Significant Effect of Sample Pretreatment on Ara h1 Extraction and Improved Sensitive SWCNT-Based Detection through Optimization
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
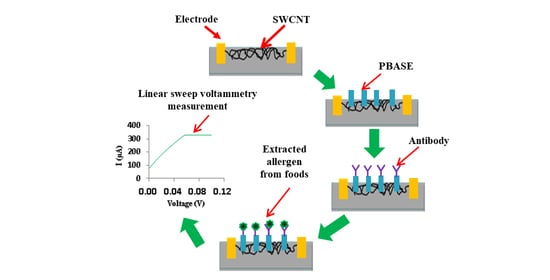
2.2. SWCNT-Based Nanobiosensor for the Detection of Ara h1 Protein
2.3. Analysis of Inhibitory Effects of Antibodies
2.4. Pretreatment of Ara h1 Protein from the Processed Food
2.5. Statistical Analysis
3. Results and Discussion
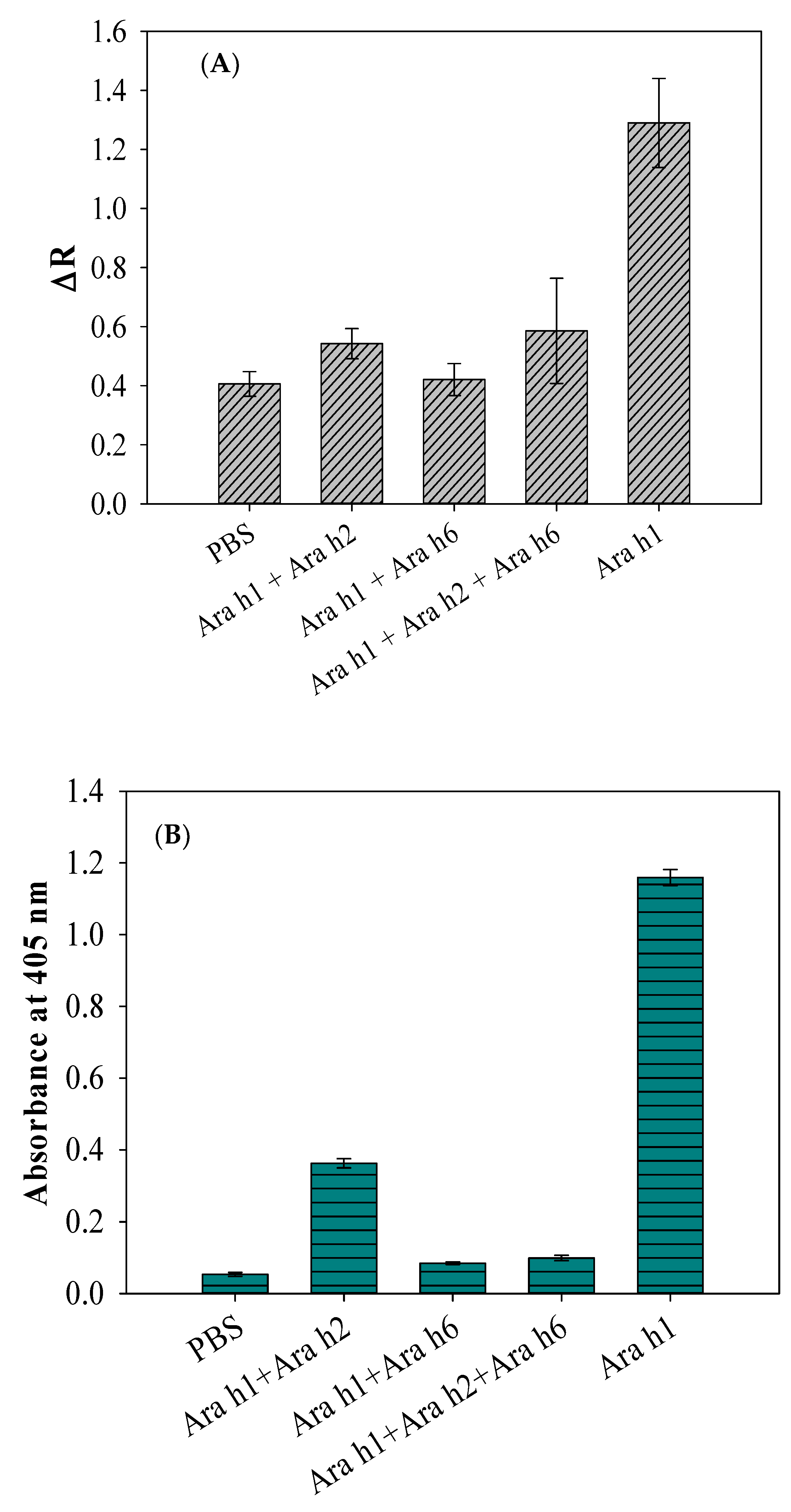
Inhibition Analysis of Antibodies
4. Conclusions
Funding
Conflicts of Interest
References
- Sobhan, A.; Oh, J.H.; Park, M.-K.; Lee, J. Detection of peanut allergen Ara h 6 in commercially processed foods using a single-walled carbon nanotube–based biosensor. J. AOAC Int. 2018, 101, 1558–1565. [Google Scholar] [CrossRef] [PubMed]
- Kagan, R.S.; Joseph, L.; Dufresne, C.; Gray-Donald, K.; Turnbull, E.; St Pierre, Y.; Clarke, A.E. Prevalence of peanut allergy in primary-school children in Montreal, Canad. J. Allergy Clin. Immunol. 2003, 112, 1223–1228. [Google Scholar] [CrossRef] [PubMed]
- Velusamy, V.; Arshak, K.; Korostynska, O.; Oliwa, K.; Adley, C. An overview of foodborne pathogen detection: In the perspective of biosensors. Biotechnol. Adv. 2010, 28, 232–254. [Google Scholar] [CrossRef]
- Sobhan, A.; Oh, J.-H.; Park, M.-K.; Kim, S.W.; Park, C.; Lee, J. Assessment of peanut allergen Ara h1 in processed foods using a SWCNTs-based nanobiosensor. Biosci. Biotechnol. Biochem. 2018, 82, 1134–1142. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Guan, L.; Shan, X.; Zhang, Y.; Li, Z. Electrochemical detection of peanut allergen Ara h 1 using a sensitive DNA biosensor based on stem–loop probe. J. Agric. Food Chem. 2012, 60, 10979–10984. [Google Scholar] [CrossRef]
- Wen, H.W.; Borejsza-Wysocki, W.; DeCory, T.R.; Durst, R.A. Peanut allergy, peanut allergens, and methods for the detection of peanut contamination in food products. Compr. Rev. Food Sci. Food Saf. 2007, 6, 47–58. [Google Scholar] [CrossRef]
- Alves, R.C.; Pimentel, F.B.; Nouws, H.P.A.; Silva, T.H.B.; Oliveira, M.B.P.P.; Delerue-Matos, C. Improving the extraction of Ara h 6 (a peanut allergen) from a chocolate-based matrix for immunosensing detection: Influence of time, temperature and additives. Food Chem. 2017, 218, 242–248. [Google Scholar] [CrossRef]
- Sobhan, A.; Oh, J.H.; Park, M.K.; Lee, J. Reusability of a single-walled carbon nanotube-based biosensor for detecting peanut allergens and Y. enterocolitica. J. Microelectron. Eng. 2020, 225, 111281. [Google Scholar] [CrossRef]
- Yamada, K.; Kim, C.-T.; Kim, J.-H.; Chung, J.-H.; Lee, H.G.; Jun, S. Single walled carbon nanotube-based junction biosensor for detection of Escherichia coli. PLoS ONE 2014, 9, e105767. [Google Scholar] [CrossRef]
- Sobhan, A.; Lee, J.; Park, M.K.; Oh, J.H. Rapid detection of Yersinia enterocolitica using a single–walled carbon nanotube-based biosensor for Kimchi product. LWT 2019, 108, 48–54. [Google Scholar] [CrossRef]
- Sobhan, A.; Oh, J.; Lee, J. Rapid Detection of Ara h2 using single walled carbon nanotube based biosensor for peanut allergen control. Appl. Mech. Mater. 2018, 878, 286–290. [Google Scholar] [CrossRef]
- Malorny, B.; Paccassoni, E.; Fach, P.; Bunge, C.; Martin, A.; Helmuth, R. Diagnostic real-time PCR for detection of salmonella in food. Appl. Environ. Microbiol. 2004, 70, 7046–7052. [Google Scholar] [CrossRef] [PubMed]
- García-Aljaro, C.; Cella, L.N.; Shirale, D.J.; Park, M.; Muñoz, F.J.; Yates, M.V.; Mulchandani, A. Carbon nanotubes-based chemiresistive biosensors for detection of microorganisms. Biosens. Bioelectron. 2010, 26, 1437–1441. [Google Scholar] [CrossRef]
- Sobhan, A.; Muthukumarappan, K.; Cen, Z.; Wei, L. Characterization of nanocellulose and activated carbon nanocomposite films’ biosensing properties for smart packaging. Carbohydr. Polym. 2019, 225, 115189. [Google Scholar] [CrossRef] [PubMed]
- Allen, B.L.; Kichambare, P.D.; Star, A. Carbon nanotube field-effect-transistor-based biosensors. Adv. Mater. 2007, 19, 1439–1451. [Google Scholar] [CrossRef]
- Iijima, S. Carbon nanotubes: Past, present, and future. Phys. B Condens. Matter. 2002, 323, 1–5. [Google Scholar] [CrossRef]
- Katz, E.; Willner, I. Biomolecule-functionalized carbon nanotubes: Applications in nanobioelectronics. Chem. Phys. Chem. 2004, 5, 1084–1104. [Google Scholar] [CrossRef]
- Kang, I.; Yeo, Y.; Kim, J.H.; Won, J.; Gollapudi, R.; Subramaniam, S.; Narasimhadevara, S.; Hurd, D.; Boerio, J.; Mall, S.; et al. Introduction to carbon nanotube and nanofiber smart materials. Compos. Part B Eng. 2006, 37, 382–394. [Google Scholar] [CrossRef]
- Barone, P.W. Near-infrared optical sensors based on single-walled carbon nanotubes. Nat. Mater. 2005, 4, 86–92. [Google Scholar] [CrossRef]
- Sobhan, A.; Muthukumarappan, K.; Wei, L.; Van Den Top, T.; Zhou, R. Development of an activated carbon-based nanocomposite film with antibacterial property for smart food packaging. Mater. Today Commun. 2020, 23, 101124. [Google Scholar] [CrossRef]
- Heller, I.; Janssens, A.M.; Ma, J.; Minot, E.D.; Lemay, S.G.; Dekker, C. Identifying the mechanism of biosensing with carbon nanotube transistors. Nano Lett. 2008, 8, 591–595. [Google Scholar] [CrossRef] [PubMed]
- Maroto, A.; Balasubramanian, K. functionalized metallic carbon nanotube devices for pH sensing. Chem. Phys. Chem. 2007, 8, 220–223. [Google Scholar] [CrossRef]
- HocineL, L.; Pitre, M. Quantitative and qualitative optimization of allergen extraction from peanut and selected tree nuts. Part 2. Optimization of buffer and ionic strength using a full factorial experimental design. Food Chem. 2016, 194, 820–827. [Google Scholar]
- Jayasena, S.; Smits, M.; de Jong, A.; Nordlee, J.; Baumert, J.; Taylor, S.L.; Pieters, R.H.; Koppelman, S.J. Comparison of six commercial ELISA kits for their specificity and sensitivity in detecting different major peanut allergens. J. Agric. Food Chem. 2015, 63, 1849–1855. [Google Scholar] [CrossRef]
- Sobhan, A.; Oh, J.-H.; Park, M.-K.; Kim, S.W.; Park, C.; Lee, J. Single walled carbon nanotube based biosensor for detection of peanut allergy-inducing protein Ara h1. J. Korean J. Chem. Eng. 2018, 34, 172–178. [Google Scholar] [CrossRef]
- Pomés, A.; Vinton, R.; Chapman, M.D. Peanut allergen (Ara h 1) detection in foods containing chocolate. J. Food Prot. 2004, 67, 793–798. [Google Scholar] [CrossRef]
- Hurst, W.J.; Krout, E.R.; Burks, W.R. A comparison of commercially available peanut ELISA test kits on the analysis of samples of dark and milk chocolate. Immunoass. Immunochem. 2002, 23, 451–459. [Google Scholar] [CrossRef]
- Pal, S.; Ying, W.; Alocilja, E.C.; Downes, F.P. Sensitivity and specificity performance of a direct-charge transfer biosensor for detecting Bacillus cereus in selected food matrices. Biosyst. Eng. 2008, 99, 461–468. [Google Scholar] [CrossRef]
- Jin, M.; Lang, J.; Shen, Z.Q.; Chen, Z.L.; Qiu, Z.G.; Wang, X.W.; Li, J.W. A rapid subtractive immunization method to prepare discriminatory monoclonal antibodies for food E. coli O157:H7 contamination. PLoS ONE 2012, 7, e31352. [Google Scholar] [CrossRef]
- Alves, R.C.; Pimentel, F.B.; Nouws, H.P.A.; Correr, W.; González-García, M.B.; Oliveira, M.B.P.P.; Delerue-Matos, C. Detection of the peanut allergen Ara h 6 in foodstuffs using a voltammetric biosensing approach. Anal. Bioanal. Chem. 2015, 407, 7157–7163. [Google Scholar] [CrossRef]
- Maier, I.; Morgan, M.R.A.; Lindner, W.; Pittner, F. Optical resonance-enhanced absorption-based near-field immunochip biosensor for allergen detection. Anal. Chem. 2008, 80, 2694–2703. [Google Scholar] [CrossRef]
- Lu, Y.; Ohshima, T.; Ushio, H. Rapid Detection of fish major allergen parvalbumin by surface plasmon resonance biosensor. J. Food Sci. 2004, 69, C652–C658. [Google Scholar] [CrossRef]
- Pollet, J.; Delport, F.; Janssen, K.P.F.; Tran, D.T.; Wouters, J.; Verbiest, T. Fast and accurate peanut allergen detection with nanobead enhanced optical fiber SPR biosensor. J. Lammertyn Talanta 2011, 83, 1436–1441. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Han, J.; Wu, Y.; Yuan, F.; Chen, Y.; Ge, Y. Simultaneous detection of eight food allergens using optical thin-film biosensor chips. J. Agric. Food Chem. 2011, 59, 6889–6894. [Google Scholar] [CrossRef]



| Extract No. | Extraction Condition (60 °C, 15 min) | Ara h1 Concentration (ng/mL Sample) | CV (%) | Extraction Condition (60 °C, 60 min) | Ara h1 Concentration (ng/mL Sample) | CV (%) |
|---|---|---|---|---|---|---|
| 1 | PBS | NA | NA | PBS | NA | NA |
| 2 | B | 14.62 ± 1.30 | 8.81 | B | 7.30 ± 1.46 | 20.08 |
| 3 | B/0.1S | 21.95 ± 3.35 | 15.28 | B/0.1S | 12.35 ± 0.98 | 7.94 |
| 4 | B/1S | 27.74 ± 1.33 | 4.81 | B/1S | 19.15 ± 3.61 | 18.88 |
| 5 | B/1M | 9.03 ± 1.80 | 19.97 | B/1S | 5.68 ± 1.65 | 29.21 |
| 6 | B/2M | 3.22 ± 0.40 | 19.97 | B/2M | 3.17 ± 2.10 | 66.23 |
| 7 | B/0.1S/1M | 2.61 ± 0.11 | 12.47 | B/0.1S/1M | 15.67 ± 2.23 | 14.26 |
| 8 | B/0.1S/2M | 2.35 ± 1.81 | 4.37 | B/0.1S/2M | 15.53 ± 0.79 | 5.08 |
| 9 | B/1S/1M | 21.67 ± 1.23 | 76.85 | B/1S/1M | 11.56 ± 1.18 | 10.25 |
| 10 | B/1S/2M | 5.06 ± 1.16 | 22.92 | B/1S/2M | 2.45 ± 0.23 | 9.38 |
| Extract No. | Extraction Condition (21 °C, 15 min) | Ara h1 Concentration (ng/mL Sample) | CV (%) | Extraction Condition (21 °C, 60 min) | Ara h1 Concentration (ng/mL Sample) | CV (%) |
|---|---|---|---|---|---|---|
| 1 | PBS | NA | NA | PBS | NA | NA |
| 2 | B | 16.28 ± 0.42 | 9.05 | B | 62.74 ± 5.63 | 8.98 |
| 3 | B/0.1S | 29.29 ± 2.65 | 2.58 | B/0.1S | 49.81 ± 5.63 | 11.29 |
| 4 | B/1S | 29.59 ± 2.57 | 8.69 | B/1S | 58.18 ± 7.05 | 12.11 |
| 5 | B/1M | 7.23 ± 4.93 | 68.19 | B/1M | 84.60 ± 7.50 | 8.86 |
| 6 | B/2M | 2.37 ± 0.11 | 29.73 | B/2M | 76.12 ± 1.85 | 2.43 |
| 7 | B/0.1S/1M | 3.19 ± 3.05 | 95.61 | B/0.1S/1M | 77.50 ± 10.81 | 13.95 |
| 8 | B/0.1S/2M | 3.18 ± 0.03 | 2.54 | B/0.1S/2M | 73.77 ± 7.98 | 10.83 |
| 9 | B/1S/1M | 19.45 ± 0.21 | 1.07 | B/1S/1M | 25.94 ± 1.38 | 5.32 |
| 10 | B/1S/2M | 2.31 ± 0.12 | 5.19 | B/1S/2M | 22.85 ± 3.09 | 13.50 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, J. Significant Effect of Sample Pretreatment on Ara h1 Extraction and Improved Sensitive SWCNT-Based Detection through Optimization. Processes 2020, 8, 1420. https://doi.org/10.3390/pr8111420
Lee J. Significant Effect of Sample Pretreatment on Ara h1 Extraction and Improved Sensitive SWCNT-Based Detection through Optimization. Processes. 2020; 8(11):1420. https://doi.org/10.3390/pr8111420
Chicago/Turabian StyleLee, Jinyoung. 2020. "Significant Effect of Sample Pretreatment on Ara h1 Extraction and Improved Sensitive SWCNT-Based Detection through Optimization" Processes 8, no. 11: 1420. https://doi.org/10.3390/pr8111420
APA StyleLee, J. (2020). Significant Effect of Sample Pretreatment on Ara h1 Extraction and Improved Sensitive SWCNT-Based Detection through Optimization. Processes, 8(11), 1420. https://doi.org/10.3390/pr8111420