Mathematical Modeling of RNA Virus Sensing Pathways Reveals Paracrine Signaling as the Primary Factor Regulating Excessive Cytokine Production
Abstract
1. Introduction
2. Materials and Methods
2.1. Model Construction
2.2. Data Sources
2.3. ODE Simulation and Sensitivity Analysis
2.4. Model Parameterization
2.5. Structural Identifiability
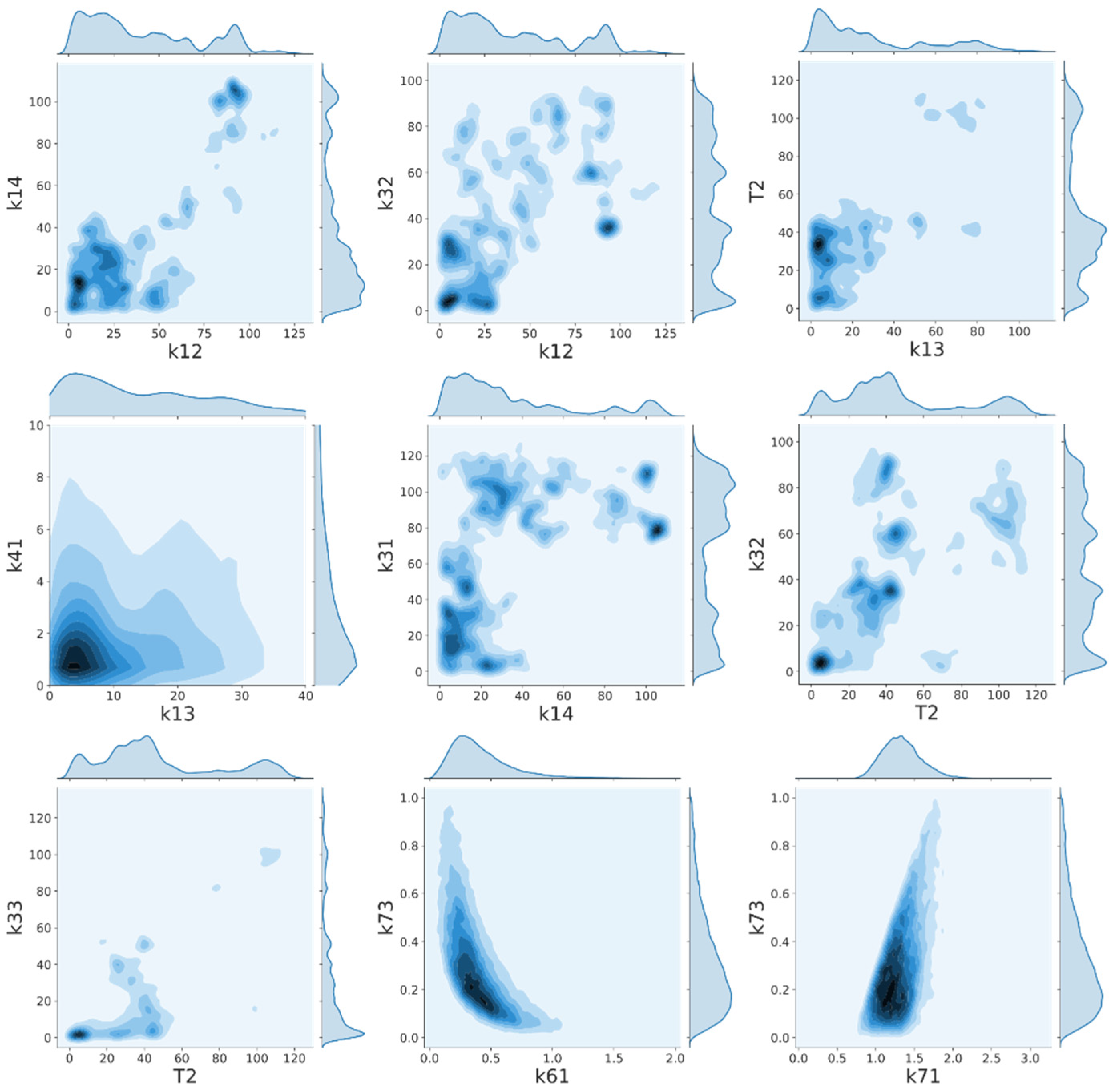
2.6. Interparameter Correlation
3. Results
3.1. ODE Model
3.2. MCMC Parameterization
3.3. Model Validation by Predicting Response to Infection Using a NSI Knockout Influenza Virus
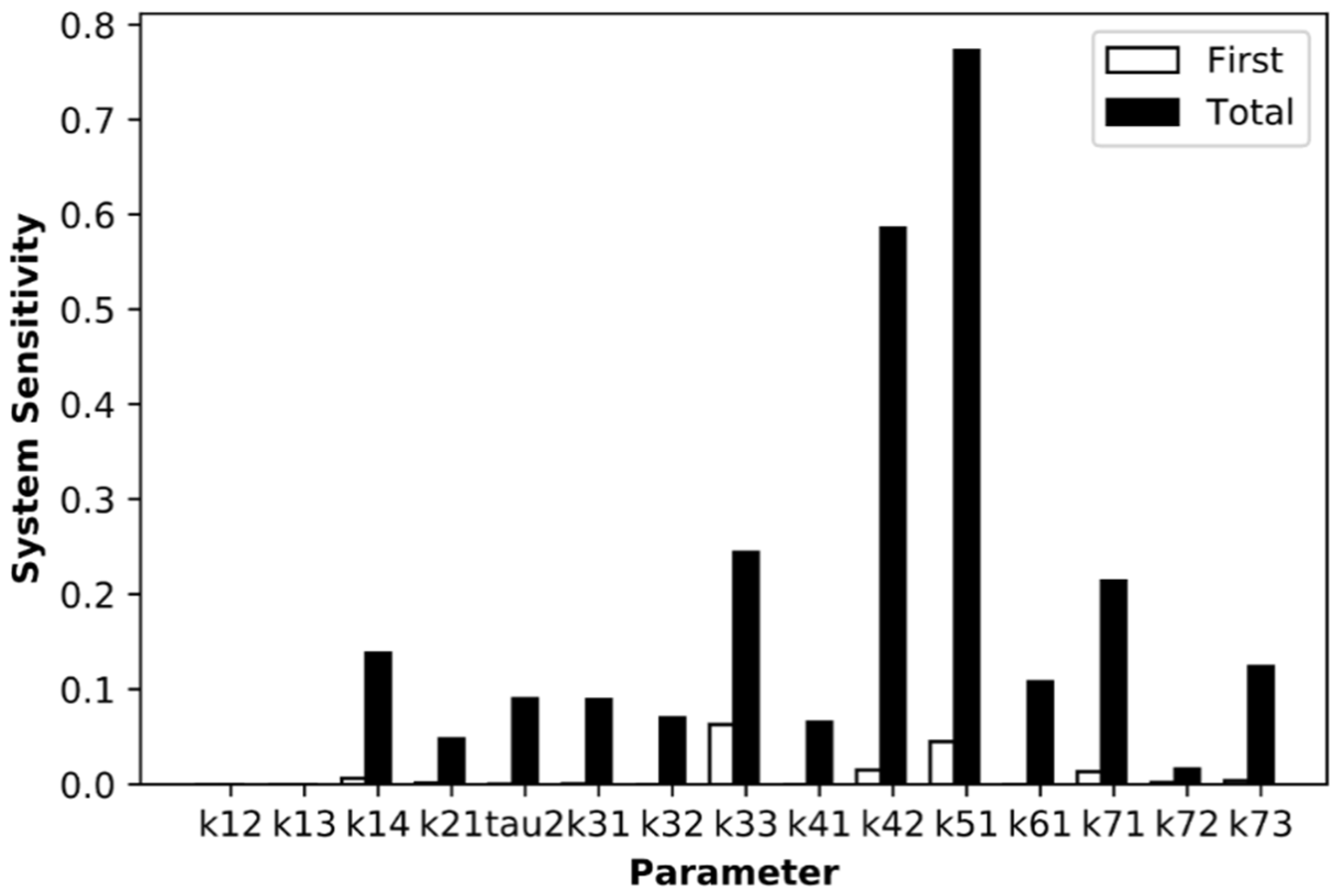
3.4. Sensitivity Analysis Reveals IRF7 Phosphorylation as Critical Step
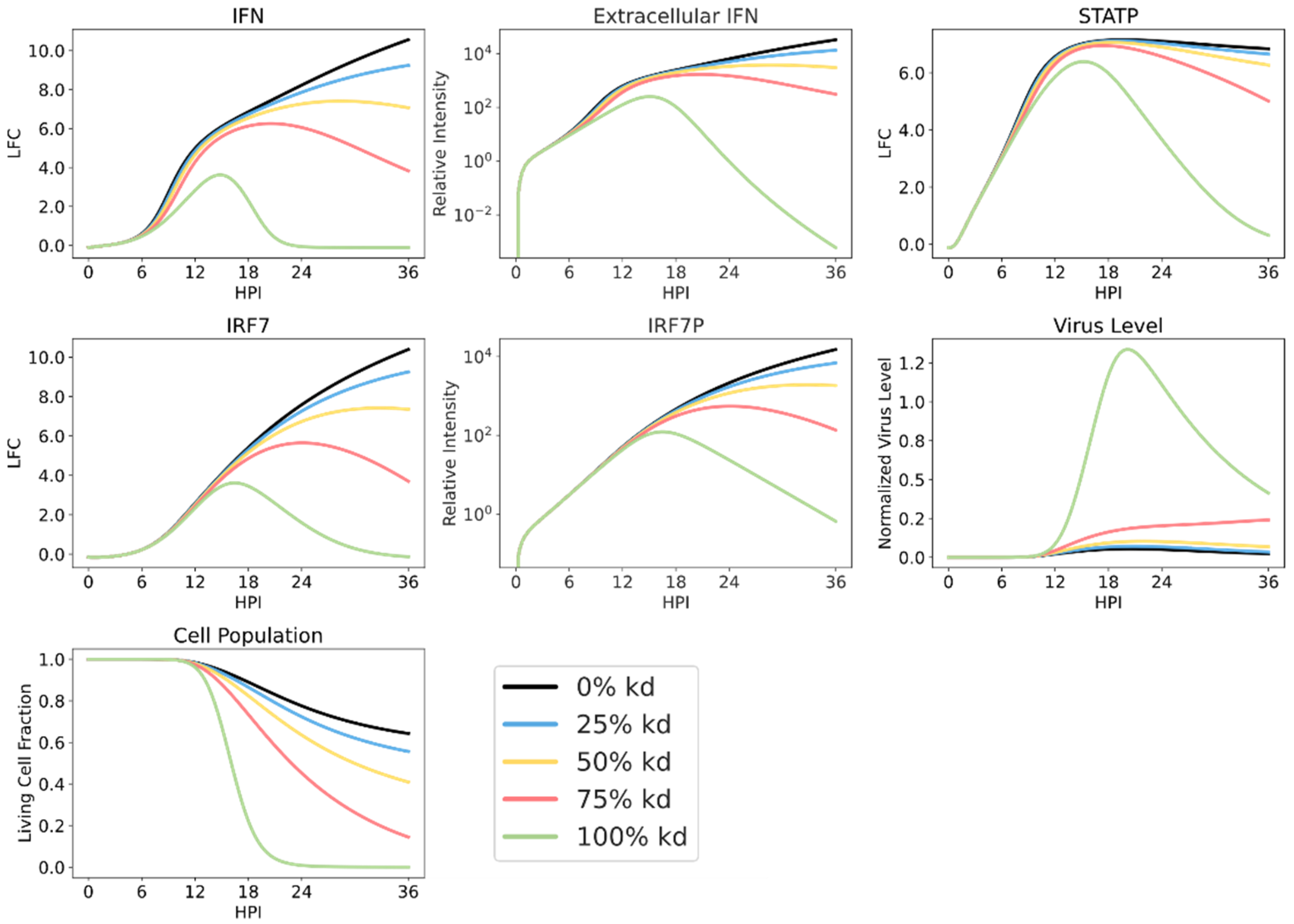
3.5. Simulating Varying Levels of RIG-I Antagonism Reveals Robust Sensor Protein Action
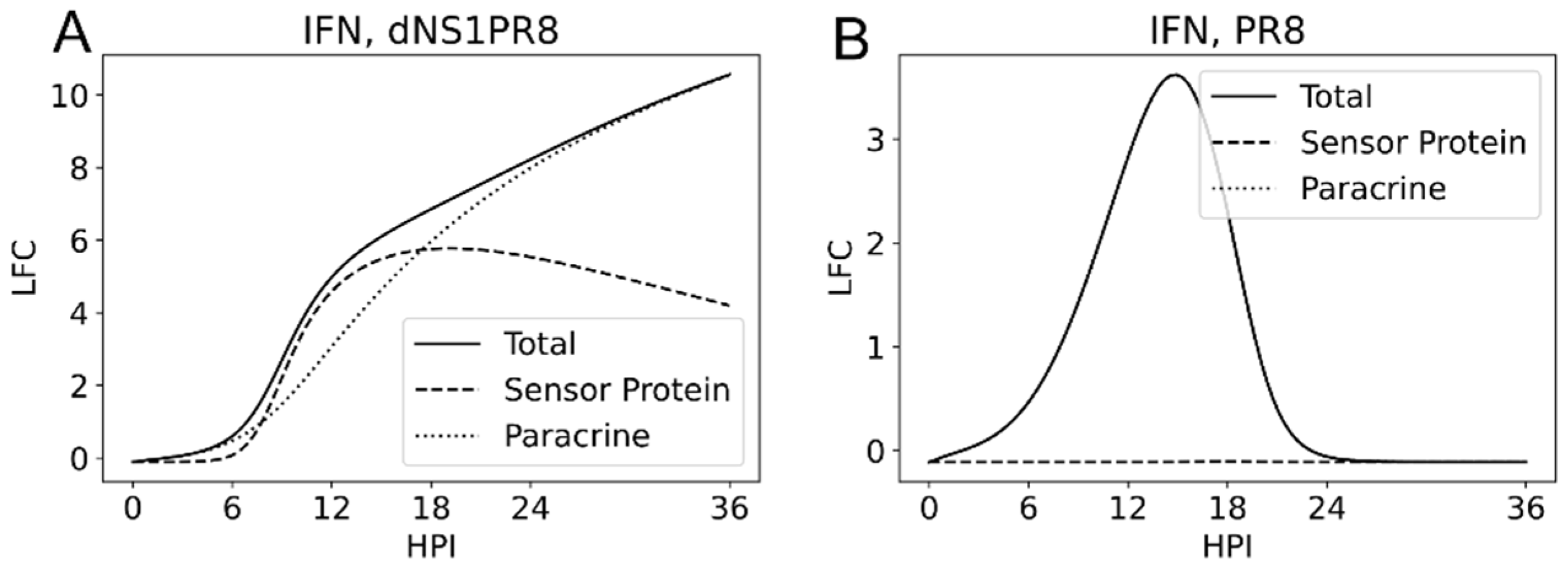
3.6. Sensor Protein and JAK/STAT Originated Interferon Production
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A. Structural Identifiability
Appendix B
| k13 | k14 | k21 | tau2 | k31 | k32 | k33 | k41 | k42 | k51 | k61 | k71 | k72 | k73 | |
| 0.17 | 0.77 | 0.3 | 0.02 | 0.31 | 0.53 | −0.05 | −0.01 | 0.23 | −0.15 | 0.01 | 0.09 | 0.35 | 0 | k12 |
| 1 | 0 | 0.09 | 0.7 | −0.1 | 0.28 | 0.3 | 0.63 | −0.02 | −0.02 | −0.04 | −0.03 | 0.43 | 0 | k13 |
| 1 | −0.06 | −0.04 | 0.5 | 0.39 | 0.12 | −0.17 | 0.23 | −0.34 | 0.01 | 0.1 | 0.28 | 0 | k14 | |
| 1 | 0.16 | 0 | 0.36 | −0.2 | 0.09 | 0.01 | 0.23 | −0.01 | −0.01 | 0.06 | 0 | k21 | ||
| 1 | 0.13 | 0.5 | 0.66 | 0.45 | −0.01 | 0.03 | −0.05 | −0.13 | 0.1 | 0 | t2 | |||
| 1 | 0.48 | 0.44 | −0.47 | 0.14 | −0.18 | 0.03 | 0.08 | 0.09 | 0 | k31 | ||||
| 1 | 0.47 | −0.05 | 0.3 | −0.06 | −0.03 | 0 | 0.06 | 0 | k32 | |||||
| 1 | 0.04 | 0.17 | −0.13 | −0.06 | −0.13 | −0.29 | 0.1 | k33 | ||||||
| 1 | 0.05 | −0.05 | −0.06 | −0.1 | 0.29 | 0 | k41 | |||||||
| 1 | −0.35 | −0.01 | −0.05 | −0.07 | 0 | k42 | ||||||||
| 1 | 0.03 | 0.07 | −0.06 | 0 | k51 | |||||||||
| 1 | −0.44 | 0.03 | −0.5 | k61 | ||||||||||
| 1 | 0.17 | 0.6 | k71 | |||||||||||
| 1 | −0.1 | k72 | ||||||||||||
| 1 | k73 |
References
- Influenza (Seasonal). Available online: https://www.who.int/news-room/fact-sheets/detail/influenza-(seasonal) (accessed on 17 September 2019).
- Lafond, K.E.; Nair, H.; Rasooly, M.H.; Valente, F.; Booy, R.; Rahman, M.; Kitsutani, P.; Yu, H.; Guzman, G.; Coulibaly, D.; et al. Global Role and Burden of Influenza in Pediatric Respiratory Hospitalizations, 1982–2012: A Systematic Analysis. PLoS Med. 2016, 13, e1001977. [Google Scholar] [CrossRef] [PubMed]
- AbdelMassih, A.F.; Ramzy, D.; Nathan, L.; Aziz, S.; Ashraf, M.; Youssef, N.H.; Hafez, N.; Saeed, R.; Agha, H. Possible molecular and paracrine involvement underlying the pathogenesis of COVID-19 cardiovascular complications. Cardiovasc. Endocrinol. Metab. 2020. [Google Scholar] [CrossRef]
- Cilloniz, C.; Pantin-Jackwood, M.J.; Ni, C.; Goodman, A.G.; Peng, X.; Proll, S.C.; Carter, V.S.; Rosenzweig, E.R.; Szretter, K.J.; Katz, J.M.; et al. Lethal Dissemination of H5N1 Influenza Virus Is Associated with Dysregulation of Inflammation and Lipoxin Signaling in a Mouse Model of Infection. J. Virol. 2010, 84, 7613–7624. [Google Scholar] [CrossRef] [PubMed]
- Peiris, J.S.M.; Cheung, C.Y.; Leung, C.Y.H.; Nicholls, J.M. Innate immune responses to influenza A H5N1: Friend or foe? Trends Immunol. 2009, 30, 574–584. [Google Scholar] [CrossRef]
- Sun, L.; Liu, S.; Chen, Z.J. SnapShot: Pathways of Antiviral Innate Immunity. Cell 2010, 140, 436. [Google Scholar] [CrossRef]
- Trinchieri, G. Type I interferon: Friend or foe? J. Exp. Med. 2010, 207, 2053–2063. [Google Scholar] [CrossRef]
- Prchal, M.; Pilz, A.; Simma, O.; Lingnau, K.; Strobl, B.; Müller, M.; Decker, T. Type I interferons as mediators of immune adjuvants for T- and B cell-dependent acquired immunity. Vaccine 2009, 27, G17–G20. [Google Scholar] [CrossRef]
- Opitz, B.; Rejaibi, A.; Dauber, B.; Eckhard, J.; Vinzing, M.; Schmeck, B.; Hippenstiel, S.; Suttorp, N.; Wolff, T. IFN? induction by influenza A virus is mediated by RIG-I which is regulated by the viral NS1 protein. Cell. Microbiol. 2007, 9, 930–938. [Google Scholar] [CrossRef]
- Wu, W.; Zhang, W.; Duggan, E.S.; Booth, J.L.; Zou, M.H.; Metcalf, J.P. RIG-I and TLR3 are both required for maximum interferon induction by influenza virus in human lung alveolar epithelial cells. Virology 2015, 482, 181–188. [Google Scholar] [CrossRef]
- Fujita, T.; Onoguchi, K.; Onomoto, K.; Hirai, R.; Yoneyama, M. Triggering antiviral response by RIG-I-related RNA helicases. Biochimie 2007, 89, 754–760. [Google Scholar] [CrossRef]
- Gack, M.U.; Albrecht, R.A.; Urano, T.; Inn, K.-S.; Huang, I.-C.; Carnero, E.; Farzan, M.; Inoue, S.; Jung, J.U.; García-Sastre, A. Influenza A Virus NS1 Targets the Ubiquitin Ligase TRIM25 to Evade Recognition by the Host Viral RNA Sensor RIG-I. Cell Host Microbe 2009, 5, 439–449. [Google Scholar] [CrossRef] [PubMed]
- Rajsbaum, R.; Albrecht, R.A.; Wang, M.K.; Maharaj, N.P.; Versteeg, G.A.; Nistal-Villán, E.; García-Sastre, A.; Gack, M.U. Species-Specific Inhibition of RIG-I Ubiquitination and IFN Induction by the Influenza A Virus NS1 Protein. PLoS Pathog. 2012, 8, e1003059. [Google Scholar] [CrossRef] [PubMed]
- Diebold, S.S.; Kaisho, T.; Hemmi, H.; Akira, S.; Reis e Sousa, C. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science 2004, 303, 1529–1531. [Google Scholar] [CrossRef]
- Takeda, K.; Kaisho, T.; Akira, S. TOLL-LIKERECEPTORS. Annu. Rev. Immunol. 2003, 21, 335–376. [Google Scholar] [CrossRef]
- Hu, Y.; Li, W.; Gao, T.; Cui, Y.; Jin, Y.; Li, P.; Ma, Q.; Liu, X.; Cao, C. The Severe Acute Respiratory Syndrome Coronavirus Nucleocapsid Inhibits Type I Interferon Production by Interfering with TRIM25-Mediated RIG-I Ubiquitination. J. Virol. 2017, 91. [Google Scholar] [CrossRef]
- Lu, X.; Pan, J.; Tao, J.; Guo, D. SARS-CoV nucleocapsid protein antagonizes IFN-β response by targeting initial step of IFN-β induction pathway, and its C-terminal region is critical for the antagonism. Virus Genes 2011, 42, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Le Page, C.; Génin, P.; Baines, M.G.; Hiscott, J. Interferon activation and innate immunity. Rev. Immunogenet. 2000, 2, 374–386. [Google Scholar] [PubMed]
- De Veer, M.J.; Holko, M.; Frevel, M.; Walker, E.; Der, S.; Paranjape, J.M.; Silverman, R.H.; Williams, B.R.G. Functional classification of interferon-stimulated genes identified using microarrays. J. Leukoc. Biol. 2001, 69, 912–920. [Google Scholar]
- Atkin-Smith, G.K.; Duan, M.; Chen, W.; Poon, I.K.H. The induction and consequences of Influenza A virus-induced cell death. Cell Death Dis. 2018, 9, 1002. [Google Scholar] [CrossRef]
- Schneider, W.M.; Chevillotte, M.D.; Rice, C.M. Interferon-stimulated genes: A complex web of host defenses. Annu. Rev. Immunol. 2014, 32, 513–545. [Google Scholar] [CrossRef]
- Alberts, B.; Johnson, A.; Lewis, J.; Raff, M.; Roberts, K.; Walter, P. Molecular Biology of the Cell; Garland Science: New York, NY, USA, 2002; ISBN 0815332181. [Google Scholar]
- Matsuoka, Y.; Matsumae, H.; Katoh, M.; Eisfeld, A.J.; Neumann, G.; Hase, T.; Ghosh, S.; Shoemaker, J.E.; Lopes, T.J.S.; Watanabe, T.; et al. A comprehensive map of the influenza A virus replication cycle. BMC Syst. Biol. 2013, 7, 97. [Google Scholar] [CrossRef]
- Qiao, L.; Phipps-Yonas, H.; Hartmann, B.; Moran, T.M.; Sealfon, S.C.; Hayot, F. Immune response modeling of interferon beta-pretreated influenza virus-infected human dendritic cells. Biophys. J. 2010, 98, 505–514. [Google Scholar] [CrossRef] [PubMed]
- Fribourg, M.; Hartmann, B.; Schmolke, M.; Marjanovic, N.; Albrecht, R.A.; García-Sastre, A.; Sealfon, S.C.; Jayaprakash, C.; Hayot, F. Model of influenza A virus infection: Dynamics of viral antagonism and innate immune response. J. Theor. Biol. 2014, 351, 47–57. [Google Scholar] [CrossRef] [PubMed]
- Kitano, H. Biological robustness. Nat. Rev. Genet. 2004, 5, 826–837. [Google Scholar] [CrossRef] [PubMed]
- Schoeberl, B.; Eichler-Jonsson, C.; Gilles, E.D.; Müller, G. Computational modeling of the dynamics of the MAP kinase cascade activated by surface and internalized EGF receptors. Nat. Biotechnol. 2002, 20, 370–375. [Google Scholar] [CrossRef] [PubMed]
- Shoemaker, J.E.; Gayen, K.; Garcia-Reyero, N.; Perkins, E.J.; Villeneuve, D.L.; Liu, L.; Doyle, F.J. Fathead minnow steroidogenesis: In silico analyses reveals tradeoffs between nominal target efficacy and robustness to cross-talk. BMC Syst. Biol. 2010, 4, 89. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, A.; Levchenko, A.; Scott, M.L.; Baltimore, D. The IκB-NF-κB Signaling Module: Temporal Control and Selective Gene Activation. Science 2002, 298, 1241–1245. [Google Scholar] [CrossRef]
- Ferrell, J.E.; Machleder, E.M.; Ma, W.; Pomerening, J.R.; Tang, C.; Ferrell, J.E.; Ryan, S.; Spiller, D.G.; Unitt, J.F.; Broomhead, D.S.; et al. The biochemical basis of an all-or-none cell fate switch in Xenopus oocytes. Science 1998, 280, 895–898. [Google Scholar] [CrossRef]
- Ho, D.D.; Neumann, A.U.; Perelson, A.S.; Chen, W.; Leonard, J.M.; Markowitz, M. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature 1995, 373, 123–126. [Google Scholar] [CrossRef]
- Baccam, P.; Beauchemin, C.; Macken, C.A.; Hayden, F.G.; Perelson, A.S. Kinetics of influenza A virus infection in humans. J. Virol. 2006, 80, 7590–7599. [Google Scholar] [CrossRef]
- Ramos, I.; Carnero, E.; Bernal-Rubio, D.; Seibert, C.W.; Westera, L.; García-Sastre, A.; Fernandez-Sesma, A. Contribution of double-stranded RNA and CPSF30 binding domains of influenza virus NS1 to the inhibition of type I interferon production and activation of human dendritic cells. J. Virol. 2013, 87, 2430–2440. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Skiena, S.; Futcher, B.; Mueller, S.; Wimmer, E. Deliberate reduction of hemagglutinin and neuraminidase expression of influenza virus leads to an ultraprotective live vaccine in mice. Proc. Natl. Acad. Sci. USA 2013, 110, 9481–9486. [Google Scholar] [CrossRef]
- Shapira, S.D.; Gat-Viks, I.; Shum, B.O.V.; Dricot, A.; de Grace, M.M.; Wu, L.; Gupta, P.B.; Hao, T.; Silver, S.J.; Root, D.E.; et al. A Physical and Regulatory Map of Host-Influenza Interactions Reveals Pathways in H1N1 Infection. Cell 2009, 139, 1255–1267. [Google Scholar] [CrossRef] [PubMed]
- Dou, D.; Revol, R.; Östbye, H.; Wang, H.; Daniels, R. Influenza A Virus Cell Entry, Replication, Virion Assembly and Movement. Front. Immunol. 2018, 9, 1581. [Google Scholar] [CrossRef] [PubMed]
- White, D.O.; Cheyne, I.M. Early events in the eclipse phase of influenza and parainfluenza virus infection. Virology 1966, 29, 49–59. [Google Scholar] [CrossRef]
- Foster, K.A.; Oster, C.G.; Mayer, M.M.; Avery, M.L.; Audus, K.L. Characterization of the A549 cell line as a type II pulmonary epithelial cell model for drug metabolism. Exp. Cell Res. 1998, 243, 359–366. [Google Scholar] [CrossRef] [PubMed]
- Cambridge, S.B.; Gnad, F.; Nguyen, C.; Bermejo, J.L.; Krüger, M.; Mann, M. Systems-wide proteomic analysis in mammalian cells reveals conserved, functional protein turnover. J. Proteome Res. 2011, 10, 5275–5284. [Google Scholar] [CrossRef]
- Sharova, L.V.; Sharov, A.A.; Nedorezov, T.; Piao, Y.; Shaik, N.; Ko, M.S.H. Database for mRNA half-life of 19 977 genes obtained by DNA microarray analysis of pluripotent and differentiating mouse embryonic stem cells. DNA Res. 2009, 16, 45–58. [Google Scholar] [CrossRef]
- Prakash, A.; Levy, D.E. Regulation of IRF7 through cell type-specific protein stability. Biochem. Biophys. Res. Commun. 2006, 342, 50–56. [Google Scholar] [CrossRef]
- Geyer, C.J. Markov Chain Monte Carlo Maximum Likelihood, Computing Science and Statistics. Proc. 23rd Symp. Interface 1991, 1991, 156–163. [Google Scholar]
- Bellman, R.; Åström, K.J. On structural identifiability. Math. Biosci. 1970, 7, 329–339. [Google Scholar] [CrossRef]
- Villaverde, A.F.; Barreiro, A.; Papachristodoulou, A. Structural Identifiability of Dynamic Systems Biology Models. PLoS Comput. Biol. 2016, 12. [Google Scholar] [CrossRef] [PubMed]
- Schober, P.; Boer, C.; Schwarte, L.A. Correlation Coefficients. Anesth. Analg. 2018, 126, 1763–1768. [Google Scholar] [CrossRef] [PubMed]
- Mistry, B.A.; D’Orsogna, M.R.; Chou, T. The Effects of Statistical Multiplicity of Infection on Virus Quantification and Infectivity Assays. Biophys. J. 2018, 114, 2974–2985. [Google Scholar] [CrossRef]
- Roberts, G.O.; Gelman, A.; Gilks, W.R. Weak convergence and optimal scaling of random walk Metropolis algorithms. Ann. Appl. Probab. 1997, 7, 110–120. [Google Scholar] [CrossRef]
- Shinya, K.; Okamura, T.; Sueta, S.; Kasai, N.; Tanaka, M.; Ginting, T.E.; Makino, A.; Eisfeld, A.J.; Kawaoka, Y. Toll-like receptor pre-stimulation protects mice against lethal infection with highly pathogenic influenza viruses. Virol. J. 2011, 8, 97. [Google Scholar] [CrossRef]
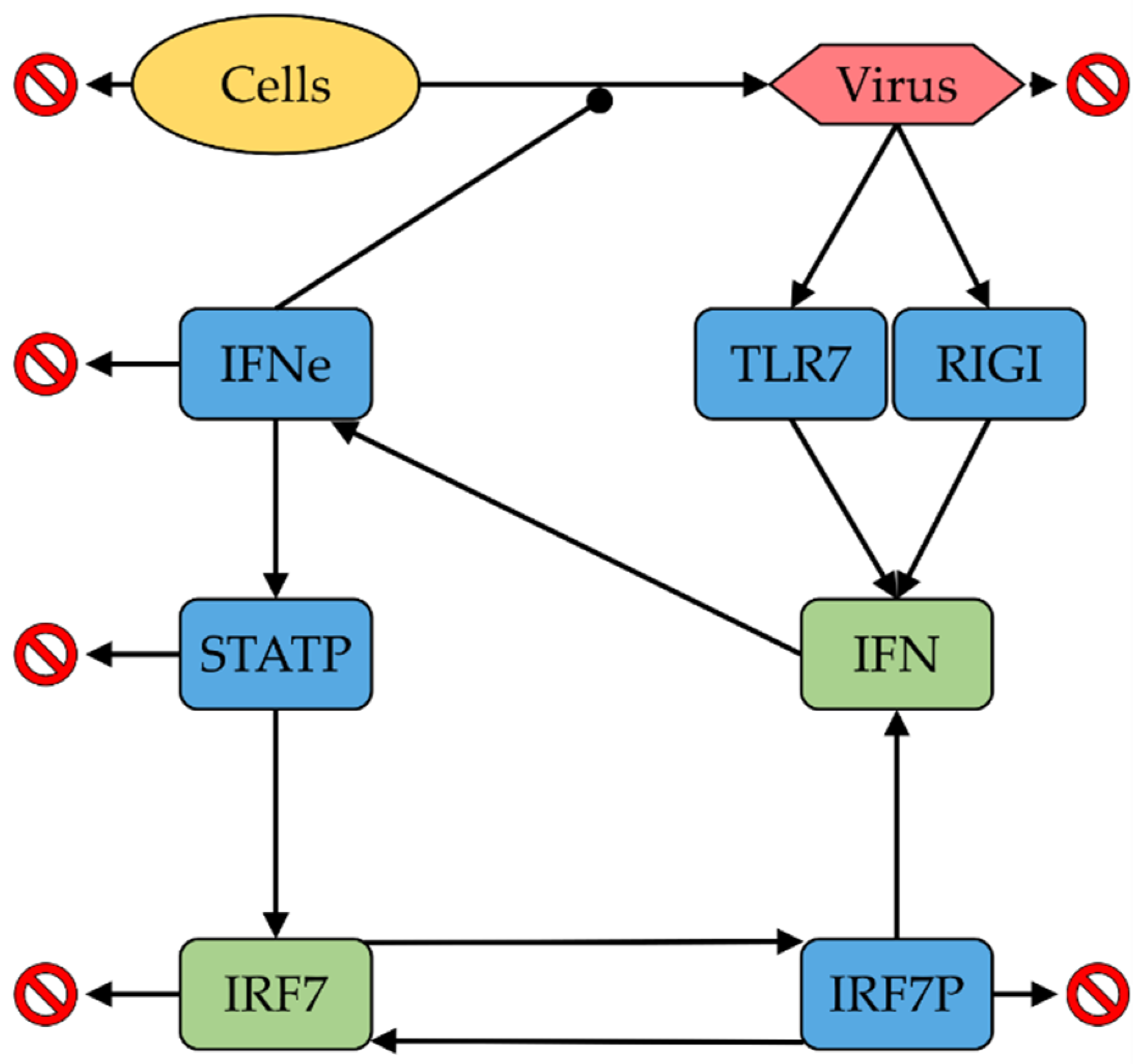
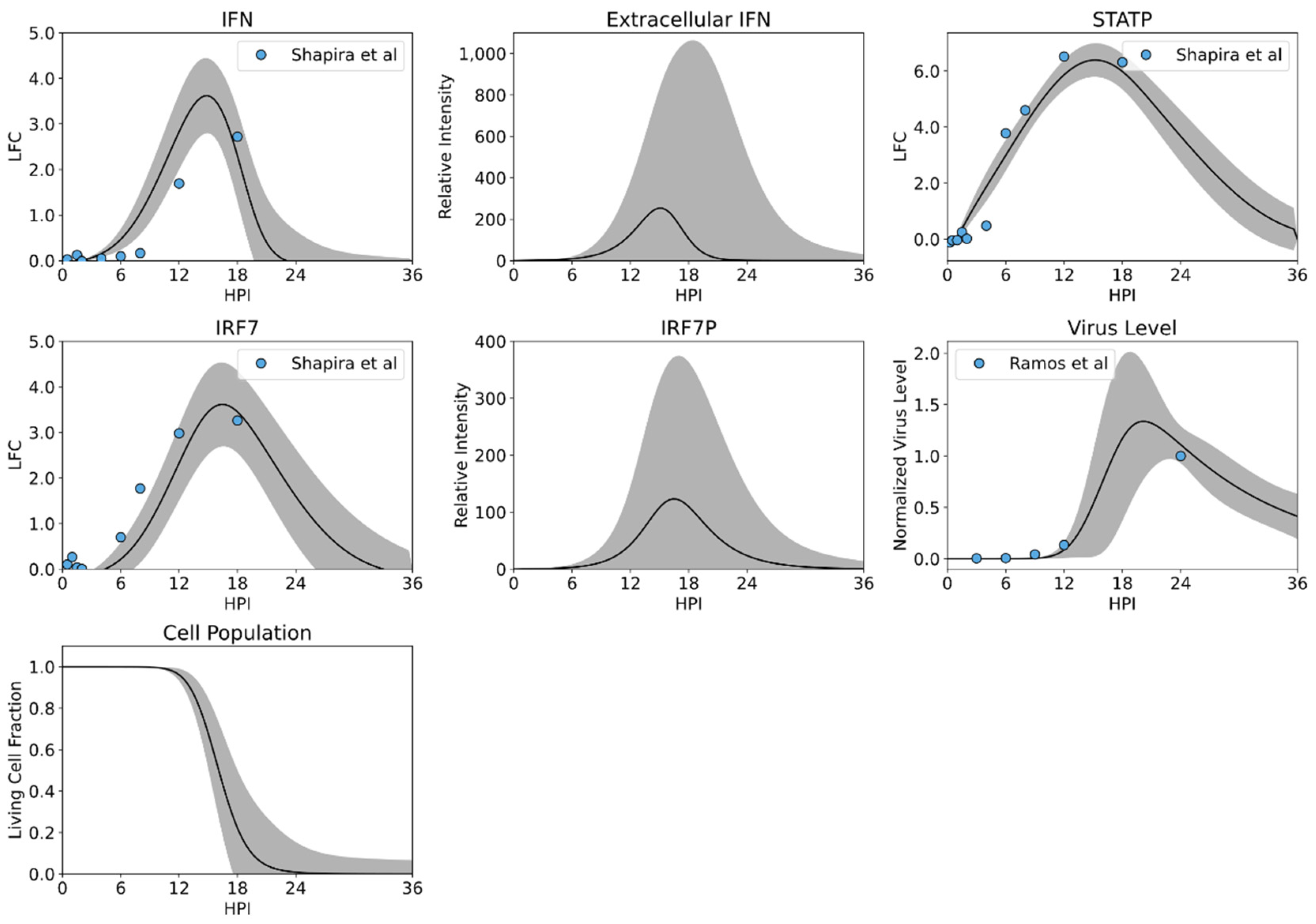
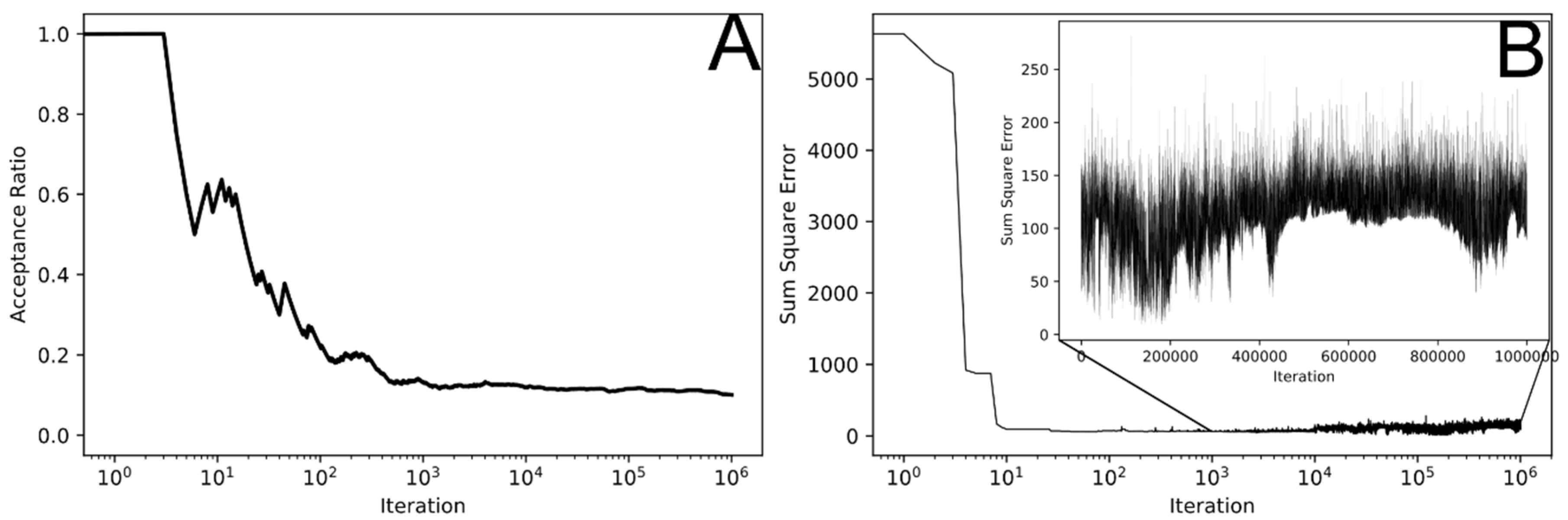
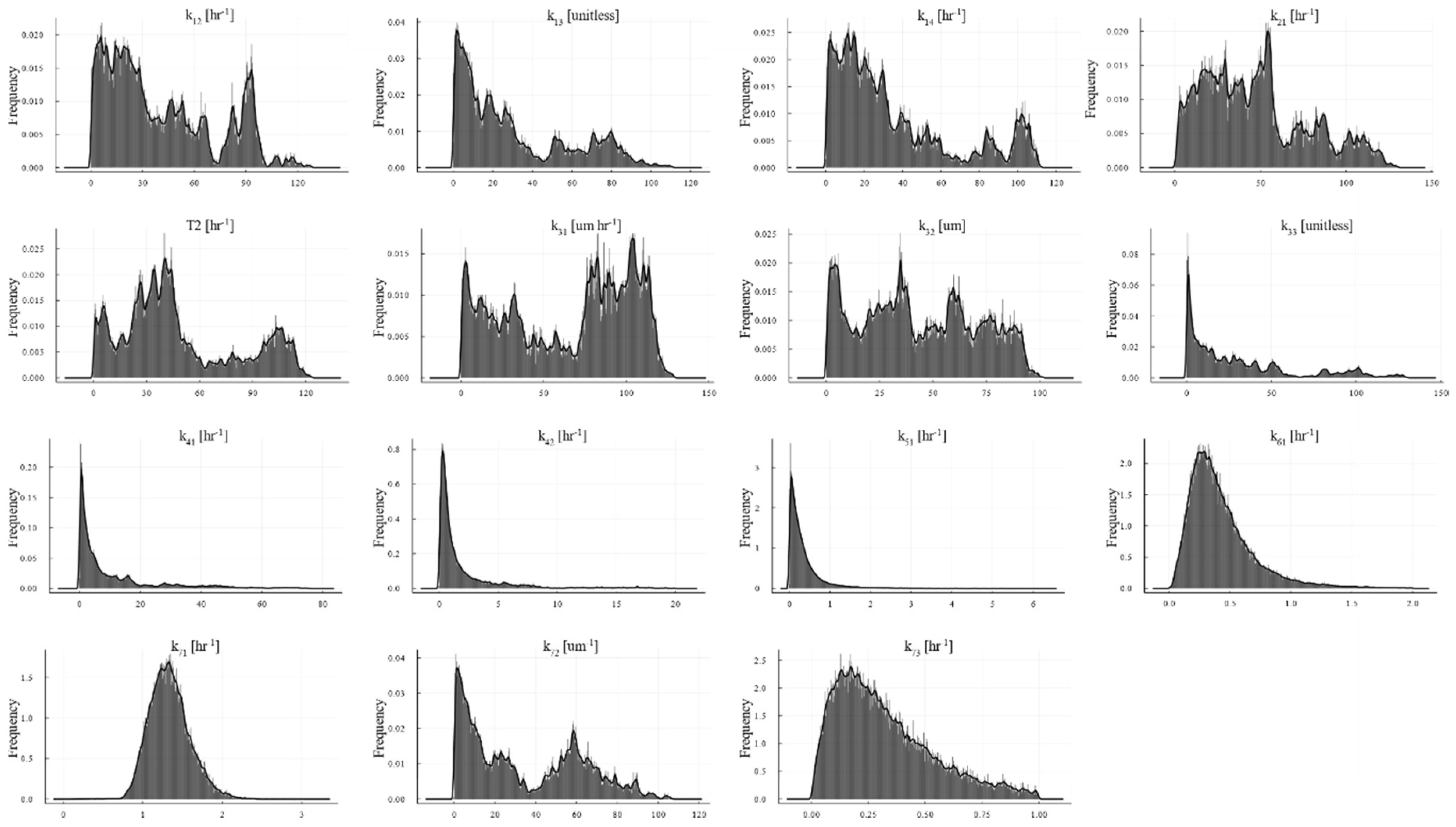

| Process | Parameter | Units | Value | Remarks |
|---|---|---|---|---|
| IFN induction via RIG-I | k11 | μM h−1 | 1e5 | Fit manually |
| IFN induction via TLR7 | k12 | h−1 | 9.746 | Fit via MCMC |
| k13 | [unitless] | 12.511 | Fit via MCMC | |
| IFN induction via IRF7P | k14 | h−1 | 13.562 | Fit via MCMC |
| Transport of IFN | k21 | h−1 | 10.385 | Fit via MCMC |
| IFNe degradation | 2 | h−1 | 3.481 | Fit via MCMC |
| STAT phosphorylation | k31 | μM h−1 | 45.922 | Fit via MCMC |
| k32 | μM | 5.464 | Fit via MCMC | |
| k33 | [unitless] | 0.068 | Fit via MCMC | |
| STATP dephosphorylation | 3 | h−1 | 0.3 | [39] |
| IRF7 induction via STATP | k41 | h−1 | 0.115 | Fit via MCMC |
| IRF7 induction via IRF7P | k42 | h−1 | 1.053 | Fit via MCMC |
| IRF7 mRNA degradation | 4 | h−1 | 0.3 | [40] |
| IRF7 phosphorylation | k51 | h−1 | 0.202 | Fit via MCMC |
| IRF7P Dephosphorylation | 5 | h−1 | 0.3 | [41] |
| Cell death | k61 | h−1 | 0.635 | Fit via MCMC |
| Viral replication | k71 | h−1 | 1.537 | Fit via MCMC |
| IFN effect on virus | k72 | μM−1 | 47.883 | Fit via MCMC |
| Nonspecific viral clearance | k73 | h−1 | 0.197 | Fit via MCMC |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Weaver, J.J.A.; Shoemaker, J.E. Mathematical Modeling of RNA Virus Sensing Pathways Reveals Paracrine Signaling as the Primary Factor Regulating Excessive Cytokine Production. Processes 2020, 8, 719. https://doi.org/10.3390/pr8060719
Weaver JJA, Shoemaker JE. Mathematical Modeling of RNA Virus Sensing Pathways Reveals Paracrine Signaling as the Primary Factor Regulating Excessive Cytokine Production. Processes. 2020; 8(6):719. https://doi.org/10.3390/pr8060719
Chicago/Turabian StyleWeaver, Jordan J. A., and Jason E. Shoemaker. 2020. "Mathematical Modeling of RNA Virus Sensing Pathways Reveals Paracrine Signaling as the Primary Factor Regulating Excessive Cytokine Production" Processes 8, no. 6: 719. https://doi.org/10.3390/pr8060719
APA StyleWeaver, J. J. A., & Shoemaker, J. E. (2020). Mathematical Modeling of RNA Virus Sensing Pathways Reveals Paracrine Signaling as the Primary Factor Regulating Excessive Cytokine Production. Processes, 8(6), 719. https://doi.org/10.3390/pr8060719






