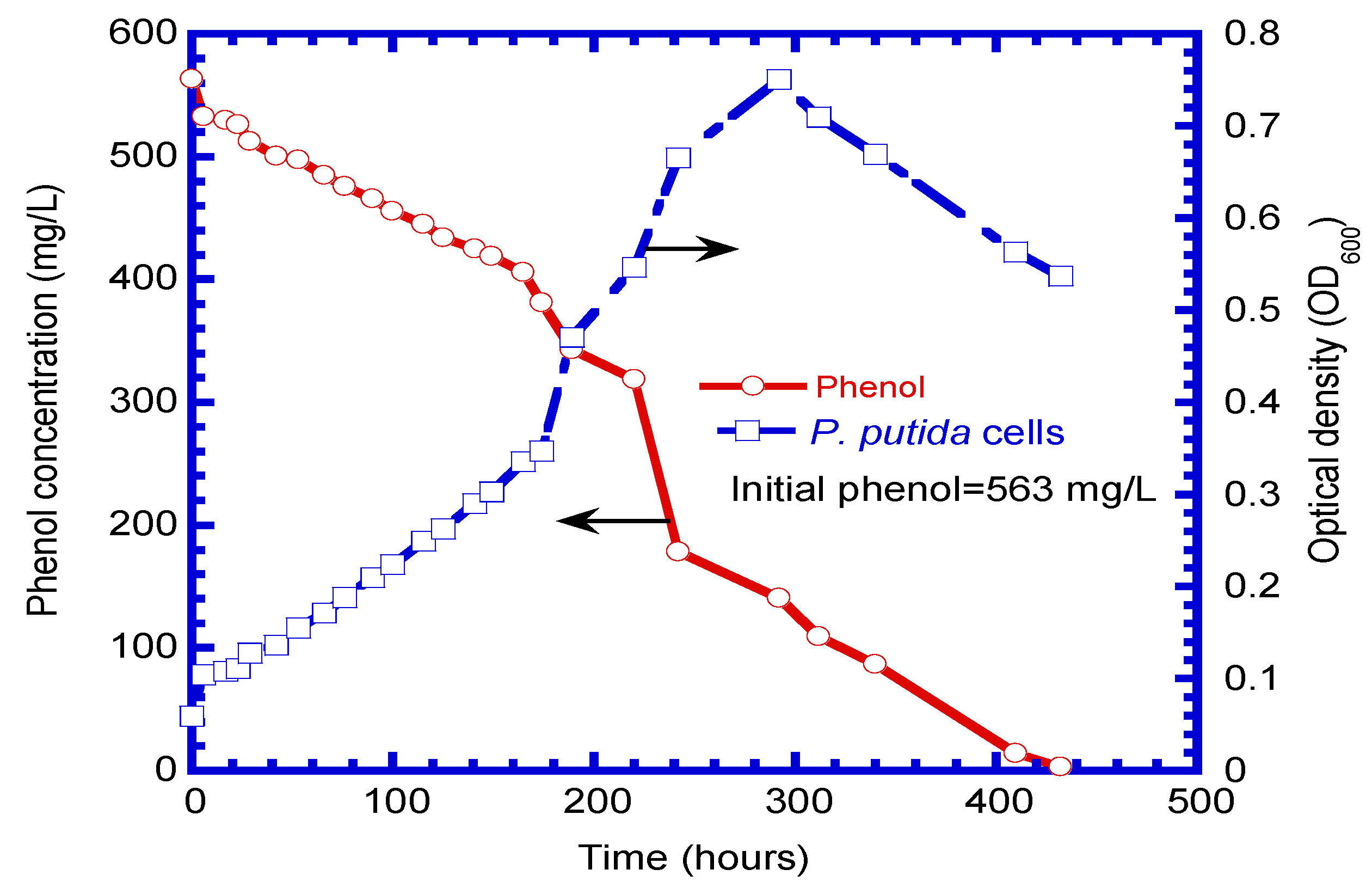
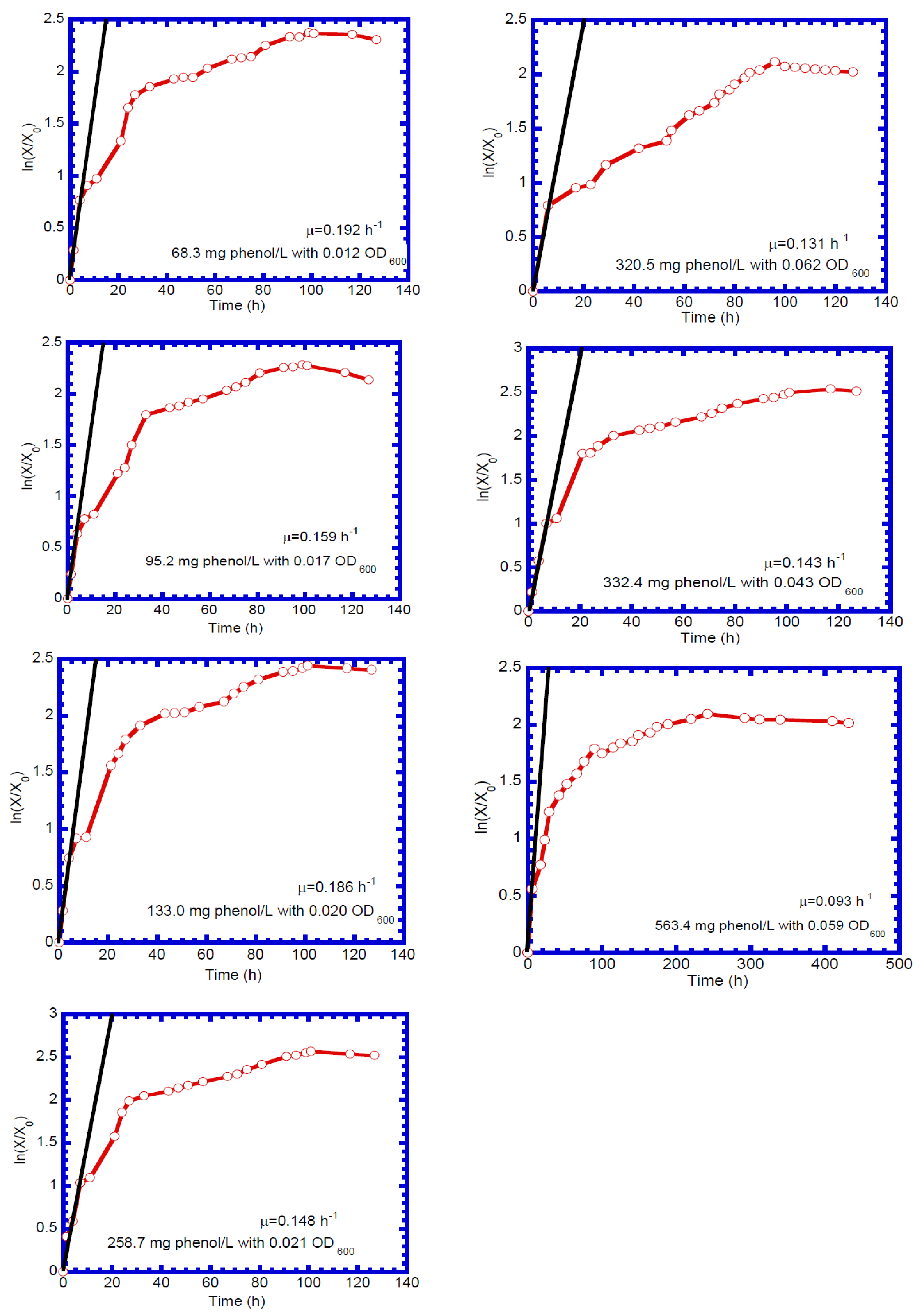
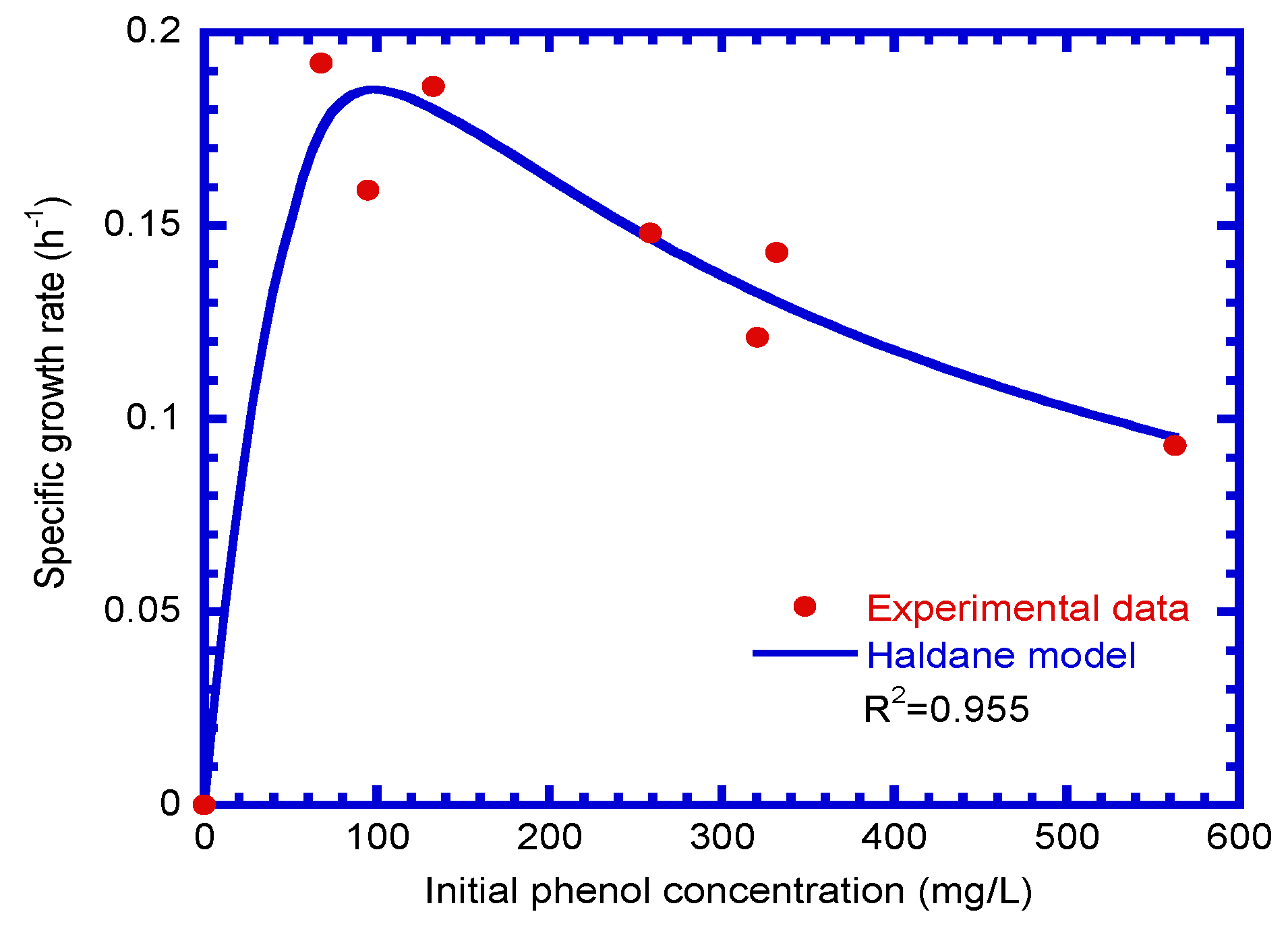

| Run No. | Temp. (°C) | pH | Initial Phenol Conc. (mg L−1) | Initial Cell Conc. (mg cell L−1) | Culture Duration (h) |
|---|---|---|---|---|---|
| 1 | 30 | 7.0 | 68.3 | 4.1 | 95 |
| 2 | 30 | 7.0 | 95.2 | 5.8 | 99 |
| 3 | 30 | 7.0 | 133.0 | 6.9 | 101 |
| 4 | 30 | 7.0 | 258.7 | 7.3 | 101 |
| 5 | 30 | 7.0 | 320.5 | 21.4 | 117 |
| 6 | 30 | 7.0 | 332.4 | 14.9 | 117 |
| 7 | 30 | 7.0 | 563.4 | 20.4 | 432 |
| Strain | Condition | Initial Phenol Conc. (mg L−1) | μmax (h−1) | KS (mg L−1) | KI (mg L−1) | References |
|---|---|---|---|---|---|---|
| P. putida (ATCC 49451) | Aerobic | 25–800 | 0.900 | 6.93 | 284.3 | Wang and Loh [25] |
| P. putida (DSM 548) | Aerobic | 1–100 | 0.436 | 6.19 | 54.1 | Monteiro et al. [44] |
| P. putida F1 (ATCC 700007) | Aerobic | 10–200 | 0.051 | 18 | 430 | Abuhamed et al. [45] |
| P. putida (MTCC 1194) | Aerobic | 244.4–996.4 | 0.06 | 190.8 | 799.0 | Banerjee et al. [46] |
| P. putida (CCRC 14365) | Aerobic | 25–600 | 0.33 | 13.9 | 669 | Chung et al. [36] |
| P. putida (MTCC 1194) | Aerobic | 10–1000 | 0.305 | 36.33 | 129.79 | Kumar et al. [30] |
| Mixed culture | Aerobic | 23.5–659 | 0.3095 | 74.65 | 648.13 | Bajaj et al. [43] |
| Mixed culture | Aerobic | 250–1000 | - | 692 | 231 | Kamali et al. [6] |
| Mixed culture | Anaerobic | 400–1000 | - | 1599.5 | 16,500 | Angelucci et al. [40] |
| P. putida (BCRC 14365) | Aerobic | 68.3–563.4 | 0.31 | 26.2 | 255.0 | This study |
| Run No. | Initial Phenol Concentration (mg L−1) | Biokinetic Parameters | |
|---|---|---|---|
| Yx/s, (mg mg−1) | b (d−1) | ||
| 1 | 68.3 | 0.600 | 5.52 × 10−2 |
| 2 | 95.2 | 0.591 | 5.65 × 10−2 |
| 3 | 133.0 | 0.597 | 5.88 × 10−2 |
| 4 | 258.7 | 0.590 | 5.72 × 10−2 |
| 5 | 321.5 | 0.594 | 5.77 × 10−2 |
| 6 | 332.4 | 0.590 | 5.65 × 10−2 |
| 7 | 563.4 | 0.585 | 5.72 × 10−2 |
| mean | 0.592 | 5.70 × 10−2 | |
| standard deviation | − | 4.995 × 10−3 | 1.122 × 10−3 |
| Symbol | Parameters Description (Unit) | Value | Remarks |
|---|---|---|---|
| ε | reactor porosity (dimensionless) | 0.72 | measured |
| A | surface area of gel beads (cm2) | 3.522 × 104 | calculated |
| b | decay coefficient of cell in gel beads (d−1) | 5.70 × 10−2 | measured |
| De | effective diffusivity of phenol in the beads (cm2 d−1) | 0.236 | calculated |
| kf | mass-transfer coefficient of phenol (cm d−1) | 107.1 | calculated |
| KI | inhibition constant of phenol (mg L−1) | 255.0 | measured |
| KS | half-saturation constant of phenol (mg L−1) | 26.2 | measured |
| Q | influent flow rate (mL d−1) | 6.272 × 103 | measured |
| SP0 | concentration of phenol in feed (mg L−1) | 25.2 | measured |
| V | effective working volume (mL) | 1.568 × 103 | measured |
| Yx/s | growth yield of cell [mg cell (mg phenol)−1] | 0.592 | measured |
| μmax | maximum specific growth rate of cell (d−1) | 7.44 | measured |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, Y.-H.; Cheng, Y.-S. Phenol Degradation Kinetics by Free and Immobilized Pseudomonas putida BCRC 14365 in Batch and Continuous-Flow Bioreactors. Processes 2020, 8, 721. https://doi.org/10.3390/pr8060721
Lin Y-H, Cheng Y-S. Phenol Degradation Kinetics by Free and Immobilized Pseudomonas putida BCRC 14365 in Batch and Continuous-Flow Bioreactors. Processes. 2020; 8(6):721. https://doi.org/10.3390/pr8060721
Chicago/Turabian StyleLin, Yen-Hui, and Yu-Siang Cheng. 2020. "Phenol Degradation Kinetics by Free and Immobilized Pseudomonas putida BCRC 14365 in Batch and Continuous-Flow Bioreactors" Processes 8, no. 6: 721. https://doi.org/10.3390/pr8060721
APA StyleLin, Y.-H., & Cheng, Y.-S. (2020). Phenol Degradation Kinetics by Free and Immobilized Pseudomonas putida BCRC 14365 in Batch and Continuous-Flow Bioreactors. Processes, 8(6), 721. https://doi.org/10.3390/pr8060721






