Optimising Brewery-Wastewater-Supported Acid Mine Drainage Treatment vis-à-vis Response Surface Methodology and Artificial Neural Network
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemical Reagents
2.2. Bacterial Inoculum
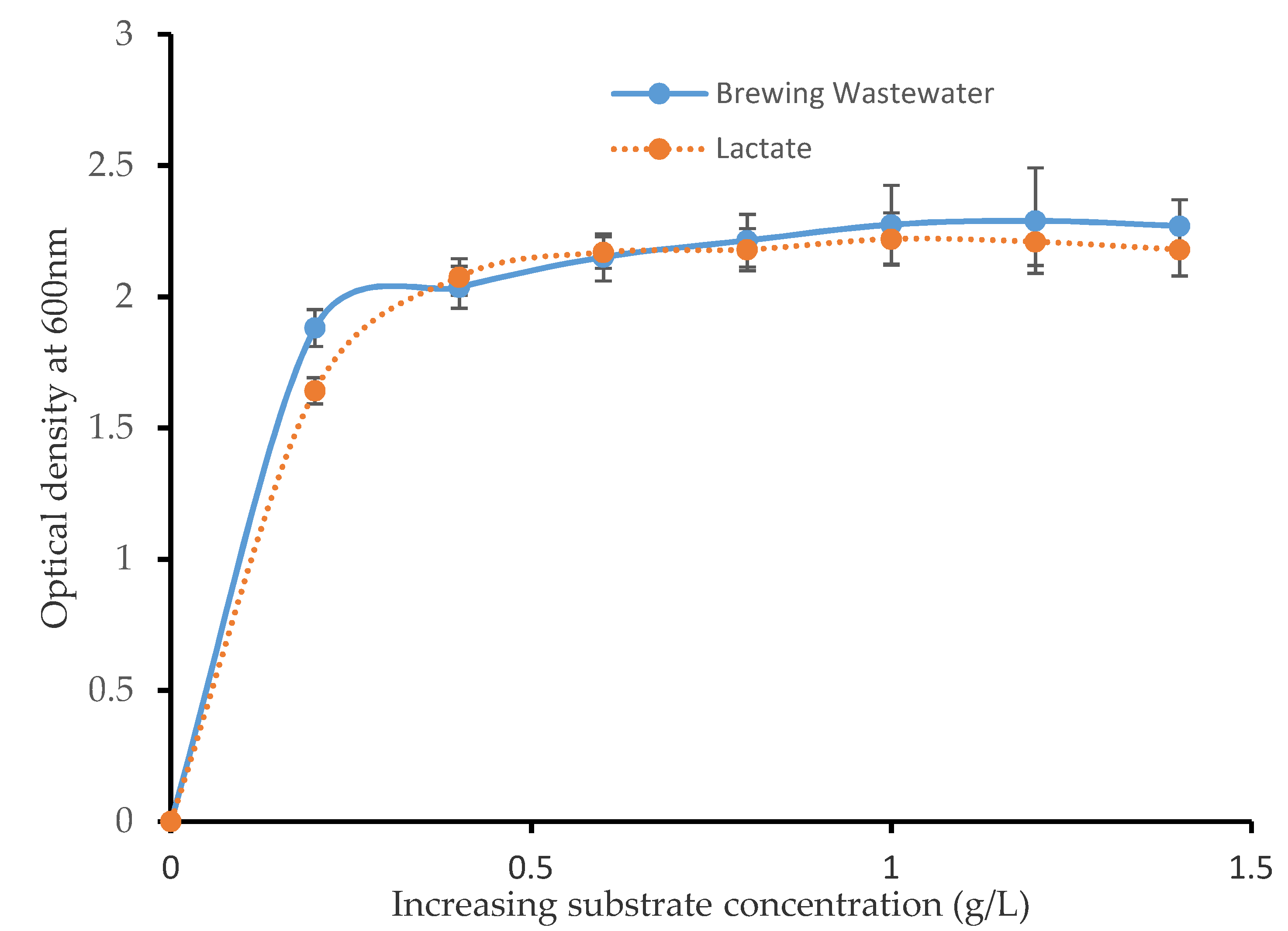
2.3. Carbon Source Limiting Growth Test
2.4. Experimental Set-Up
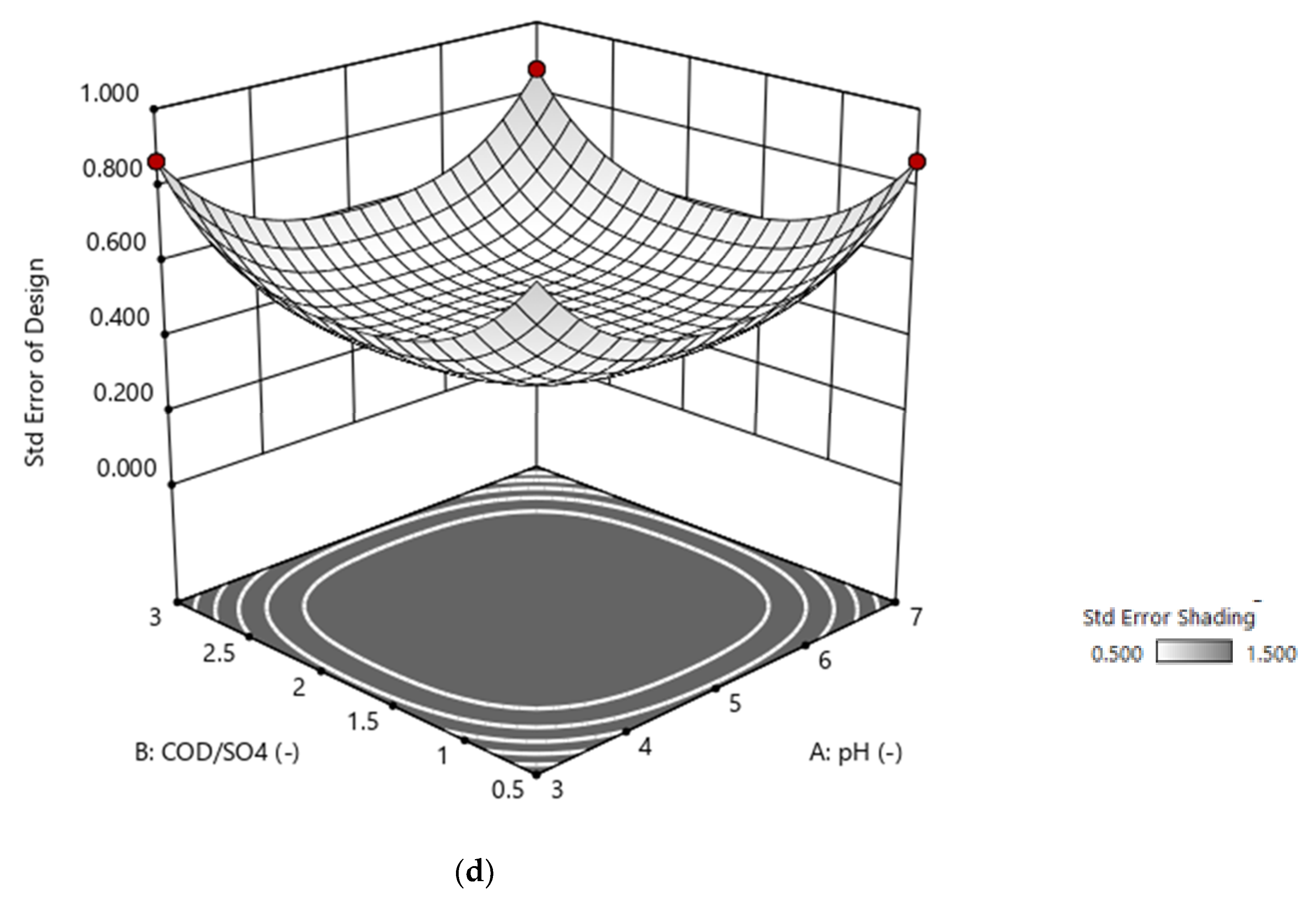
2.5. Design of Experiment—Box–Behnken
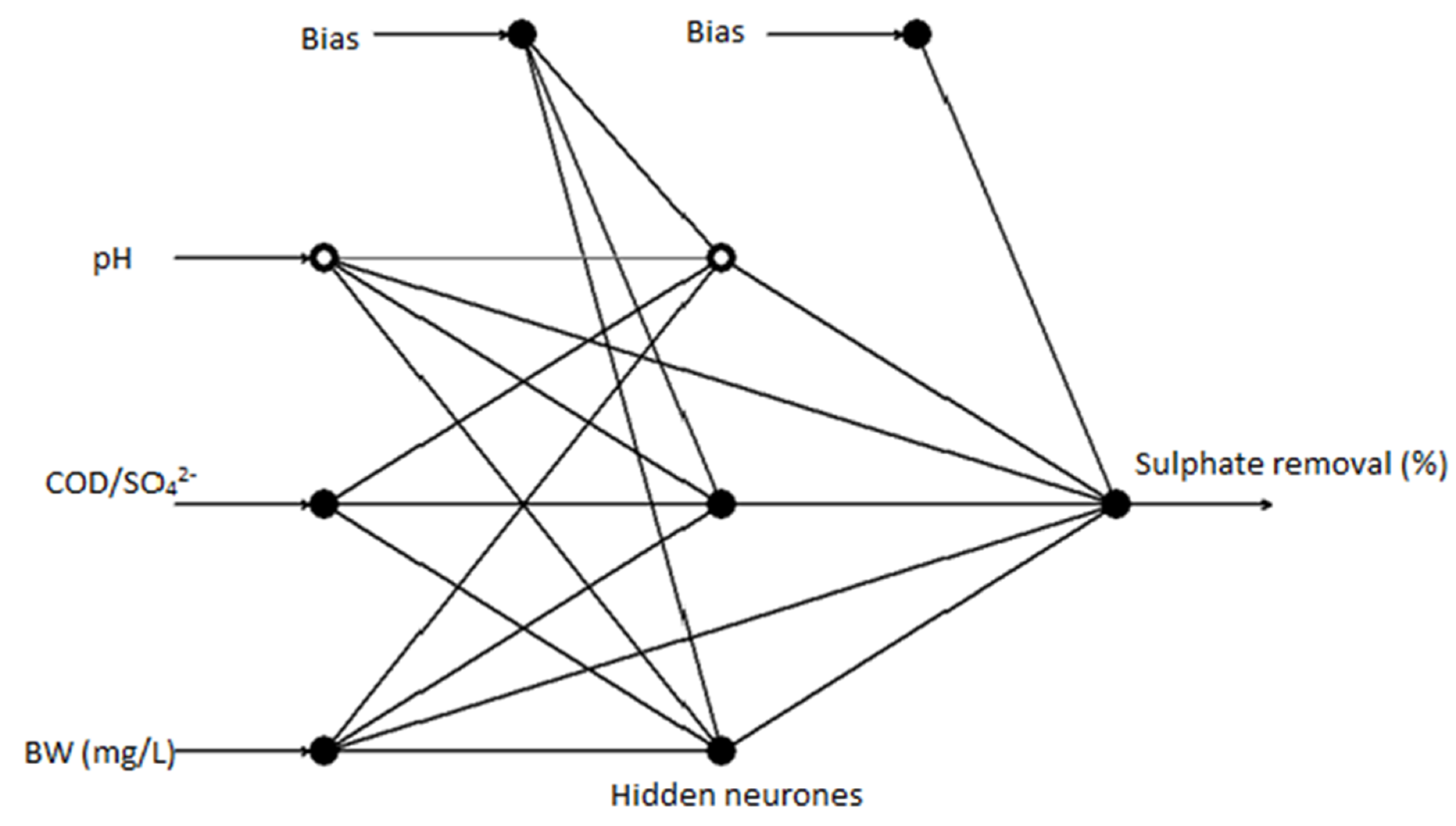
2.6. Artificial Neural Network (ANN) Analysis
2.7. Appraisal of Artificial Neural Network Predictability
3. Results and Discussion
3.1. Effect of Carbon Substrate Limitation on the SRB Consortium
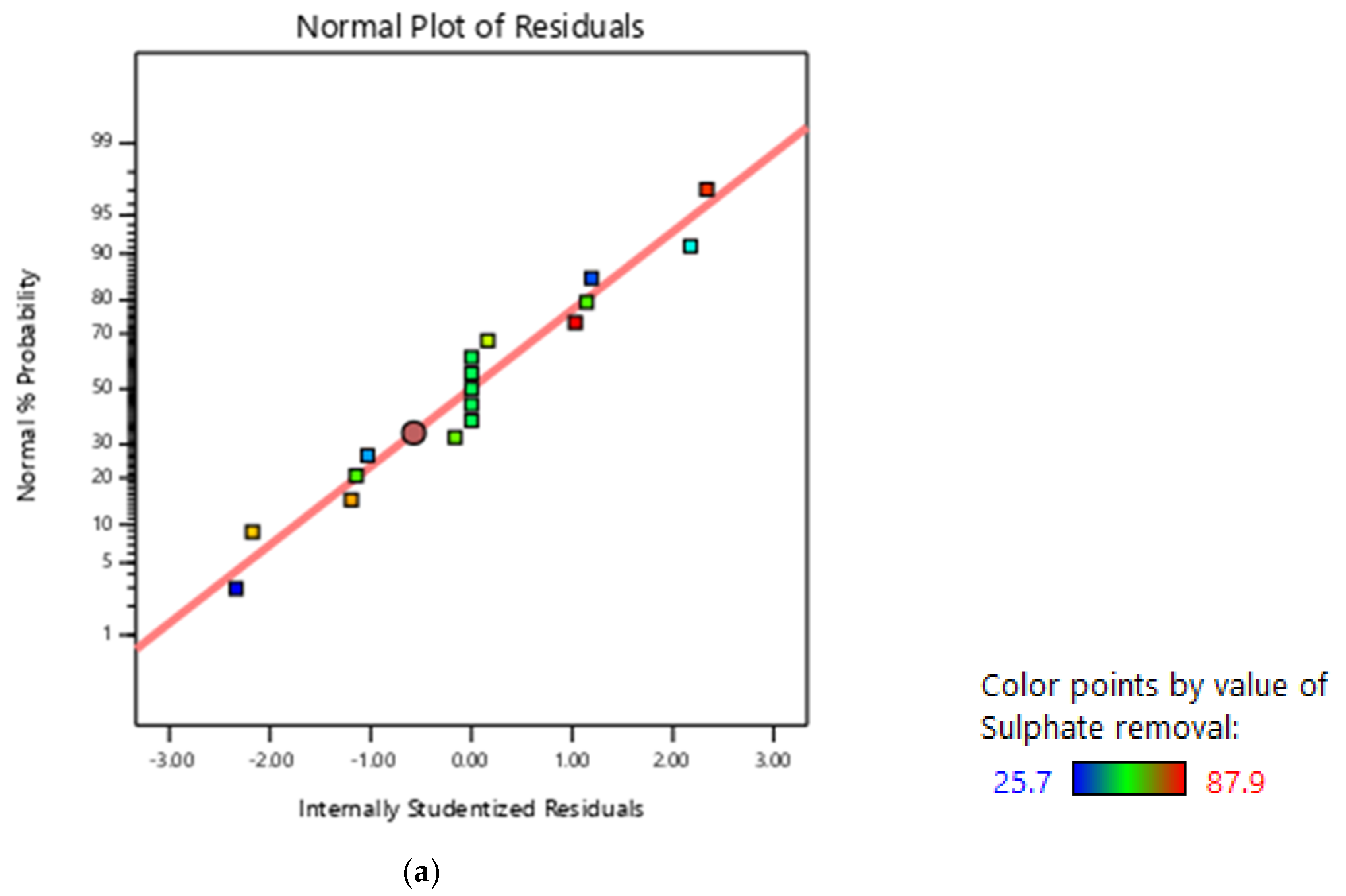
3.2. RSM Modelling: Box–Behnken Design
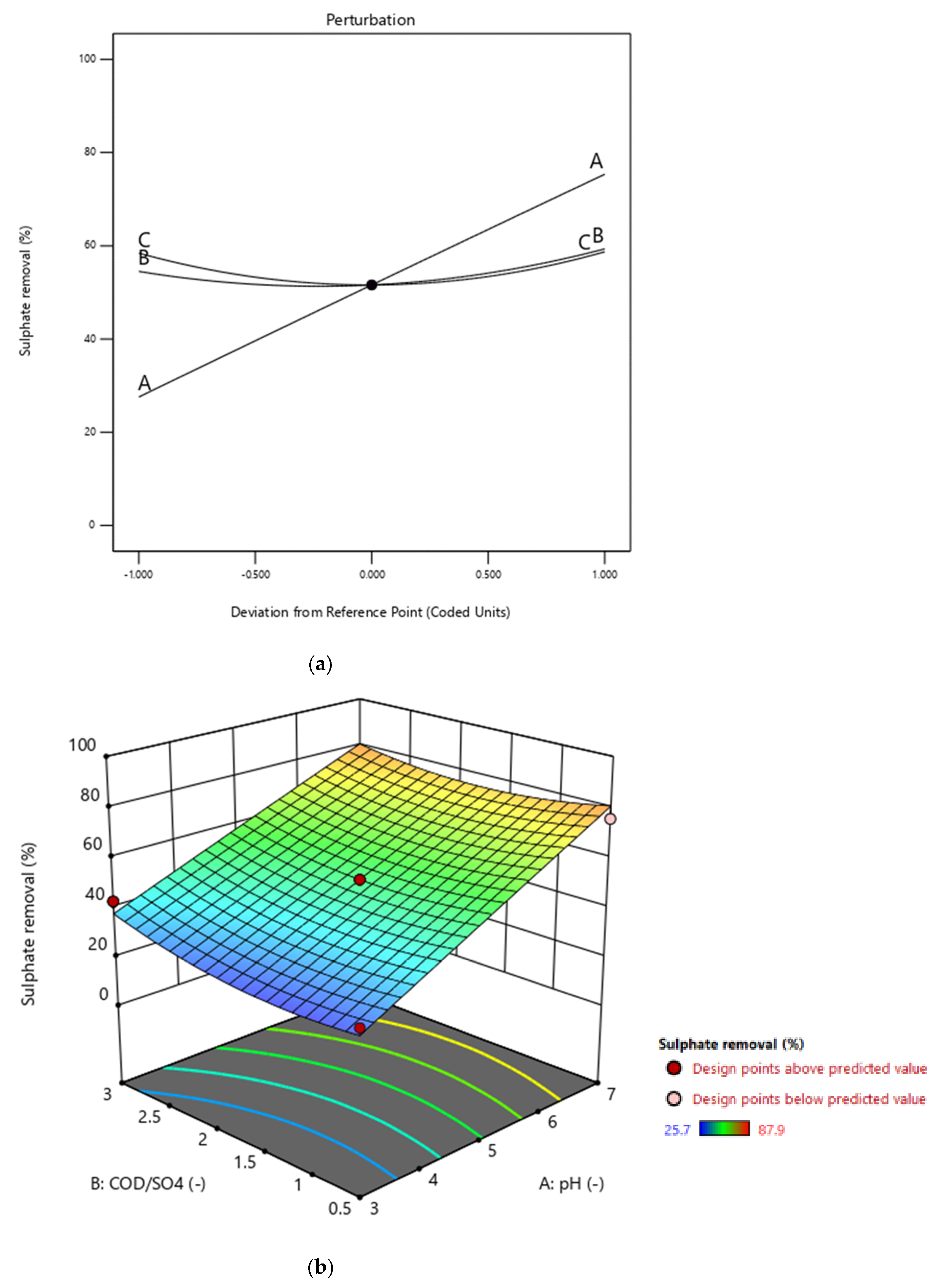
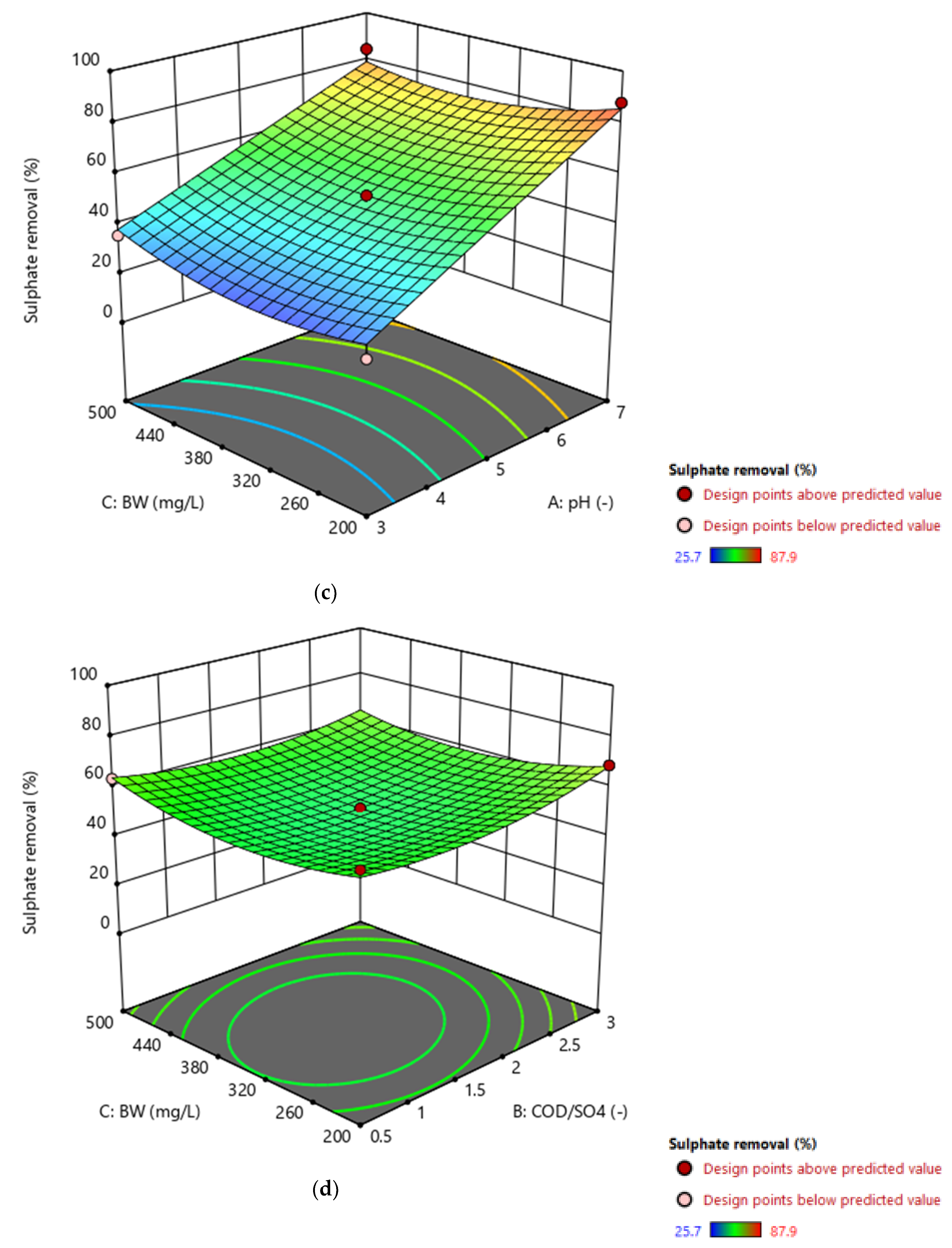
3.3. Graphical Representation of the Model
3.4. Artificial Neural Network Analysis of Sulphate Reduction Using Brewery Wastewater
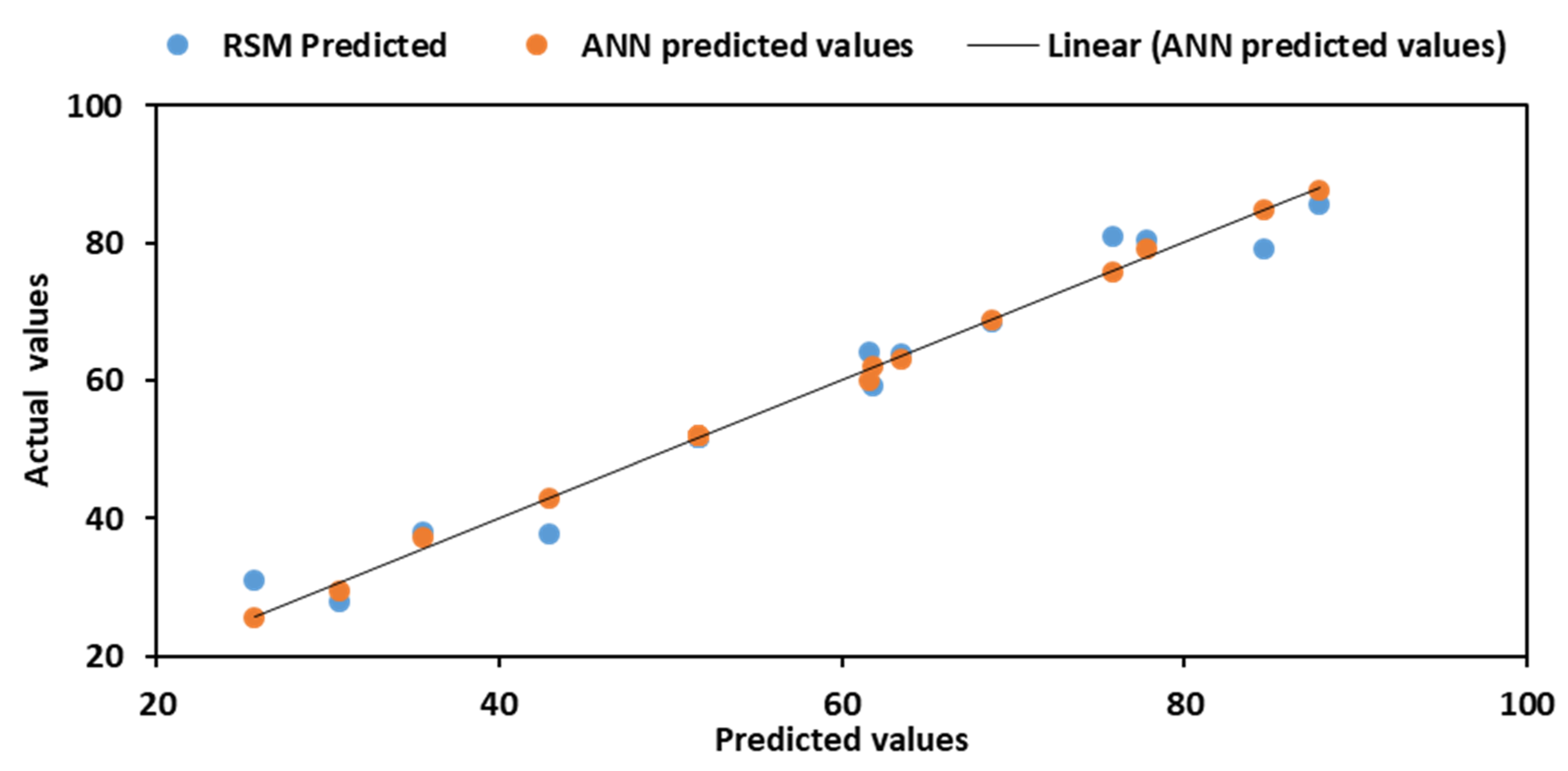
3.5. Optimum Comparison of RSM and ANN
3.6. Overall Effect of Individual Parameters
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- SAB. South African Breweries Heritage 2020. Available online: http://www.sab.co.za/heritage (accessed on 30 July 2020).
- Seluy, L.G.; Isla, M.A. A process to treat high-strength brewery wastewater via ethanol recovery and vinasse fermentation. Ind. Eng. Chem. Res. 2014, 53, 17043–17050. [Google Scholar] [CrossRef]
- Ma, L.; Liang, J.; Liu, Y.; Zhang, Y.; Ma, P.; Pan, Z.; Jiang, W. Production of a bioflocculant from Enterobacter sp. P3 using brewery wastewater as substrate and its application in fracturing flowback water treatment. Environ. Sci. Pollut. Res. 2020, 27, 18242–18253. [Google Scholar] [CrossRef] [PubMed]
- Hultberg, M.; Bodin, H. Fungi-based treatment of brewery wastewater—Biomass production and nutrient reduction. Appl. Microbiol. Biotechnol. 2017, 101, 4791–4798. [Google Scholar] [CrossRef] [PubMed]
- Nassary, E.K.; Nasolwa, E.R. Unravelling disposal benefits derived from underutilized brewing spent products in Tanzania. J. Environ. Manag. 2019, 242, 430–439. [Google Scholar] [CrossRef]
- Karlović, A.; Jurić, A.; Ćorić, N.; Habschied, K.; Krstanović, V.; Mastanjević, K. By-Products in the malting and brewing industries—Re-usage possibilities. Fermentation 2020, 6, 82. [Google Scholar] [CrossRef]
- Pham, T.-L.; Bui, M.H. Removal of nutrients from fertilizer plant wastewater using Scenedesmus sp.: Formation of bioflocculation and enhancement of removal efficiency. J. Chem. 2020, 2020, 8094272.C. [Google Scholar] [CrossRef]
- Hawrot-Paw, M.; Koniuszy, A.; Gałczyńska, M.; Zając, G.; Szyszlak-Bargłowicz, J. Production of microalgal biomass using aquaculture wastewater as growth medium. Water 2020, 12, 106. [Google Scholar] [CrossRef]
- Simate, G.S.; Cluett, J.; Iyuke, S.E.; Musapatika, E.T.; Ndlovu, S.; Walubita, L.F.; Alvarez, A.E. The treatment of brewery wastewater for reuse: State of the art. Desalination 2011, 273, 235–247. [Google Scholar] [CrossRef]
- Riera-Vila, I.; Anderson, N.O.; Hodge, C.F.; Rogers, M. Anaerobically-digested brewery wastewater as a nutrient solution for substrate-based food production. Horticulture 2019, 5, 43. [Google Scholar] [CrossRef]
- Nicholson, S.E.; Funk, C.; Fink, A.H. Rainfall over the African continent from the 19th through the 21st century. Glob. Planet Chang. 2018, 165, 114–127. [Google Scholar] [CrossRef]
- Auditor-General SA. Report of the Auditor-General to Parliament on a Performance Audit of the Rehabilitation of Abandoned Mines at the Department of Minerals and Energy; Auditor-General: Pretoria, South Africa, 2009. [Google Scholar]
- La, H.-J.; Kim, K.-H.; Quan, Z.-X.; Cho, Y.-G.; Lee, S.-T. Enhancement of sulfate reduction activity using granular sludge in anaerobic treatment of acid mine drainage. Biotechnol. Lett. 2003, 25, 503–508. [Google Scholar] [CrossRef] [PubMed]
- Valenzuela, E.I.; García-Figueroa, A.C.; Amábilis-Sosa, L.E.; Molina-Freaner, F.E.; Pat-Espadas, A.M. Stabilization of potentially toxic elements contained in mine waste: A microbiological approach for the environmental management of mine tailings. J. Environ. Manag. 2020, 270, 110873. [Google Scholar] [CrossRef] [PubMed]
- Sivamani, S.; Selvakumar, S.; Rajendran, K.; Muthusamy, S. Artificial neural network-genetic algorithm-based optimization of biodiesel production from Simarouba glauca. Biofuels 2019, 10, 393–401. [Google Scholar] [CrossRef]
- Betiku, E.; Taiwo, A.E. Modeling and optimization of bioethanol production from breadfruit starch hydrolyzate vis-à-vis response surface methodology and artificial neural network. Renew. Energy 2015, 74, 87–94. [Google Scholar] [CrossRef]
- Akinpelu, E.A.; Fosso-Kankeu, E.; Waanders, F.; Angadam, J.O.; Ntwampe, S.K.O. Diversity and Performance of sulphate-reducing bacteria in acid mine drainage remediation systems. In Frontiers in Water-Energy-Nexus—Nature-Based Solutions, Advanced Technologies and Best Practices for Environmental Sustainability; Naddeo, V., Balakrishnan, M., Choo, K.-H., Eds.; Springer: Cham, Switzerland, 2020; pp. 121–123. [Google Scholar]
- Kozik, V.; Barbusinski, K.; Thomas, M.; Sroda, A.; Jampilek, J.; Sochanik, A.; Smolinski, A.; Bak, A. Taguchi method and Response Surface Methodology in the treatment of highly contaminated tannery wastewater using commercial potassium ferrate. Materials 2019, 12, 3784. [Google Scholar] [CrossRef] [PubMed]
- Najib, T.; Solgi, M.; Farazmand, A.; Heydarian, S.M.; Nasernejad, B. Optimization of sulfate removal by sulfate reducing bacteria using response surface methodology and heavy metal removal in a sulfidogenic UASB reactor. J. Environ. Chem. Eng. 2017, 5, 3256–3265. [Google Scholar] [CrossRef]
- Akinpelu, E.A.; Ntwampe, S.K.O.; Mekuto, L.; Tombo, E.F.I. Optimizing the bioremediation of free cyanide containing wastewater by Fusarium oxysporum grown on beetroot waste using response surface methodology. In Lecture Notes in Engineering and Computer Science, Proceedings of the World Congress on Engineering and Computer Science, San Francisco, CA, USA, 22–24 October 2019; Ao, S.I., Douglas, C., Grundfest, W.S., Eds.; Newswood Limited: San Francisco, CA, USA, 2016; pp. 664–670. [Google Scholar]
- Mona, S.; Kaushik, A.; Kaushik, C.P. Biosorption of chromium(VI) by spent cyanobacterial biomass from a hydrogen fermentor using Box-Behnken model. Int. Biodeterior. Biodegrad. 2011, 65, 656–663. [Google Scholar] [CrossRef]
- Montgomery, D.C. Design and Analysis of Experiments; John Wiley & Sons: Hoboken, NJ, USA, 2008. [Google Scholar]
- Martins, M.; Faleiro, M.L.; Barros, R.J.; Veríssimo, A.R.; Costa, M.C. Biological sulphate reduction using food industry wastes as carbon sources. Biodegradation 2009, 20, 559–567. [Google Scholar] [CrossRef]
- Ghaffari, A.; Abdollahi, H.; Khoshayand, M.R.; Bozchalooi, I.S.; Dadgar, A.; Rafiee-Tehrani, M. Performance comparison of neural network training algorithms in modeling of bimodal drug delivery. Int. J. Pharm. 2006, 327, 126–138. [Google Scholar] [CrossRef]
- Taiwo, A.E.; Madzimbamuto, T.N.; Ojumu, T.V. Optimization of corn steep liquor dosage and other fermentation parameters for ethanol production by Saccharomyces cerevisiae type 1 and anchor instant yeast. Energies 2018, 11, 1740. [Google Scholar] [CrossRef]
- Sin, H.N.; Yusof, S.; Hamid, N.S.A.; Rahman, R.A. Optimization of enzymatic clarification of sapodilla juice using response surface methodology. J. Food Eng. 2006, 73, 313–319. [Google Scholar] [CrossRef]
- Nath, A.; Chattopadhyay, P.K. Optimization of oven toasting for improving crispness and other quality attributes of ready to eat potato-soy snack using response surface methodology. J. Food. Eng. 2007, 80, 1282–1292. [Google Scholar] [CrossRef]
- White, C.; Gadd, G.M. A comparison of carbon/energy and complex nitrogen sources for bacterial sulphate-reduction: Potential applications to bioprecipitation of toxic metals as sulphides. J. Ind. Microbiol. 1996, 17, 116–123. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.C.; Papirio, S.; Luongo, V.; Nancharaiah, Y.V.; Cennamo, P.; Esposito, G.; van Hullebusch, E.D.; Lens, P.N.L. Comparative performance of anaerobic attached biofilm and granular sludge reactors for the treatment of model mine drainage wastewater containing selenate, sulfate and nickel. Chem. Eng. J. 2018, 345, 545–555. [Google Scholar] [CrossRef]
- Weijma, J.; Bots, E.A.A.; Tandlinger, G.; Stams, A.J.M.; Pol, L.W.H.; Lettinga, G. Optimisation of sulphate reduction in a methanol-fed thermophilic bioreactor. Water Res. 2002, 36, 1825–1833. [Google Scholar] [CrossRef]
- Shafi, J.; Sun, Z.; Ji, M.; Gu, Z.; Ahmad, W. ANN and RSM based modelling for optimization of cell dry mass of Bacillus sp. strain B67 and its antifungal activity against Botrytis cinerea. Biotechnol. Biotechnol. Equip. 2018, 32, 58–68. [Google Scholar] [CrossRef]
- Sánchez-Andrea, I.; Sanz, J.L.; Bijmans, M.F.M.; Stams, A.J.M. Sulfate reduction at low pH to remediate acid mine drainage. J. Hazard. Mater. 2014, 269, 98–109. [Google Scholar] [CrossRef]
- Senko, J.M.; Zhang, G.; McDonough, J.T.; Bruns, M.A.; Burgos, W.D. Metal Reduction at Low pH by a Desulfosporosinus species: Implications for the biological treatment of acidic mine drainage. Geomicrobiol. J. 2009, 26, 71–82. [Google Scholar] [CrossRef]
- Sánchez-Andrea, I.; Stams, A.J.M.; Amils, R.; Sanz, J.L. Enrichment and isolation of acidophilic sulfate-reducing bacteria from Tinto River sediments. Environ. Microbiol. Rep. 2013, 5, 672–678. [Google Scholar] [CrossRef]
- Alazard, D.; Joseph, M.; Battaglia-Brunet, F.; Cayol, J.-L.; Ollivier, B. Desulfosporosinus acidiphilus sp. nov.: A moderately acidophilic sulfate-reducing bacterium isolated from acid mining drainage sediments. Extremophiles 2010, 14, 305–312. [Google Scholar] [CrossRef]
- Bijmans, M.F.M.; Dopson, M.; Peeters, T.W.T.; Lens, P.N.L.; Buisman, C.J.N. Sulfate reduction at pH 5 in a high-rate membrane bioreactor: Reactor performance and microbial community analyses. J. Microbiol. Biotechnol. 2009, 19, 698–708. [Google Scholar] [CrossRef] [PubMed]
- Mohan, S.V.; Rao, N.C.; Prasad, K.K.; Sarma, P.N. Bioaugmentation of an anaerobic sequencing batch biofilm reactor (AnSBBR) with immobilized sulphate reducing bacteria (SRB) for the treatment of sulphate bearing chemical wastewater. Process Biochem. 2005, 40, 2849–2857. [Google Scholar] [CrossRef]
- Nagpal, S.; Chuichulcherm, S.; Peeva, L.; Livingston, A. Microbial sulfate reduction in a liquid–solid fluidized bed reactor. Biotechnol. Bioeng. 2000, 70, 370–380. [Google Scholar] [CrossRef]
- Choi, E.; Rim, J.M. Competition and inhibition of sulfate reducers and methane producers in anaerobic treatment. Water Sci. Technol. 1991, 23, 1259–1264. [Google Scholar] [CrossRef]
- Kousi, P.; Remoundaki, E.; Hatzikioseyian, A.; Battaglia-Brunet, F.; Joulian, C.; Kousteni, V.; Tsezos, M. Metal precipitation in an ethanol-fed, fixed-bed sulphate-reducing bioreactor. J. Hazard. Mater. 2011, 189, 677–684. [Google Scholar] [CrossRef]

| Variable | Code | Unit | Coded Factor Level | ||
|---|---|---|---|---|---|
| 1 | 0 | −1 | |||
| pH | A | - | 7 | 5 | 3 |
| COD/SO42− | B | - | 3 | 1.75 | 0.5 |
| Brewing wastewater | C | mg/L | 500 | 350 | 200 |
| Run | Variables | Sulphate Removal (%) | ||||
|---|---|---|---|---|---|---|
| A | B | C | Actual | RSM Predicted | ANN Predicted | |
| 1 | 1 | 1 | 0 | 77.80 | 80.56 | 79.27 |
| 2 | −1 | −1 | 0 | 30.70 | 27.94 | 29.40 |
| 3 | 0 | 1 | −1 | 68.80 | 68.43 | 68.81 |
| 4 | 0 | 1 | 1 | 61.60 | 64.25 | 60.02 |
| 5 | 1 | 0 | 1 | 84.60 | 79.19 | 84.94 |
| 6 | 1 | 0 | −1 | 87.90 | 85.51 | 87.58 |
| 7 | −1 | 0 | −1 | 25.70 | 31.11 | 25.63 |
| 8 | 0 | 0 | 0 | 51.62 | 51.62 | 51.95 |
| 9 | 0 | 0 | 0 | 51.62 | 51.62 | 51.93 |
| 10 | −1 | 1 | 0 | 42.90 | 37.86 | 42.95 |
| 11 | 0 | −1 | 1 | 63.50 | 63.88 | 63.21 |
| 12 | 0 | 0 | 0 | 51.62 | 51.62 | 51.93 |
| 13 | 0 | 0 | 0 | 51.62 | 51.62 | 51.93 |
| 14 | −1 | 0 | 1 | 35.60 | 37.99 | 37.19 |
| 15 | 1 | −1 | 0 | 75.80 | 80.84 | 75.82 |
| 16 | 0 | 0 | 0 | 51.62 | 51.62 | 51.93 |
| 17 | 0 | −1 | −1 | 61.80 | 59.15 | 62.08 |
| Source | Sum of Squares | df | Mean Square | F-Value | p-Value | |
|---|---|---|---|---|---|---|
| Model | 5048.90 | 9 | 560.99 | 26.12 | 0.0001 | significant |
| A—pH | 4569.68 | 1 | 4569.68 | 212.78 | <0.0001 | significant |
| B—COD/SO4 | 46.56 | 1 | 46.56 | 2.17 | 0.1844 | |
| C—BW | 0.1512 | 1 | 0.1512 | 0.0070 | 0.9355 | |
| AB | 26.01 | 1 | 26.01 | 1.21 | 0.3075 | |
| AC | 43.56 | 1 | 43.56 | 2.03 | 0.1974 | |
| BC | 19.80 | 1 | 19.80 | 0.9221 | 0.3689 | |
| A2 | 0.0916 | 1 | 0.0916 | 0.0043 | 0.9498 | |
| B2 | 119.50 | 1 | 119.50 | 5.56 | 0.0504 | |
| C2 | 204.99 | 1 | 204.99 | 9.55 | 0.0176 | significant |
| Residual | 150.33 | 7 | 21.48 | |||
| Lack of Fit | 150.33 | 3 | 50.11 | |||
| Pure Error | 0.0000 | 4 | 0.0000 | |||
| Cor Total | 5199.23 | 16 |
| ANN Model Predictive Tool | Training Set | Testing Set |
|---|---|---|
| MSE | 0.53 | 0.76 |
| RMSE | 0.73 | 0.87 |
| AAD | 0.011 | 0.0093 |
| R2 | 0.99 | 0.99 |
| Statistical Tool | RSM Whole Data Set | ANN Whole Data Set | Optimisation Variable | RSM | ANN |
|---|---|---|---|---|---|
| MSE | 8.84 | 0.57 | Sulphate reduction (%) | 91.59 | 89.56 |
| RMSE | 2.97 | 0.75 | pH | 6.99 | 6.99 |
| AAD | 0.046 | 0.011 | COD/SO42− | 2.87 | 0.50 |
| R2 | 0.97 | 0.99 | BW (mg/L) | 200.24 | 200.31 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Akinpelu, E.A.; Ntwampe, S.K.O.; Taiwo, A.E.; Nchu, F. Optimising Brewery-Wastewater-Supported Acid Mine Drainage Treatment vis-à-vis Response Surface Methodology and Artificial Neural Network. Processes 2020, 8, 1485. https://doi.org/10.3390/pr8111485
Akinpelu EA, Ntwampe SKO, Taiwo AE, Nchu F. Optimising Brewery-Wastewater-Supported Acid Mine Drainage Treatment vis-à-vis Response Surface Methodology and Artificial Neural Network. Processes. 2020; 8(11):1485. https://doi.org/10.3390/pr8111485
Chicago/Turabian StyleAkinpelu, Enoch A., Seteno K. O. Ntwampe, Abiola E. Taiwo, and Felix Nchu. 2020. "Optimising Brewery-Wastewater-Supported Acid Mine Drainage Treatment vis-à-vis Response Surface Methodology and Artificial Neural Network" Processes 8, no. 11: 1485. https://doi.org/10.3390/pr8111485
APA StyleAkinpelu, E. A., Ntwampe, S. K. O., Taiwo, A. E., & Nchu, F. (2020). Optimising Brewery-Wastewater-Supported Acid Mine Drainage Treatment vis-à-vis Response Surface Methodology and Artificial Neural Network. Processes, 8(11), 1485. https://doi.org/10.3390/pr8111485






