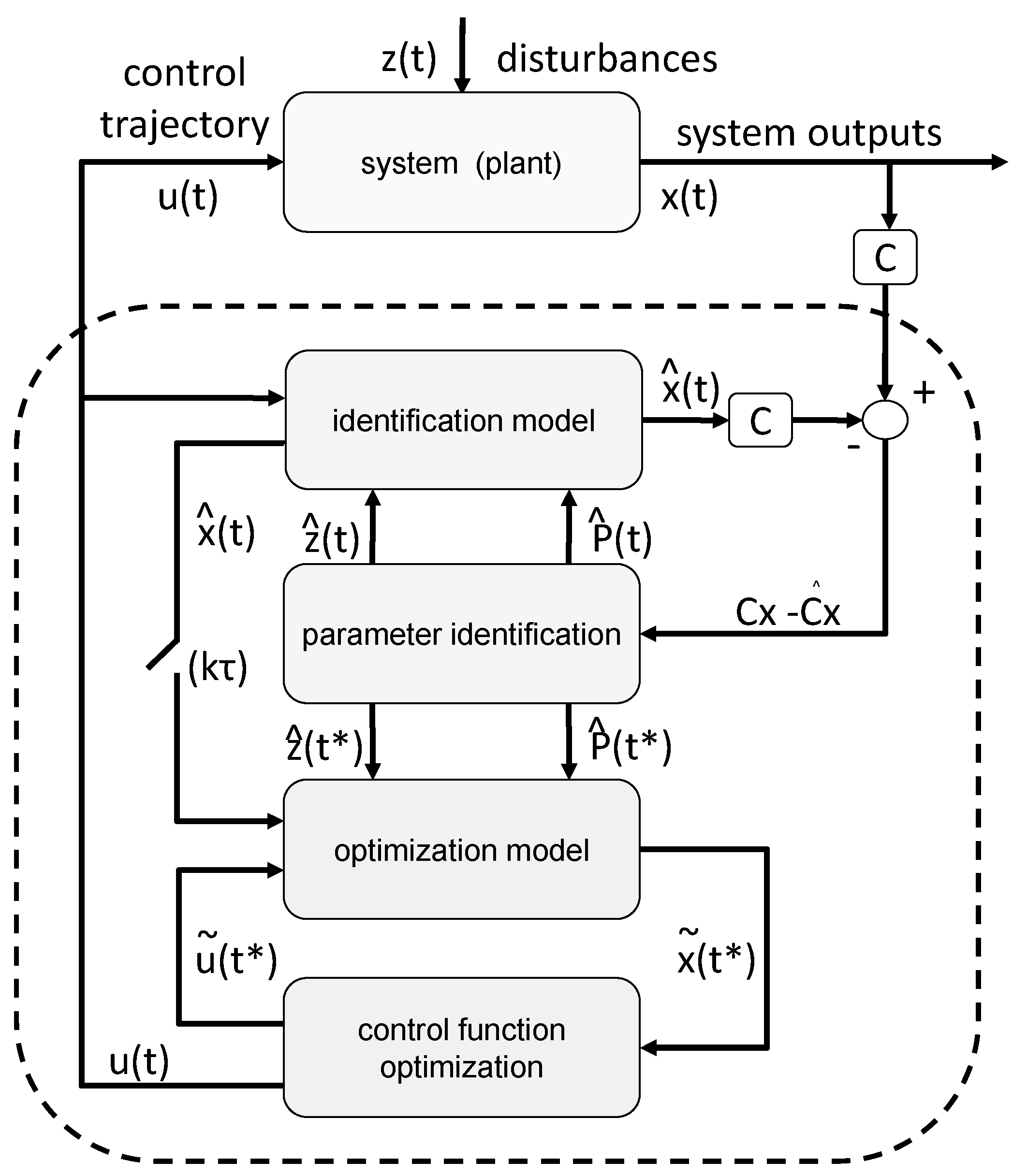
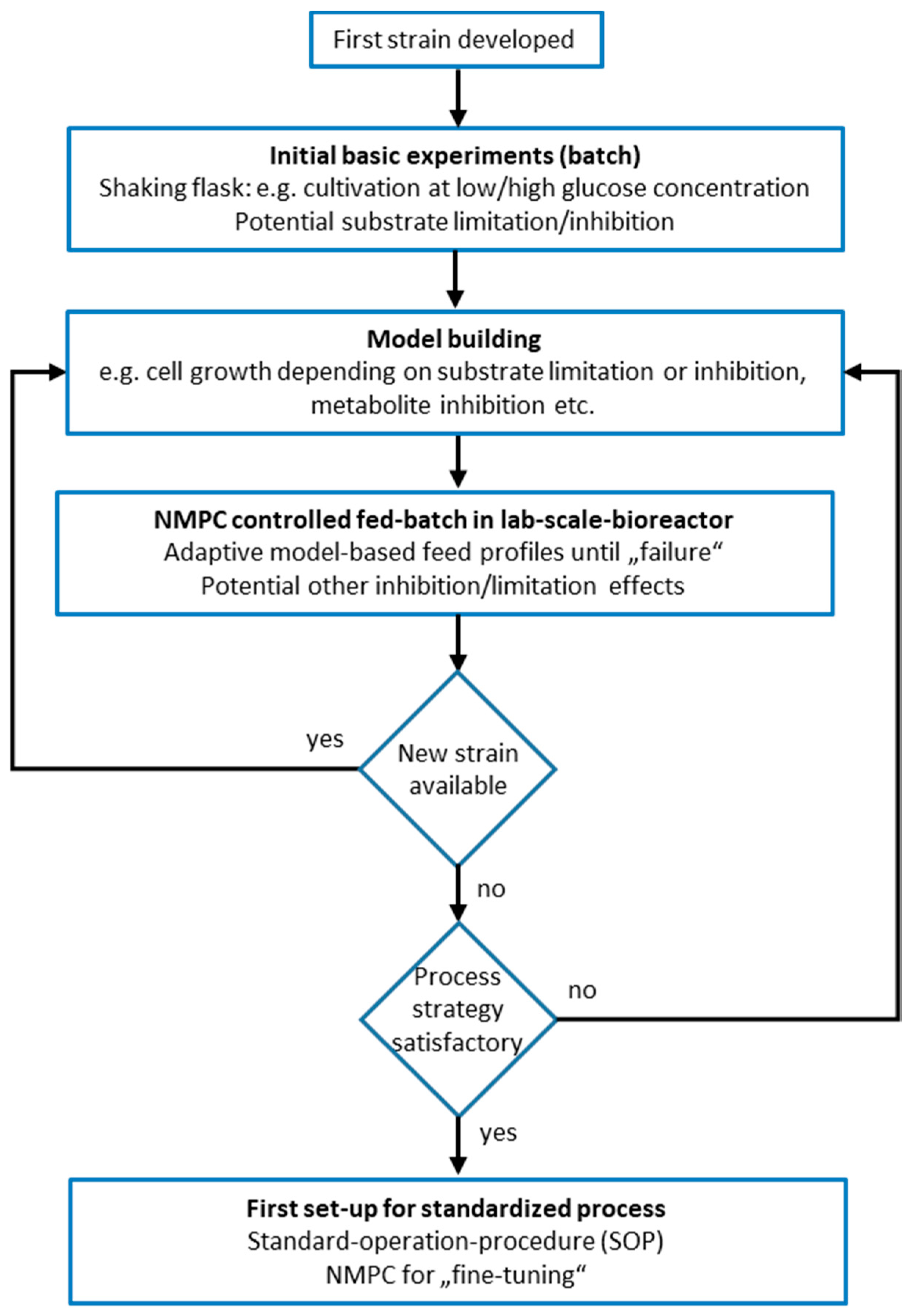
In the following, the experimental data together with the simulation results are discussed in the context of the process development workflow.
3.2.1. Experimental Setup 1-Strain 1–Batch and Fed-Batch Model A/B
Following the process development strategy (see
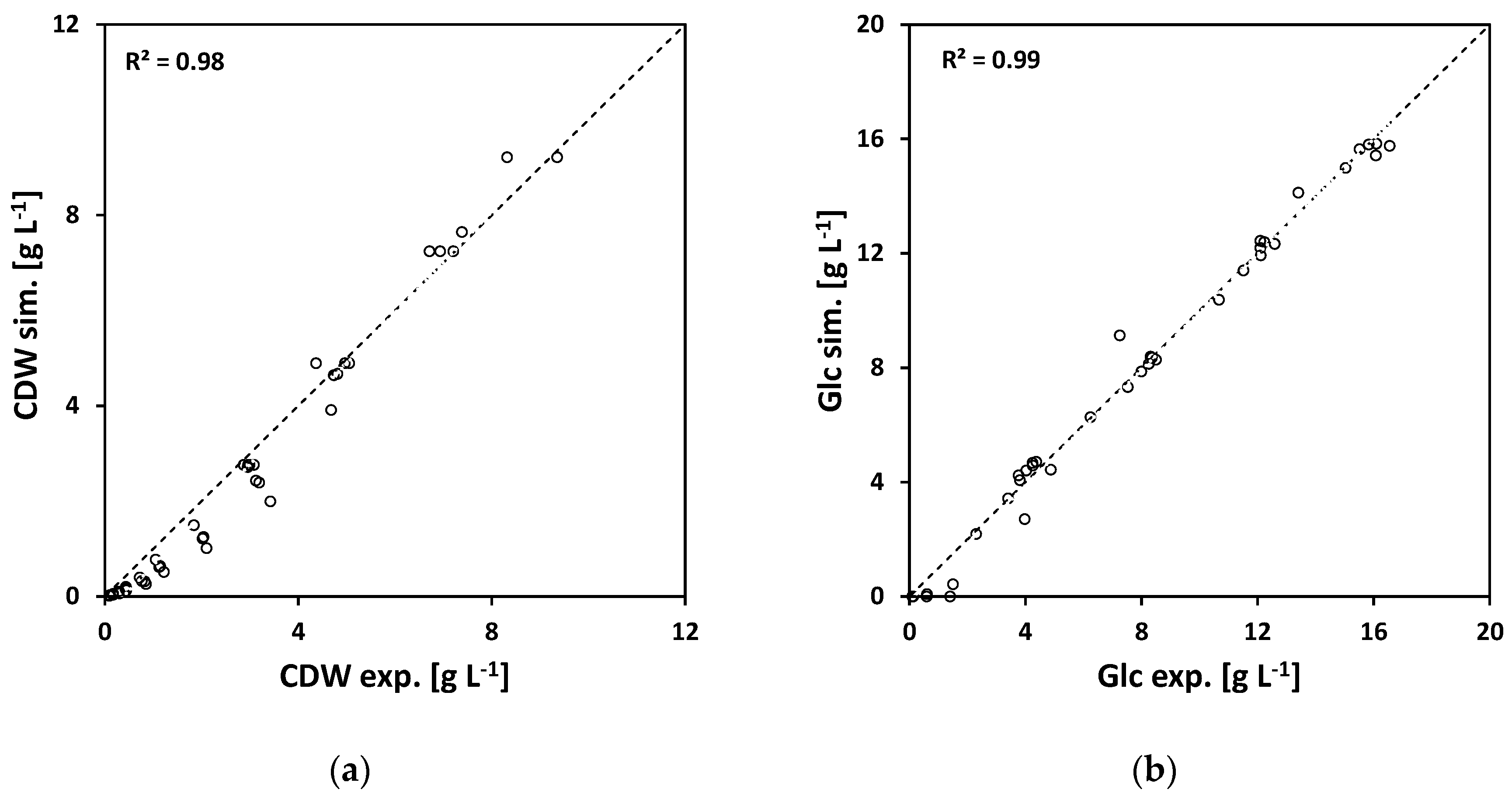
Figure 2), first initial shaking flask experiments were performed. The shaking flask experiments had varying initial glucose concentrations of 5, 8, 12, and 16 g L
−1 in order to study the substrate limiting effects on growth. Based on these data, the model parameters for Model A were estimated. The results are listed in
Table A4. In
Figure 5, the experimental and simulated results are plotted against each other. The experiment and simulation are in good agreement with each other.
The initial shaking flask experiments led to a first description of the process regarding the substrate-limiting effects. In order to control the feed trajectory with an NMPC, the underlying model also has to have some form of substrate inhibition term. Otherwise, the feed optimization would be somewhat trivial. Hence, a substrate inhibition term was added and the model was extended into Model B (
Table 2,
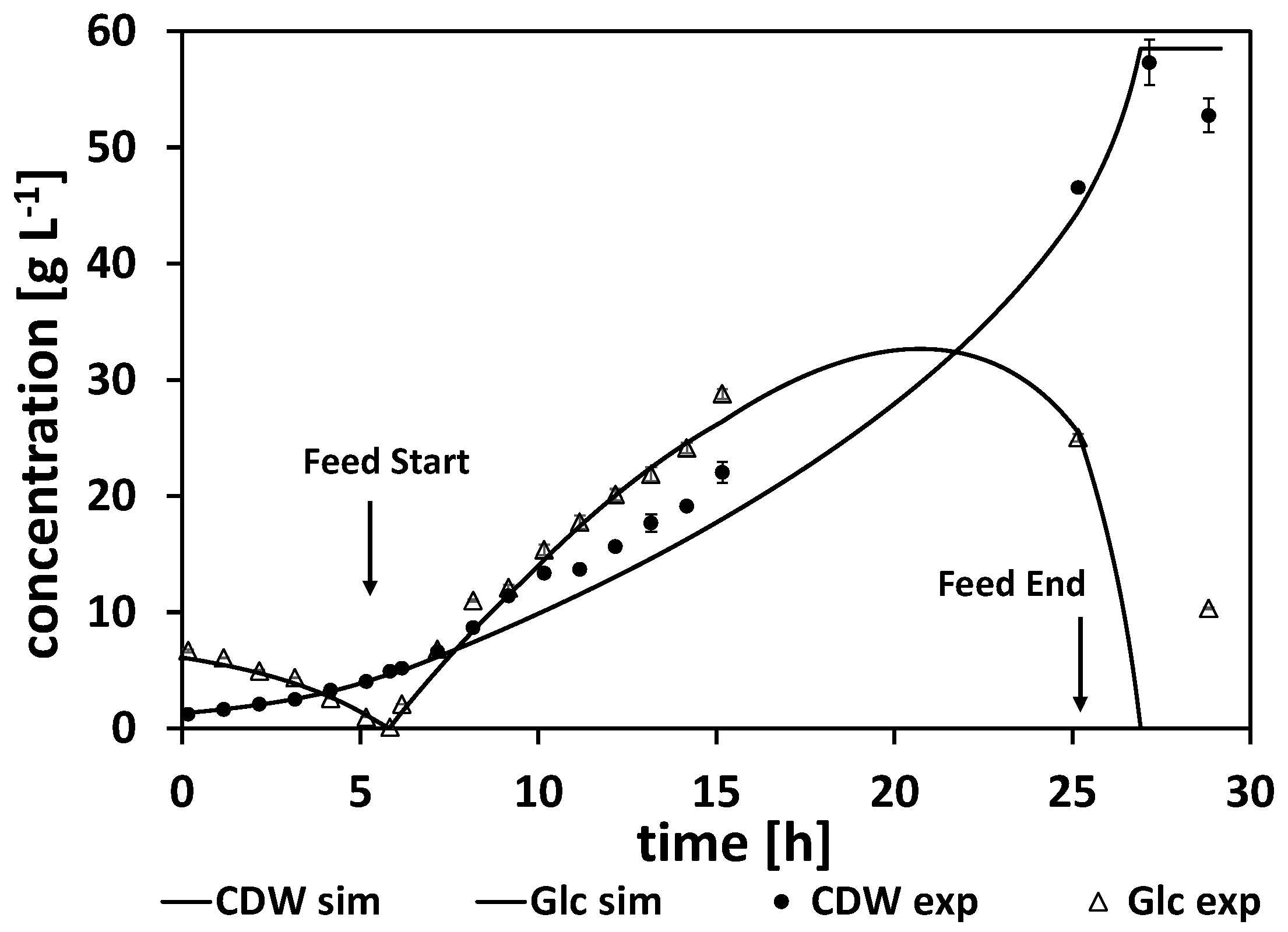
Table A7). To study the substrate-inhibiting effects, a fed-batch with a constant feeding rate was performed. The experimental and simulated cultivation courses are shown in
Figure 6. The results of the parameter estimation are listed in
Table A6. This experiment was also used to study process parameters such as inoculum density (OD ≥ 0.3), aeration rate (0.5–1.0 vvm), and stirrer speed. All the estimated parameters for these first experiments can be found in Section
Appendix A.1.
3.2.2. Experimental Setup 2-Strain 2–NMPC Fed-Batch Model B+
After the first initial experiments with strain generation 1 had been performed and the design of Model B was finished, the first two steps of the process development strategy (see
Section 1.2) had been accomplished. Step 3 of the process development strategy (
Figure 2) is the execution of an NMPC-controlled fed-batch. This step was performed with strain generation 2 and Model B+.
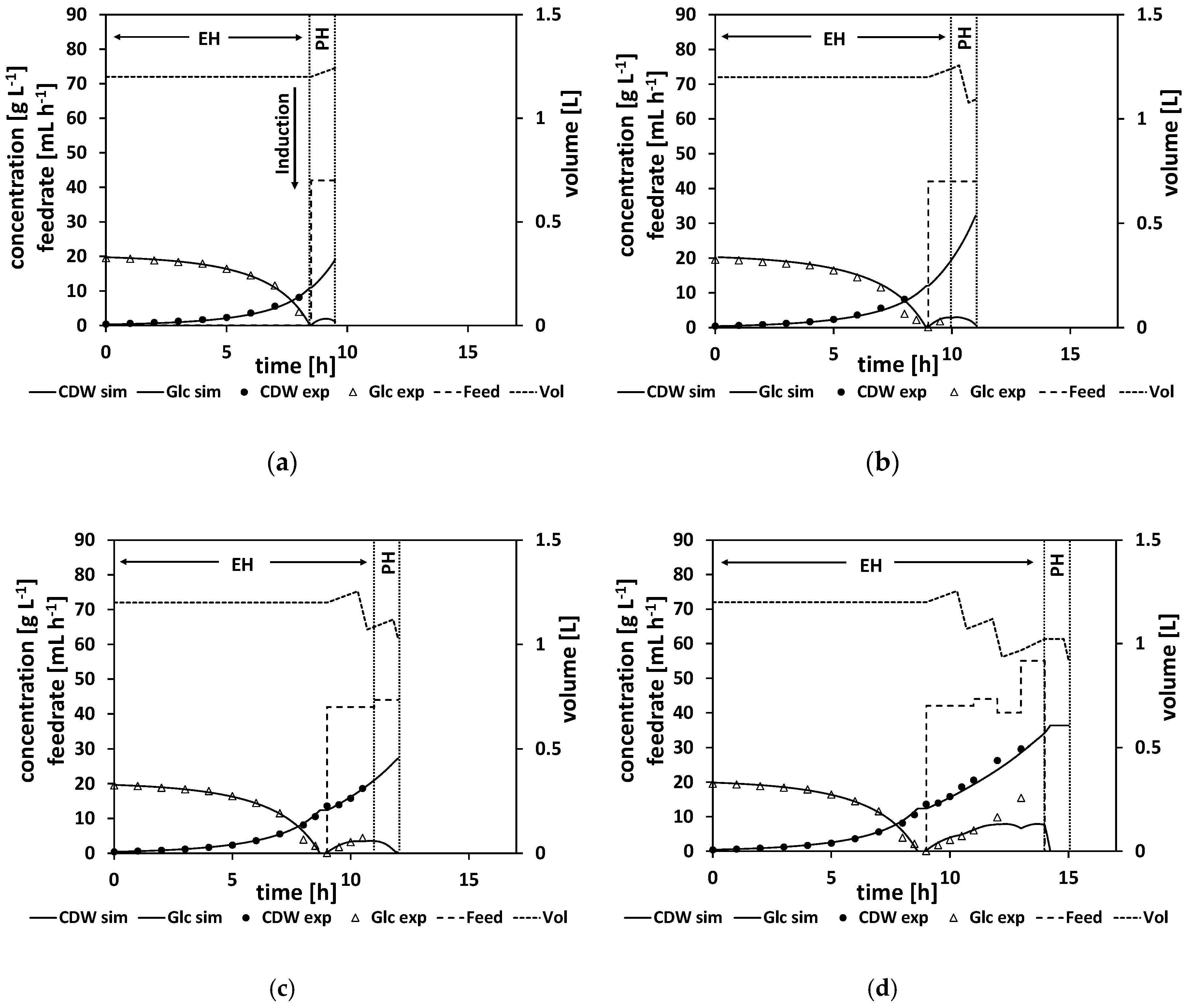
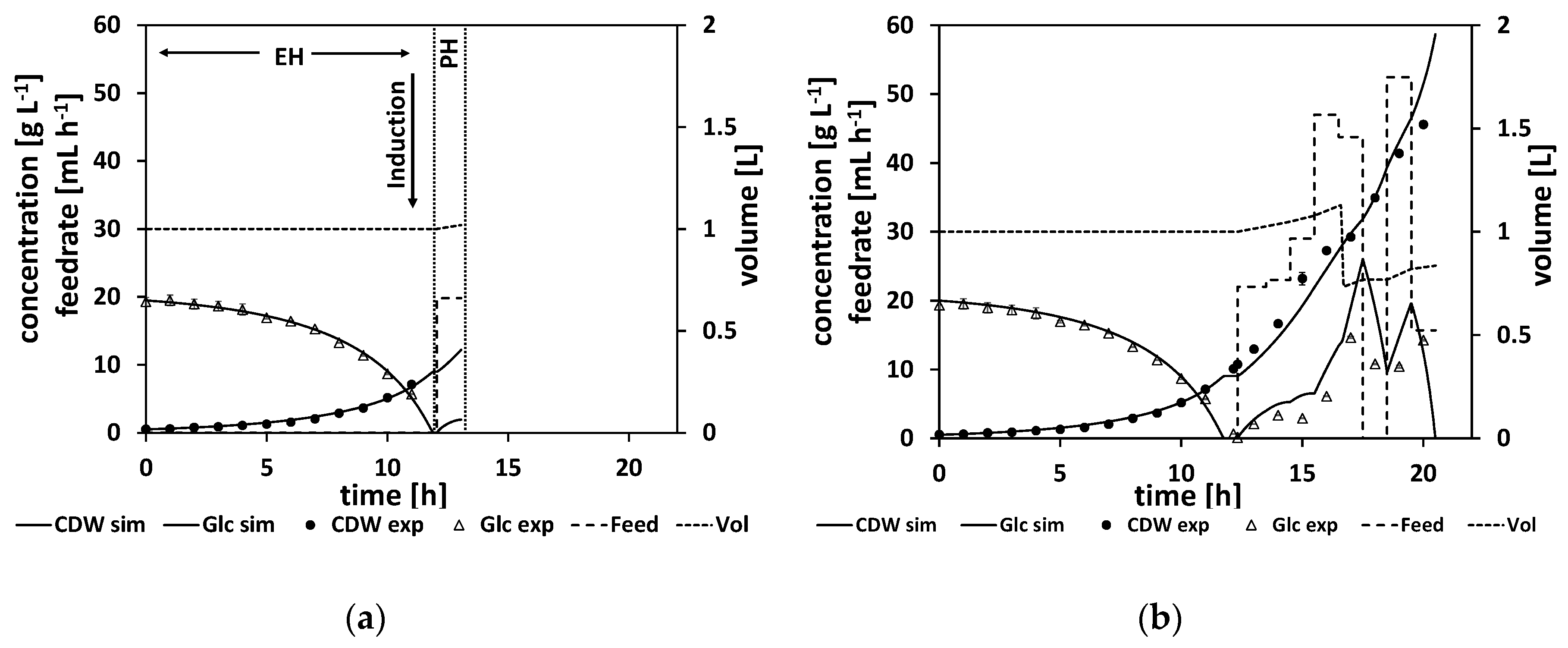
To illustrate the workflow of an NMPC-controlled fed-batch experiment, the experiment is discussed in more detail in the following. By the end of the batch phase of the experiment (when the glucose levels start to drop), the model parameters were adapted to the experimental data (see
Section 2.2.1 for a detailed explanation of the parameter estimation). For this experiment, this step was performed at a process time of about t = 8 h. Based on the adapted model, the time point for the complete consumption of glucose (time point when the glucose reaches 0 g L
−1, named “drop time” in the following) is calculated by process extrapolation with the adapted model. In the second step, the optimal feeding rate beginning at the calculated drop time is determined (see
Section 2.2.2 for a detailed explanation of the feed optimization) with the target to maximize cell growth. The result of this parameter estimation and the corresponding feed trajectory optimization is listed in
Table A9. The calculated drop time was at about τ = 8.5 h and at the calculated initial feeding rate of 42 mL h
−1. In
Figure 7a, the state of the experiment at that moment is shown. The experimental data for the cell dry weight and glucose are shown as marks and the simulated cultivation courses are shown as lines. On the left side of the vertical dashed lines is the time frame where the process data for the parameter estimation are collected from. It is denoted as the estimation horizon (EH) of the NMPC algorithm. In between the vertical dashed lines is the prediction horizon (PH) of the NMPC algorithm. Here, the calculated feed profile of F
in = 42 mL h
−1 is shown, starting at about τ = 8.5 h, and the resulting extrapolated simulation courses the for biomass and glucose concentration are displayed. In this experiment, the feed started as the DO signal increased, indicating glucose depletion. This was at process time t = 9 h, and thereby half an hour after the predicted time point.
As outlined in
Section 2.2 the whole NMPC procedure of parameter estimation and feed profile calculation is repeated every hour. Therefore, at a process time close to t = 10 h, the model parameters were readapted with regard to the new process data from sample analytics. Based on this newly adapted model, the next feed trajectory was optimized beginning at τ = 10 h. The results of this parameter estimation and the resulting optimized feed profile of F
in = 42 mL h
−1 are listed in
Table A10. The corresponding cultivation courses at that time point are shown in
Figure 7b, with the estimation horizon on the left side of the vertical dashed lines showing the experimental data and the simulated cultivation courses fitted to this data and the prediction horizon in between the vertical dashed lines, with the optimized second feeding rate and the corresponding simulated courses for the process states, biomass concentration, glucose concentration, and volume.
This whole procedure was repeated every hour. In
Figure 7c, the state of the experiment is shown when the third cycle of the NMPC algorithm was performed at a process time close to τ = 11 h. The third feeding rate was calculated based on the model with the once again, for the third time, adapted model parameter set (see
Table A10 for the adapted model parameters and the calculated feeding rate values).
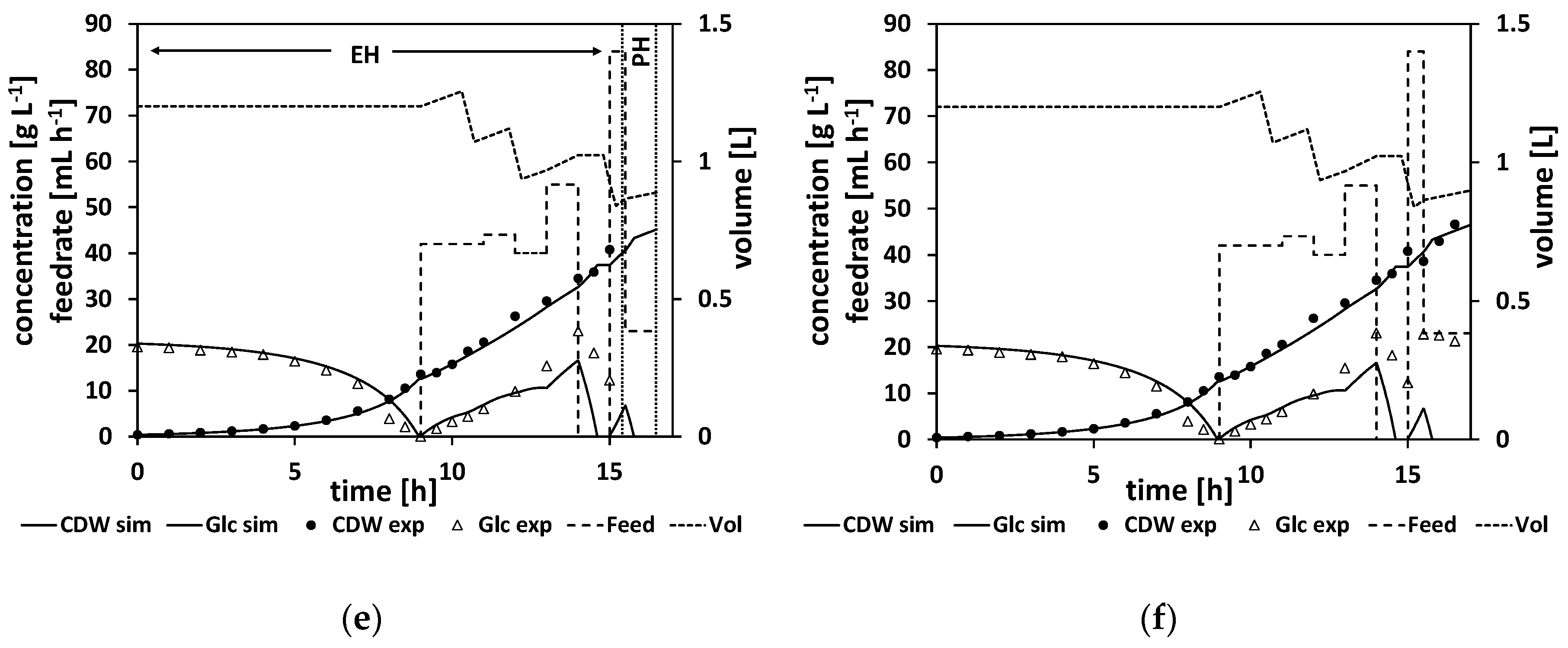
Figure 7d and e show the cultivation at even later stages of the experiment, and
Figure 7f shows the finished process. Note that the estimation horizon grows for every subsequent NMPC cycle and more and more process data become available for parameter estimation, while the prediction horizon remains a one-hour time window and is moving further. All the estimated parameters and the calculated feed trajectory for this experiment can be found in Section
Appendix A.2 in
Table A8,
Table A9 and
Table A10.
When studying the resulting feed trajectory and comparing the different feeding rates, keep in mind that the volume of fermentation broth is changing over the time course. In order to prevent foam building in the bioreactor, the fermentation volume was lowered from time to time in this and all the following experiments.
It is also noteworthy that Model B was adjusted while the process was running in the following way. After the calculation of the first feeding rate, the reaction rate
and the parameter
were separated into two parameters
and
for the batch phase, as well as
and
for the feed phase. This simple modification was made in order to take the slower growth rate in the feed phase into account. The corresponding model is Model B+ (see
Table A7). The model equations are essentially the same as in Model B. While, in the batch phase, the maximum specific growth rate was about
for most NMPC cycles it reduced its value by half in the fed-batch phase to about
(see
Table A10).
In this experiment, a biomass concentration of 42 g L
−1 was reached, which corresponds to an OD
600 of about 108, which is already a relatively high cell density. Noteworthy here are the relatively high measured glucose levels, especially at the end of the feed phase. This indicates that the model predicted too-high consumption rates, leading to too-high feeding rates. Ideally, the NMPC should keep the glucose concentration relatively low, at about
(for our parameter values, at about 1.6 g L
−1), which is the glucose level that would maximize the growth rate according to the model structure. As can be seen in
Figure 7d–f, the simulation cannot reproduce the high glucose levels measured at the later stages of the process, leading to too-high feeding rates to balance out the too-low simulated glucose levels. Additionally, the growth seems more linear than exponential in the feed phase. This linear growth cannot be simulated accordingly by the model. All in all, the NMPC predicted too-high consumption rates compared to reality, and therefore Model B, although leading to a high CDW concentration, can still be improved to get a more accurate controlled process.
3.2.3. Experimental Setup 3-Strain 3–NMPC Fed-Batch Model B+ (Weighted Sample Points)
Following the process development strategy (see
Section 1.2 step 3 → step 2 and
Figure 2), in the model the NMPC control has to be adjusted according to our results from the previous NMPC-controlled fed-batch experiment 2. As analyzed above, especially at the later stages of the experiment, the simulation could not fit the experimental data sufficiently to ensure the calculation of a reasonable feed trajectory. In order to solve this problem, the model remained unchanged, but the way the parameter estimation procedure was adapted by introduction of weights.
More precisely, in order to get a more accurate adaptation of the simulation to the experiment, at the later stages of the experiment the sample points were weighted, such that the last available sample point (the newest data) weighs 10 times as much as the first available sample point within each estimation horizon. This modification was made to get a closer adaptation of the model to the process at later stages of the experiment in order to get a more realistic extrapolation and thereby a better feed profile calculation. The weighted target function for the parameter identification is given by:
The weights
are normalized (
) and equidistant (
for all i = 2,…,n−1), and chosen such that
. The used model was unchanged (Model B+), and the third strain generation was used for this experiment. The whole NMPC cycle procedure was executed as described for the previous experiment, starting with the glucose drop time prediction by the end of the batch phase and following with the first model parameter estimation and the first feeding rate calculation (for the estimated parameter and feeding rate values, see
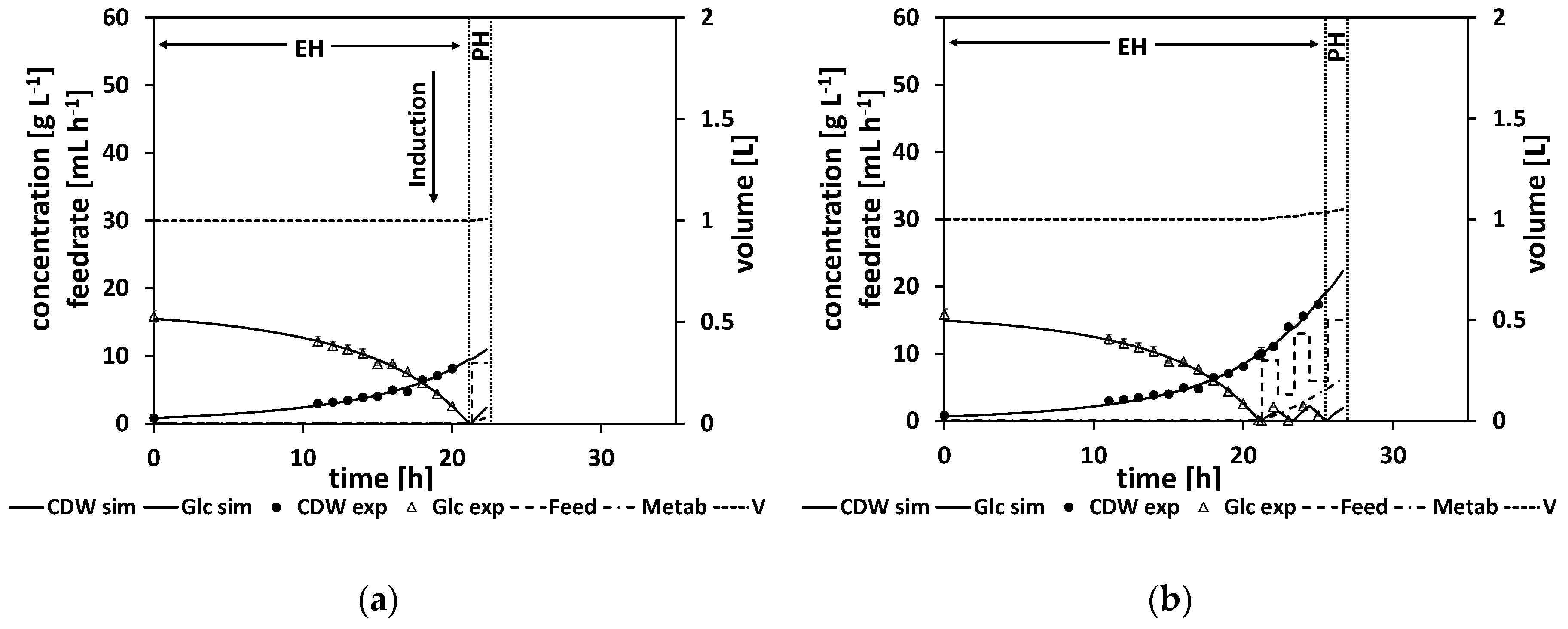
Table A13). In
Figure 8a, the state of the cultivation at this first NMPC cycle is shown. In the following, eight NMPC cycles were performed. The results of each parameter estimation and the resulting calculated feed trajectory are listed in
Table A14.
Figure 8b shows the finished process. All the estimated parameters and the calculated feed trajectories for this experiment can be found in Section
Appendix A.3 in
Table A12,
Table A13 and
Table A14.
As before, a high biomass concentration of 45 g L
−1 was reached, which corresponds to an OD
600 of about 115. This concentration is similar to the concentrations reached in the previous cultivation, despite a new strain being used. Unfortunately, the measured glucose levels at the later stages of the experiment were still too high, indicating an overfeeding of the NMPC algorithm and thereby an insufficient process control. Nonetheless, the simulation seems to fit the experimental data better compared to the last experiment (
Figure 8b).
3.2.4. Experimental Setup 4-Strain 4–NMPC Fed-Batch Model C
As a result of the previous experiments and within the framework of the process development strategy (
Section 1.2 step 3 → step 2), this time the model was adjusted accordingly (see the middle part of
Figure 2) to take the findings of the previous experiments into account.
In detail, in order to slow down the simulated growth at later stages of the feed phase even more, a not-further-characterized growth-inhibiting metabolite was added to the model. The corresponding model is denoted Model C (see
Table 2 or
Table A15). The balance equations for the substrate (4) and the growth rate (9) were adjusted accordingly. The concentration of this metabolite starts to increase at the feed start and slows down the reaction rates as it accumulates. The inhibiting effect is larger as the metabolite concentration increases. As the metabolite is not characterized any further and especially not measured in any way, the initial metabolite concentration has to be predefined (see
Table A16). The corresponding model parameters describing the metabolite and its growth-diminishing effect were received from a parameter adaptation of Model C to the previous experiment 3. The parameters values are listed in
Table A16.
The actual experiment 4 was performed in the same way as the previous two experiments following the NMPC cycle procedure discussed in
Section 2.2 and following the routine displayed in
Figure 1.
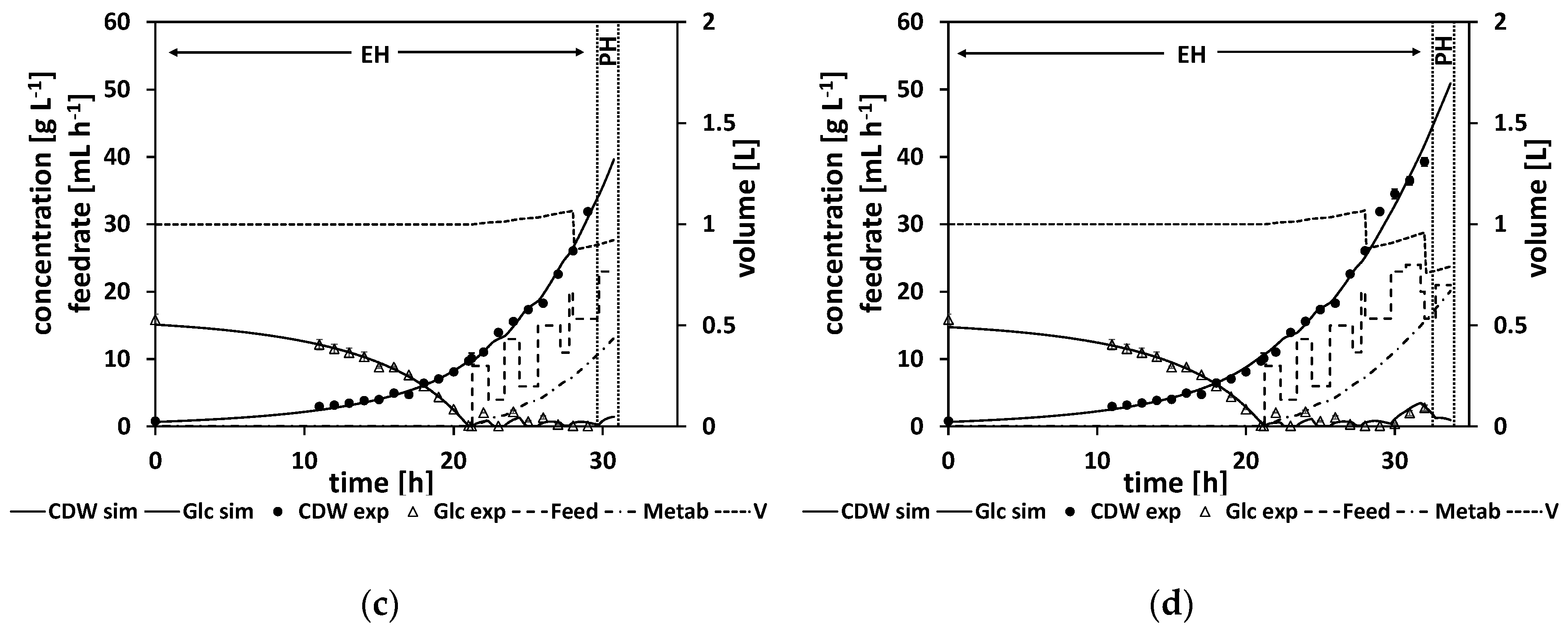
Figure 9a shows the first NMPC cycle, where the initial glucose drop time is calculated and a first parameter adaptation with a subsequent feeding rate is calculated (for the resulting parameter and feeding rate values, see
Table A17), whereas in
Figure 9b–d different NMPC cycles at later stages of the process are shown. Noteworthy in comparison to the previous experiments is the metabolite, which is presented by a dashed-dotted line. Its concentration starts to increase at the start of the feed phase. All the estimated parameters and the calculated feed trajectories for this experiment can be found in
Appendix A.4 in
Table A16,
Table A17 and
Table A18.
The predicted drop time of about τ = 21.3 h was in very good agreement with the time point of the DO signal increase at about τ = 21.25 h (see
Table A17 and
Table A18). A biomass concentration of 40 g L
−1 was reached, which corresponds to an OD
600 of about 103. This concentration is similar to the concentrations reached in previous cultivations. In general, compared to the previous strain generations, strain 4 grows slower.
In the beginning of the feed phase, the growth rate diminishes. As the first feeding rate is calculated by the extrapolation based on a model adapted to the data from the batch phase, this leads to an initial overfeed (
Figure 9a).
Figure 9b,c show how the NMPC algorithm reacts to this initial overfeed leading to a wavering feed profile.
In contrast to the former experiments, the NMPC kept the glucose at relatively low levels over the course of the fed-batch. As discussed earlier, this is desirable behavior for the NMPC algorithm, as ideally the NMPC algorithm should keep the glucose concentration relatively low at about (which is at about 1.6 g L−1), which is the glucose level that would maximize the growth rate according to the model structure. Therefore, this experiment was successful. It can already be used to derive a standardized operation procedure by smoothening the feed profile.
3.2.5. Experiment 4-Strain 4-Fed-Batch Model D–Product Formation
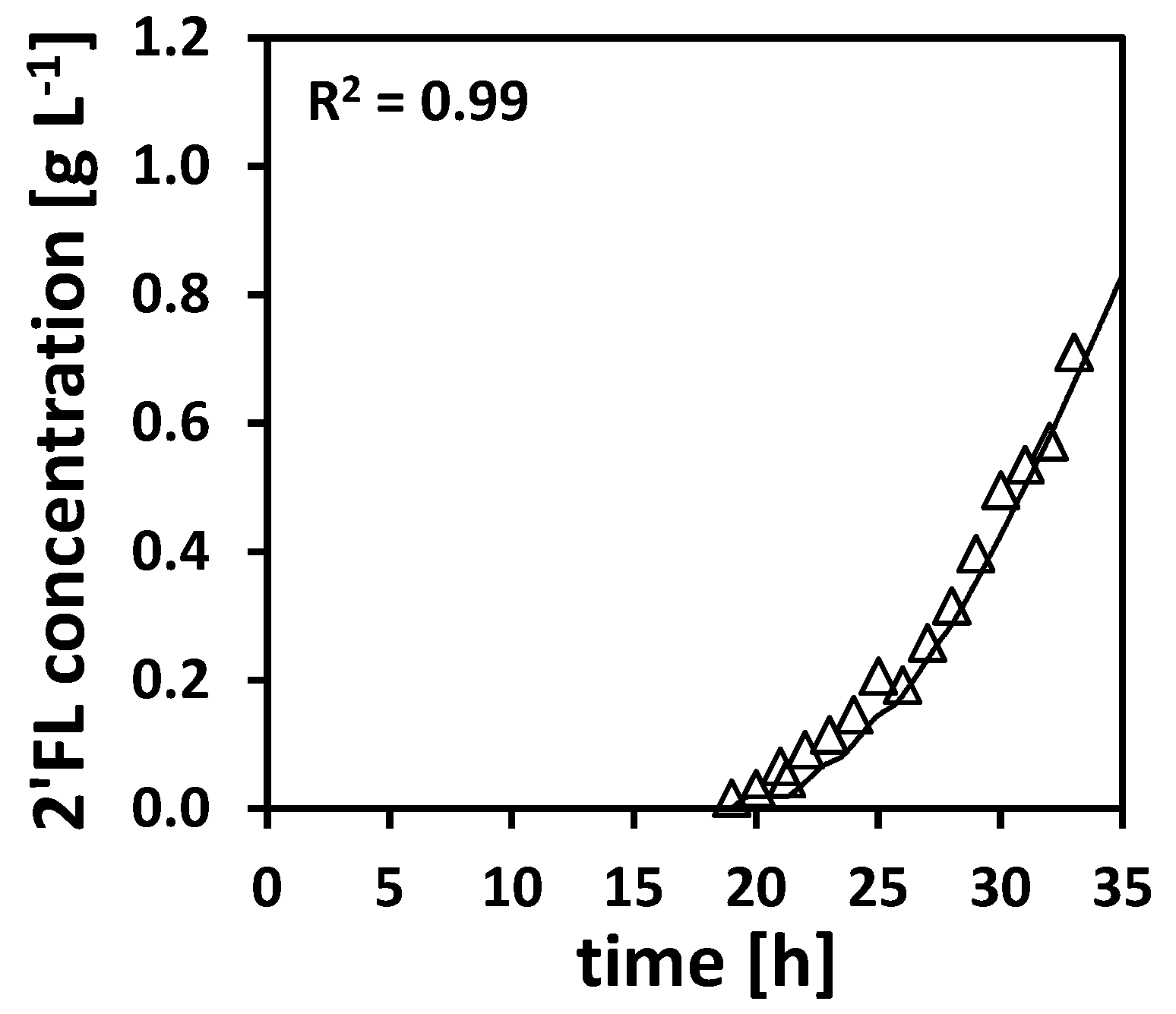
For the last experiment 4, the product concentration of 2′-Fucosyllactose (2′-FL) was analyzed. As these analytics were performed after the actual experiment, the results could not be used for the actual NMPC algorithm. It is conceivable that, in future, the approaches for the discussed NMPC algorithm could be modified in order to maximize the product building instead of cell growth. However, this might lead to the same results as our current approach, as the following analysis suggests a growth-coupled product formation.
In order to simulate the product formation, the model was extended accordingly (see
Table 2 or
Table A19). The resulting model is denoted Model D. In Model D, the states and balance equations for the inducer and the product formation are added. The product formation rate
was chosen to be proportional to the growth rate
, such that a growth-coupled product formation is simulated. The parameter estimation results are listed in
Appendix A.5 in
Table A20,
Table A21 and
Table A22.
In
Figure 10, the time course for the measured concentration of 2′-FL is shown as marks together with the simulation according to Model D. As the simulation with Model D shows a good agreement with the measured data, a growth-coupled product formation can be assumed.