Assessment of the Use of Natural Extracted Dyes and Pancreatin Enzyme for Dyeing of Four Natural Textiles: HPLC Analysis of Phytochemicals
Abstract
1. Introduction
2. Materials and Methods
2.1. Natural Dyes and Textiles
2.2. Extraction Method
2.3. Dyeing Procedures
2.4. Mordanting Procedures
2.5. Treatment of Fabrics with Natural Dyes Using Pancreatin Enzyme and Its Buffer Solution
2.6. Measurement of Color Strength and Color Differences of Dyed Fabrics
2.7. Color Difference Formula (L*a*b*)
CIE L*a*b* between two colors given in terms of L*a*b* is calculated.
2.8. HPLC Analyses of Phenolics/Flavonoids and Caffeine Contents
2.9. Statistical Analysis
3. Results
3.1. Effects of Pancreatin Enzyme Concentration, Type of Dye, and Fiber on the Properties of Naturally Dyed Fabrics
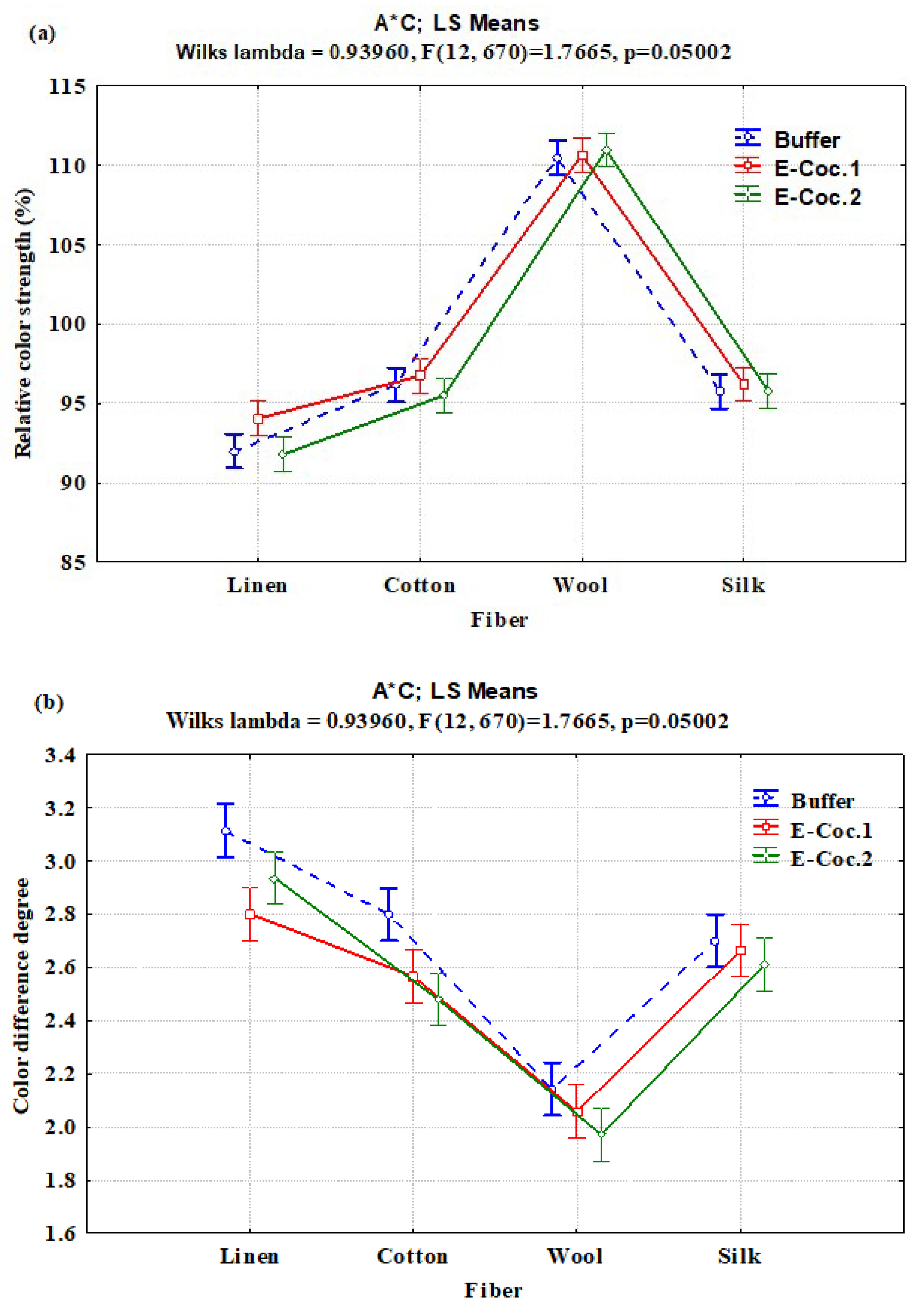
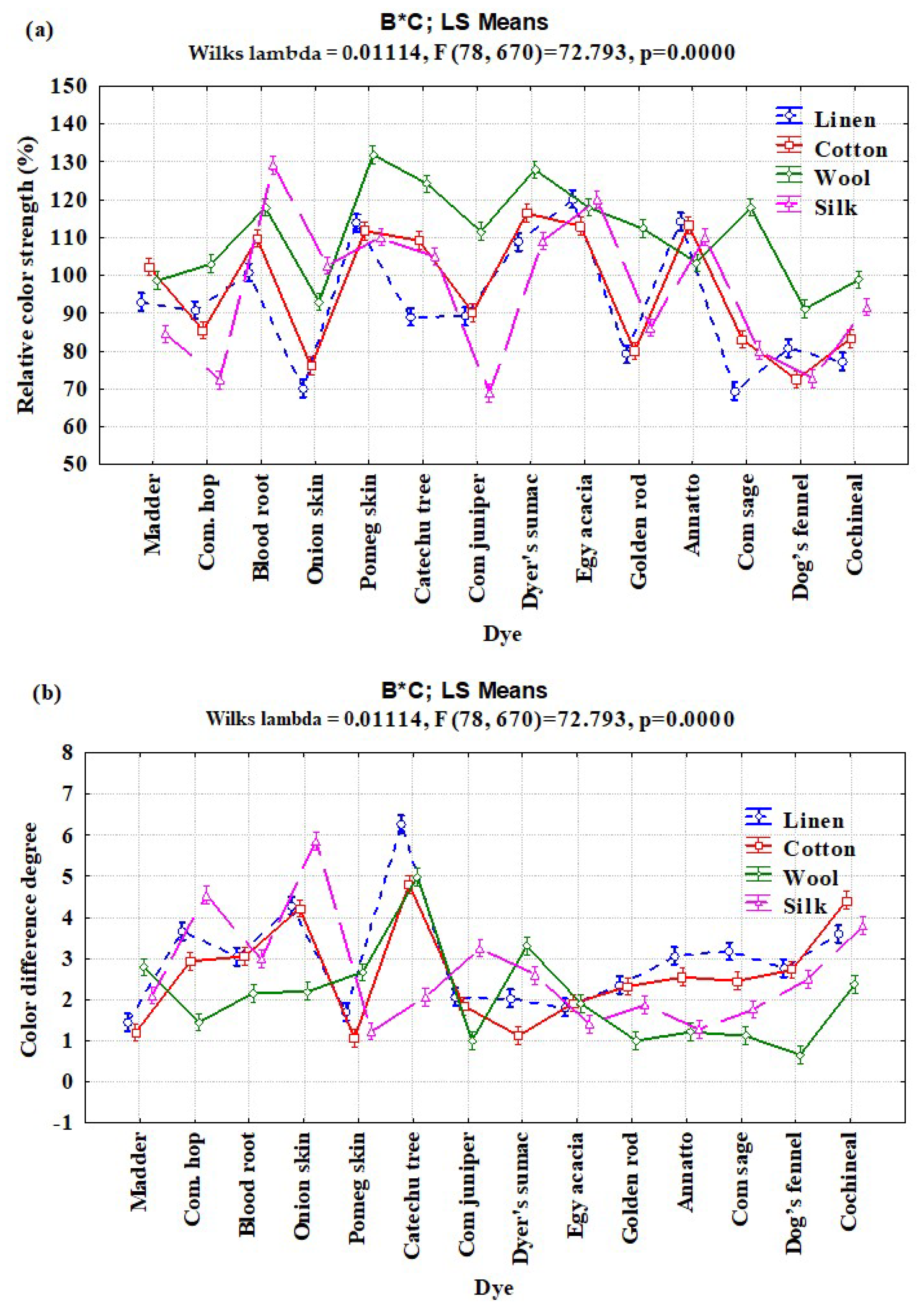
3.2. Interaction Effects of Enzyme Concentration, Dye Type, and Fiber Type on Relative Color Strength (%) and Total Color Difference
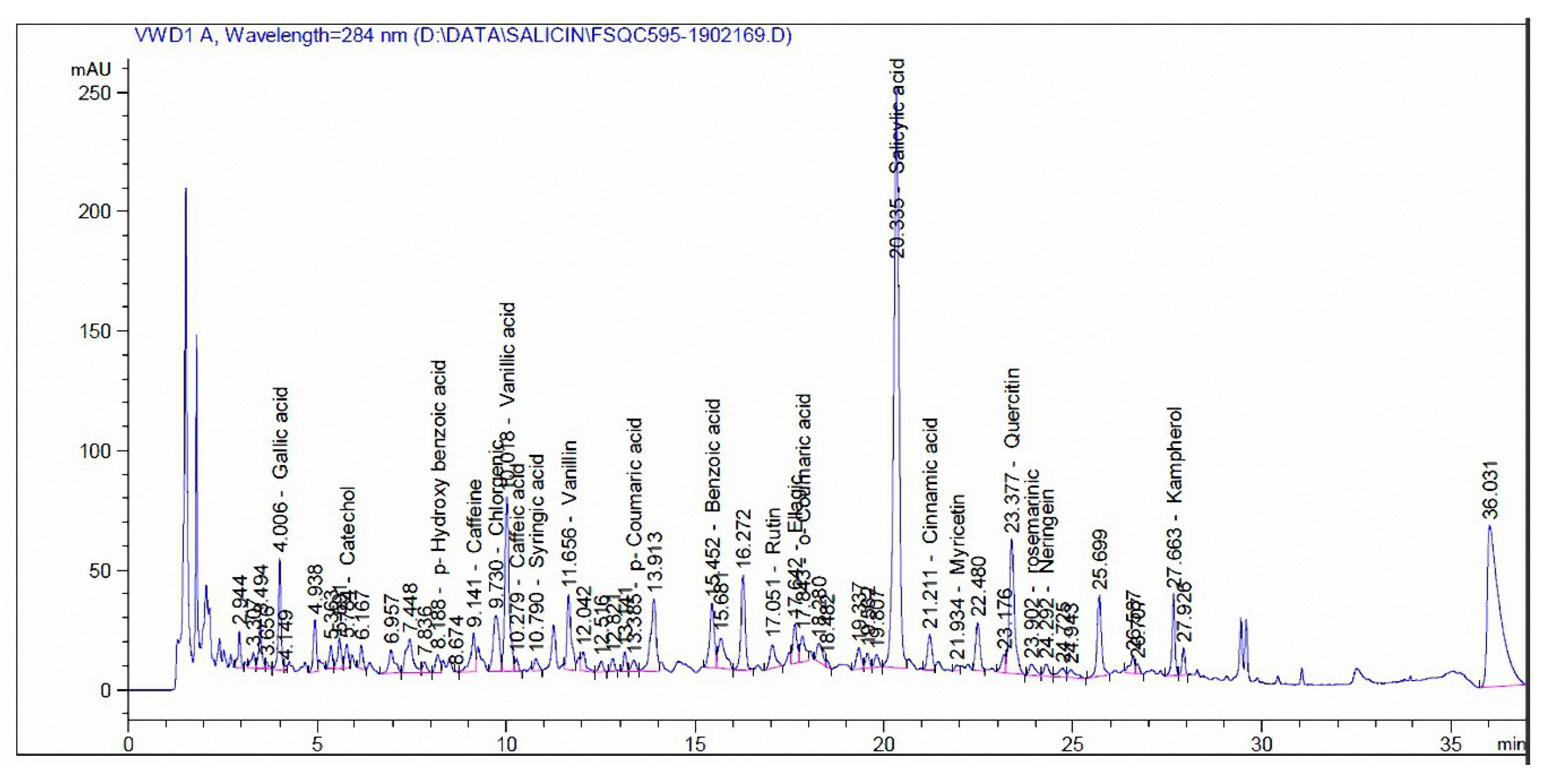
3.3. Phenolic and Flavonoid Compounds by HPLC Analysis
4. Discussion
4.1. Chemical Structures of Different Natural Dyes and Their Properties
4.2. Effects of Enzyme
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Negi, J.S.; Singh, P.; Pant, G.J.N.; Rawat, M.S.M. High-performance liquid chromatography analysis of plant saponins: An update 2005–2010. Pharmacogn. Rev. 2011, 5, 155–158. [Google Scholar] [CrossRef]
- Spáčil, Z.; Nováková, L.; Solich, P. Analysis of phenolic compounds by high performance liquid chromatography and ultra performance liquid chromatography. Talanta 2008, 76, 189–199. [Google Scholar] [CrossRef]
- Johansen, K.T.; Ebild, S.J.; Christensen, S.B.; Godejohann, M.; Jaroszewski, J.W. Alkaloid analysis by high-performance liquid chromatography-solid phase extraction-nuclear magnetic resonance: New strategies going beyond the standard. J. Chromatogr. A 2012, 1270, 171–177. [Google Scholar] [CrossRef]
- Chen, Y.; Zhang, Z.; Zhang, Y.; Zhang, X.; Zhang, Z.; Liao, Y.; Zhang, B. A new method for simultaneous determination of phenolic acids, alkaloids and limonoids in Phellodendri Amurensis Cortex. Molecules 2019, 24, 709. [Google Scholar] [CrossRef]
- Sid, N.; EL-Hawary, S. High-Performance Natural Dyes for Cellulosic Fibers: Review—part 1. J. Text. Color. Polym. Sci. 2019, 16, 21–33. [Google Scholar]
- Kim, S.; Moldes, D.; Cavaco-Paulo, A. Laccases for enzymatic colouration of unbleached cotton. Enzy. Microb. Technol. 2007, 40, 1788–1793. [Google Scholar] [CrossRef]
- Mojsov, K. Application of enzymes in the textile industry: A review. In Proceedings of the II International Congress “Engineering, Ecology and Materials in the Processing Industry”, Jahorina, Bosnia and Hercegovina, 9–11 March 2011; pp. 1–17. [Google Scholar]
- Kim, H.R.; Seo, H.Y. Enzymatic hydrolysis of polyamide fabric by using acylase. Text. Res. J. 2013, 83, 1181–1189. [Google Scholar] [CrossRef]
- Shim, E.J.; Lee, S.H.; Song, W.S.; Kim, H.R. Development of an enzyme-immobilized support using a polyester woven fabric. Text. Res. J. 2017, 87, 3–14. [Google Scholar] [CrossRef]
- Wang, F.; Gong, J.; Zhang, X.; Ren, Y.; Zhang, J. Preparation of biocolorant and eco-dyeing derived from polyphenols based on laccase-catalyzed oxidative polymerization. Polymers 2018, 10, 19. [Google Scholar] [CrossRef]
- Quandt, C.; Kuhl, B. Enzymatic processes: Operational possibilities and optimization (Enzymes Possibilités et perspectives). L’Industrie Tex. 2001, 1334–1335, 116–119. [Google Scholar]
- Korf, U.; Kohl, T.; van der Zandt, H.; Zahn, R.; Schleeger, S.; Ueberle, B.; Wandschneider, S.; Bechtel, S.; Schnolzer, M.; Ottleben, H.; et al. Large-scale protein expression for proteome research. Proteomics 2005, 5, 3571–3580. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Anumanthan, A.; Gao, X.G.; Ilangovan, K.; Suzara, V.V.; Düzgünes, N.; Renugopalakrishnan, V. Expression of recombinant proteins in Pichia pastoris. Appl. Biochem. Biotechnol. 2007, 142, 105–124. [Google Scholar] [CrossRef] [PubMed]
- Lenting, H.B.M. Enzymes in textile production. In Enzymes in Industry, Production and Applications, 3rd ed.; Aehle, W., Ed.; Wiley-VCH Verlag GmbH & Co: Weinheim, Germany, 2007; pp. 218–230. [Google Scholar]
- Ahmed, H.E.; Kolisis, F.N. An investigation into the removal of starch paste adhesives from historical textiles by using the enzyme α-amylase. J. Cult. Herit. 2011, 12, 169–179. [Google Scholar] [CrossRef]
- Chand, N.; Nateri, A.S.; Sajedi, R.H.; Mahdavi, A.; Rassa, M. Enzymatic desizing of cotton fabric using a Ca2+-independent α-amylase with acidic pH profile. J. Mol. Catal. B Enzym. 2012, 83, 46–50. [Google Scholar] [CrossRef]
- Araújo, R.; Casal, M.; Cavaco-Paulo, A. Application of enzymes for textile fibres processing. Biocatal. Biotransform. 2008, 26, 332–349. [Google Scholar] [CrossRef]
- Agnes, T.B.; Eastop, D. Chemical Principles of Textile Conservation, 1st ed.; Butterworth, Heinemann: London, UK, 1998. [Google Scholar]
- Ahmed, H.E. Protease enzyme used for artificial ageing on modern cotton fabric for historic textile preservation and restoration. Int. J. Conserv. Sci. 2013, 4, 177–188. [Google Scholar]
- Ibrahim, N.A.; El-Gamal, A.R.; Gouda, M.; Mahrous, F. A new approach for natural dyeing and functional finishing of cotton cellulose. Carbohydr. Polym. 2010, 82, 1205–1211. [Google Scholar] [CrossRef]
- El-Hennawi, H.M.; Ahmed, K.A.; Abd El-Thalouth, I. A novel bio-technique using laccase enzyme in textile printing to fix natural dyes. Ind. J. Fibre Text. Res. 2012, 37, 245–249. [Google Scholar]
- Bettiol, J.L.P.; Showell, M.S. Detergent Compositions Comprising a Mannanase and a Protease. Patent WO 99/009128, 25 February 1999. [Google Scholar]
- Nechwatal, A.; Blokesch, A.; Nicolai, M.; Krieg, M.; Kolbe, A.; Wolf, M.; Gerhardt, M. A contribution to the investigation of enzyme-catalysed hydrolysis of poly(ethylene eterephthalate) oligomers. Macromol. Mater. Eng. 2006, 291, 1486–1494. [Google Scholar] [CrossRef]
- Maurer, K.H. Detergent proteases. Curr. Opin. Biotechnol. 2004, 5, 330–334. [Google Scholar] [CrossRef]
- Nakajima, K.; Oshida, H.; Muneyuki, T.; Kakei, M. Pancrelipase: An evidence-based review of its use for treating pancreatic exocrine insufficiency. Core Evid. 2012, 7, 77–91. [Google Scholar] [CrossRef] [PubMed]
- DeLano, F.A.; Hoyt, D.B.; Schmid-Schönbein, G.W. Pancreatic digestive enzyme blockade in the intestine increases survival after experimental shock. Sci. Transl. Med. 2013, 5, 169ra11. [Google Scholar] [CrossRef] [PubMed]
- Adeel, S.; Ali, S.; Bhatti, I.A.; Zsila, F. Dyeing of cotton fabric using pomegranate (Punica granatum) Aqueous Extract. Asian J. Chem. 2009, 21, 3493–3499. [Google Scholar]
- Kalimuthu, J.; Sekar, M.; Subramanian, R.; Shanmugam, K. Natural dyeing process for recycled paper from the waste vegetables. Int. J. ChemTech Res. 2016, 9, 151–157. [Google Scholar]
- Dapson, R.W.; Frank, M.; Penney, D.P.; Kiernan, J.A. Revised procedures for the certification of carmine (C.I. 75470, Natural red 4) as a biological stain. Biotech. Histochem. 2007, 82, 13–15. [Google Scholar] [CrossRef]
- Greenhawt, M.; McMorris, M.; Baldwin, J. Carmine hypersensitivity masquerading as azithromycin hypersensitivity. Allergy Asthma Proc. 2009, 30, 95–101. [Google Scholar] [CrossRef]
- Gawish, S.M.; Farouk, R.; Ramadn, A.M.; Mashaly, H.M.; Helmy, H.M. Eco-friendly multifunctional properties of cochineal and weld for simultaneous dyeing and finishing of proteinic fabrics. Int. J. Eng. Technol. 2016, 8, 2246–2253. [Google Scholar]
- Derksen, G.C.H. Red, Redder, Madder. Analysis and Isolation of Anthraquinones from Madder Roots (Rubia tinctorum). Ph.D. Thesis, Wageningen Universiteit, Wageningen, The Netherlands, 2001. [Google Scholar]
- Marczylo, T.; Arimoto-Kobayashi, S.; Hayatsu, H. Protection against Trp-p-2 mutagenicity by purpurin: Mechanism of in vitro antimutagenesis. Mutagenesis 2000, 15, 223–228. [Google Scholar] [CrossRef]
- Westendorf, J.; Pfau, W.; Schulte, A. Carcinogenicity and DNA adduct formation observed in ACI rats after long-term treatment with madder root, Rubia tinctorum L. Carcinogenesis 1998, 19, 2163–2168. [Google Scholar] [CrossRef]
- Li, X.; Liu, Z.; Chen, Y.; Wang, L.J.; Zheng, Y.N.; Sun, G.Z.; Ruan, C.C. Rubiacordone A: A New Anthraquinone Glycoside from the Roots of Rubia cordifolia. Molecules 2009, 14, 566–572. [Google Scholar] [CrossRef]
- Wang, S.; Zhu, F. Chemical composition and biological activity of staghorn sumac (Rhus typhina). Food Chem. 2017, 237, 431–443. [Google Scholar] [CrossRef] [PubMed]
- Zarkogianni, M.; Mikropoulou, E.; Varella, E.; Tsatsaroni, E. Colour and fastness of natural dyes: Revival of traditional dyeing techniques. Color Technol. 2011, 127, 18–27. [Google Scholar] [CrossRef]
- Bechtold, T.; Mussak, R. Handbook of Natural Colorants; Wiley: Chichester, UK, 2009. [Google Scholar]
- Hussein, A.; Elhassaneen, Y. Natural dye from red onion skins and applied in dyeing cotton fabrics for the production of women’s headwear resistance to ultraviolet radiation (UVR). J. Am. Sci. 2014, 10, 129–139. [Google Scholar]
- Oancea, S.; Draghici, O. pH and thermal stability of anthocyanin-based optimised extracts of romanian red onion cultivars. Czech J. Food Sci. 2013, 31, 283–291. [Google Scholar] [CrossRef]
- Landi, S. The Textile Conservators Manual, 1st ed.; Butterworth: London, UK, 1985. [Google Scholar]
- Kamel, M.M.; Helmy, H.M.; Shakour, A.A.; Rashed, S.S. The Effects of Industrial Environment on Colour Fastness to Light of Mordanted Wool Yarns Dyed with Natural Dyes. Res. J. Text. Appar. 2012, 16, 46–57. [Google Scholar] [CrossRef]
- Hamed, I.; Zidan, Y.; Saber, N.; Ahmed, H. Effect of pepsin and trypsin enzymes used in textile conservation on natural dyed textiles samples. J. Home Econ. 2015, 25, 237–247. [Google Scholar]
- Helmy, H.M.; Kamel, M.M.; Hagag, K.; El-Hawary, N.; El-shemy, N.S. Antimicrobial activity of dyed wool fabrics with peanut red skin extract using different heating techniques. Egyp. J. Chem. 2017, 60, 103–116. [Google Scholar] [CrossRef]
- Kubelka, P. New contributions to the optics of intensely light-scattering materials Part II: Nonhomogeneous layers. J. Opt. Soc. Am. 1954, 44, 330–335. [Google Scholar] [CrossRef]
- Salem, M.Z.M.; Mansour, M.M.A.; Elansary, H.O. Evaluation of the effect of inner and outer bark extracts of Sugar Maple (Acer saccharum var. saccharum) in combination with citric acid against the growth of three common molds. J. Wood Chem. Technol. 2019, 39, 136–147. [Google Scholar] [CrossRef]
- Al-Huqail, A.A.; Behiry, S.I.; Salem, M.Z.M.; Ali, H.M.; Siddiqui, M.H.; Salem, A.Z.M. Antifungal, antibacterial, and antioxidant activities of Acacia saligna (Labill.) H. L. Wendl. flower extract: HPLC analysis of phenolic and flavonoid compounds. Molecules 2019, 24, 700. [Google Scholar] [CrossRef]
- Behiry, S.I.; Okla, M.K.; Alamri, S.A.; EL-Hefny, M.; Salem, M.Z.M.; Alaraidh, I.A.; Ali, H.M.; Al-Ghtani, S.M.; Monroy, J.C.; Salem, A.Z.M. Antifungal and antibacterial activities of Musa paradisiaca L. peel extract: HPLC analysis of phenolic and flavonoid contents. Processes 2019, 7, 215. [Google Scholar] [CrossRef]
- SAS. Users Guide: Statistics (Release 8.02); SAS Inst Inc: Cary, NC, USA, 2001. [Google Scholar]
- Lloyd, A.G. Extraction and chemistry of cochineal. Food Chem. 1980, 5, 91–107. [Google Scholar] [CrossRef]
- Chieli, A.; Sanyova, J.; Doherty, B.; Brunetti, B.G.; Miliani, C. Chromatographic and spectroscopic identification and recognition of ammoniacal cochineal dyes and pigments. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2016, 162, 86–92. [Google Scholar] [CrossRef] [PubMed]
- Balazsy, A.T.; Eastop, D. Chemical Principles of Textile Conservation, 2nd ed.; Routledge Series in Conservation and Museology; Butterworth-Heinemann: Oxford, UK, 2004. [Google Scholar]
- Shams-Nateri, A. Reusing wastewater of madder natural dye for wool dyeing. J. Clean. Prod. 2011, 19, 775–781. [Google Scholar] [CrossRef]
- Samanta, A.K.; Agarwal, P. Application of natural dyes on textiles. Indian J. Fibre Text. Res. 2009, 34, 384–399. [Google Scholar]
- Shahid, M.; Islam, S.; Mohammad, F. Recent advancements in natural dye applications: A review. J. Clean. Prod. 2013, 53, 310–331. [Google Scholar] [CrossRef]
- Yusuf, M.; Ahmad, A.; Shahid, M.; Khan, M.I.; Khan, S.A.; Manzoor, N.; Mohammad, F. Assessment of colorimetric, antibacterial and antifungal properties of woolen yarn dyed with the extract of the leaves of henna (Lawsonia inermis). J. Clean. Prod. 2012, 27, 42–50. [Google Scholar] [CrossRef]
- Romeo, F.V.; Ballistreri, G.; Fabroni, S.; Pangallo, S.; Nicosia, M.G.; Schena, L.; Rapisarda, P. Chemical Characterization of Different Sumac and Pomegranate Extracts Effective against Botrytis cinerea Rots. Molecules 2015, 20, 11941–11958. [Google Scholar] [CrossRef]
- Kosar, M.; Bozan, B.; Temelli, F.; Baser, K.H.C. Antioxidant activity and phenolic composition of sumac (Rhus coriaria L.) extracts. Food Chem. 2007, 103, 952–959. [Google Scholar] [CrossRef]
- Peng, Y.; Zhang, H.; Liu, R.; Mine, Y.; McCallum, J.; Kirby, C.; Tsao, R. Antioxidant and anti-inflammatory activities of pyranoanthocyanins and other polyphenols from staghorn sumac (Rhus hirta L.) in Caco-2 cell models. J. Funct. Foods 2016, 20, 139–147. [Google Scholar] [CrossRef]
- Kossah, R.; Nsabimana, C.; Zhao, J.; Chen, H.; Tian, F.; Zhang, H.; Chen, W. Comparative study on the chemical composition of Syrian Sumac (Rhus coriaria L.) and Chinese Sumac (Rhus typhina L.) Fruits. Pak. J. Nut. 2009, 8, 1570–1574. [Google Scholar]
- Pourahmad, J.; Eskandari, M.R.; Shakibaei, R.; Kamalinejad, M. A search for hepatoprotective activity of aqueous extract of Rhus coriaria L. against oxidative stress cytotoxicity. Food Chem. Toxicol. 2010, 8, 854–858. [Google Scholar] [CrossRef] [PubMed]
- Shabana, M.M.; El Sayed, A.M.; Yousif, M.F.; El Sayed, A.M.; Sleem, A.A. Bioactive constituents from Harpephyllum caffrum Bernh and Rhus coriaria L. Pharmacogn. Mag. 2011, 7, 298–306. [Google Scholar] [PubMed]
- Satyanarayana, D.N.; Chandra, K.R. Dyeing of Cotton Cloth with Natural Dye Extracted From Pomegranate Peel and its Fastness. IJESRT 2013, 2, 2664–2669. [Google Scholar]
- Lee, Y.H.; Hwang, E.K.; Kim, H.D. Colorimetric Assay and Antibacterial Activity of Cotton, Silk, and Wool Fabrics Dyed with Peony, Pomegranate, Clove, Coptis chinenis and Gallnut Extracts. Materials 2009, 2, 10. [Google Scholar] [CrossRef]
- Hamad, Y.K.; Abobakr, Y.; Salem, M.Z.M.; Ali, H.M.; Al-Sarar, A.S.; Al-Zabib, A.A. Activity of plant extracts/essential oils against some plant pathogenic fungi and mosquitoes: GC/MS analysis of bioactive compounds. BioResources 2019, 14, 4489–4511. [Google Scholar]
- Gawish, S.M.; Mashaly, H.M.; Helmy, H.M.; Ramadan, A.M.; Farouk, R. Effect of Mordant on UV Protection and Antimicrobial Activity of Cotton, Wool, Silk and Nylon Fabrics Dyed with Some Natural Dyes. J. Nanomed. Nanotechnol. 2017, 8, 1. [Google Scholar] [CrossRef]
- Dumitrescu, I.; Visileanu, E.; Niculescu, M. Natural dyes obtained from plants and vegetable wastes. Colourage 2004, 51, 121–129. [Google Scholar]
- Bhattacharya, S.D.; Shah, A.K. Metal ion effect on dyeing of wool fabric with catechu. Color Technol. 2000, 116, 10–12. [Google Scholar] [CrossRef]
- Slimestad, R.; Fossen, T.; Vågen, I. Onions: A Source of Unique Dietary Flavonoids. J. Agric. Food Chem. 2007, 55, 10067–10080. [Google Scholar] [CrossRef]
- Arshad, M.S.; Sohaib, M.; Nadeem, M.; Saeed, F.; Imran, A.; Javed, A.; Zaid Amjad, Z.; Batool, S.M. Status and trends of nutraceuticals from onion and onion by-products: A critical review. Cogent Food Agric. 2017, 3, 1280254. [Google Scholar] [CrossRef]
- Vankar, P.S.; Shanker, R. Ecofriendly ultrasonic natural dyeing of cotton fabric with enzyme pretreatments. Desalination 2008, 230, 62–69. [Google Scholar] [CrossRef]
- Vankar, P.S.; Shanker, R.; Verma, A. Enzymatic natural dyeing of cotton and silk fabrics without metal mordants. J. Clean. Prod. 2007, 15, 1441–1450. [Google Scholar] [CrossRef]
- Goodarzian, H.; Ekrami, E. Wool dyeing with extracted dye from pomegranate (Punica granatum L.) peel. World App. Sci. 2010, 8, 1387–1389. [Google Scholar]
- Mahmud-Ali, A.; Fitz-Binder, C.; Bechtold, T. Aluminium based dye lakes from plant extracts for textile coloration. Dyes Pigments 2012, 94, 533–540. [Google Scholar] [CrossRef]
- Ghaheh, F.S.; Mortazavi, S.M.; Alihosseini, F.; Fassihi, A.; Nateri, A.S.; Abedi, D. Assessment of antibacterial activity of wool fabrics dyed with natural dyes. J. Clean. Prod. 2014, 72, 139–145. [Google Scholar] [CrossRef]
- Bechtold, T.; Mahmud-Ali, A.; Mussak, R. Natural dyes for textile dyeing e Comparison of methods to assess quality of Canadian Golden Rod plant material. Dyes Pigments 2007, 75, 287–293. [Google Scholar] [CrossRef]
- Leitner, P.; Fitz-Binder, C.; Mahmud-Ali, A.; Bechtold, T. Production of a concentrated natural dye from Canadian Goldenrod (Solidago Canadiensis) extracts. Dyes Pigments 2012, 93, 1416–1421. [Google Scholar] [CrossRef]
- Önal, A. Extraction of dyestuff from onion (Allium cepa L.) and its application in the dyeing of wool, feathered-leather and cotton. Turk. J. Chem. 1996, 20, 194–203. [Google Scholar]
- Tera, F.M.; Elnagar, K.E.; Mohamed, S.M. Dyeing and light fastness properties of onion scale dye on different fabric types for conservative applications. J. Tex. Appar. Technol. Manag. 2012, 7, 1–6. [Google Scholar]
- Chandravanshi, S.; Upadhyay, S.K. Interaction of natural dye (Allium cepa) with ionic surfactants. J. Chem. 2012, 2013, 685679. [Google Scholar] [CrossRef]
- Ferreira, E.S.B.; Hulme, A.N.; McNab, H.; Quye, A. The natural constituents of historical textile dyes. Chem. Soc. Rev. 2004, 33, 329–336. [Google Scholar] [CrossRef] [PubMed]
- Vankar, P.S.; Shanker, R. Dyeing of cotton, wool and silk with extract of Allium cepa. Pigm. Resin Technol. 2009, 38, 242–247. [Google Scholar] [CrossRef]
- Das, D.; Maulik, S.R. Dyeing of wool and silk with Punica granatum. Indian J. Fibre Text. 2006, 31, 559–564. [Google Scholar]
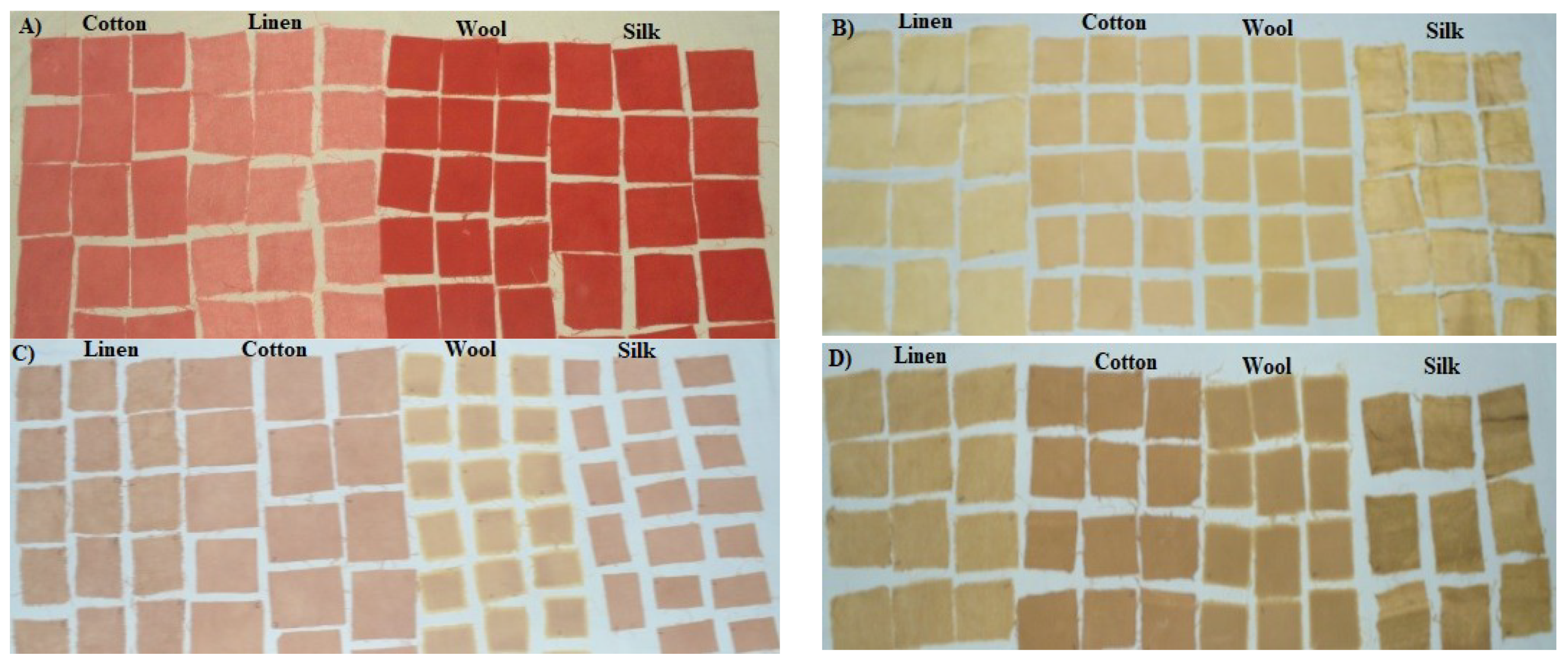

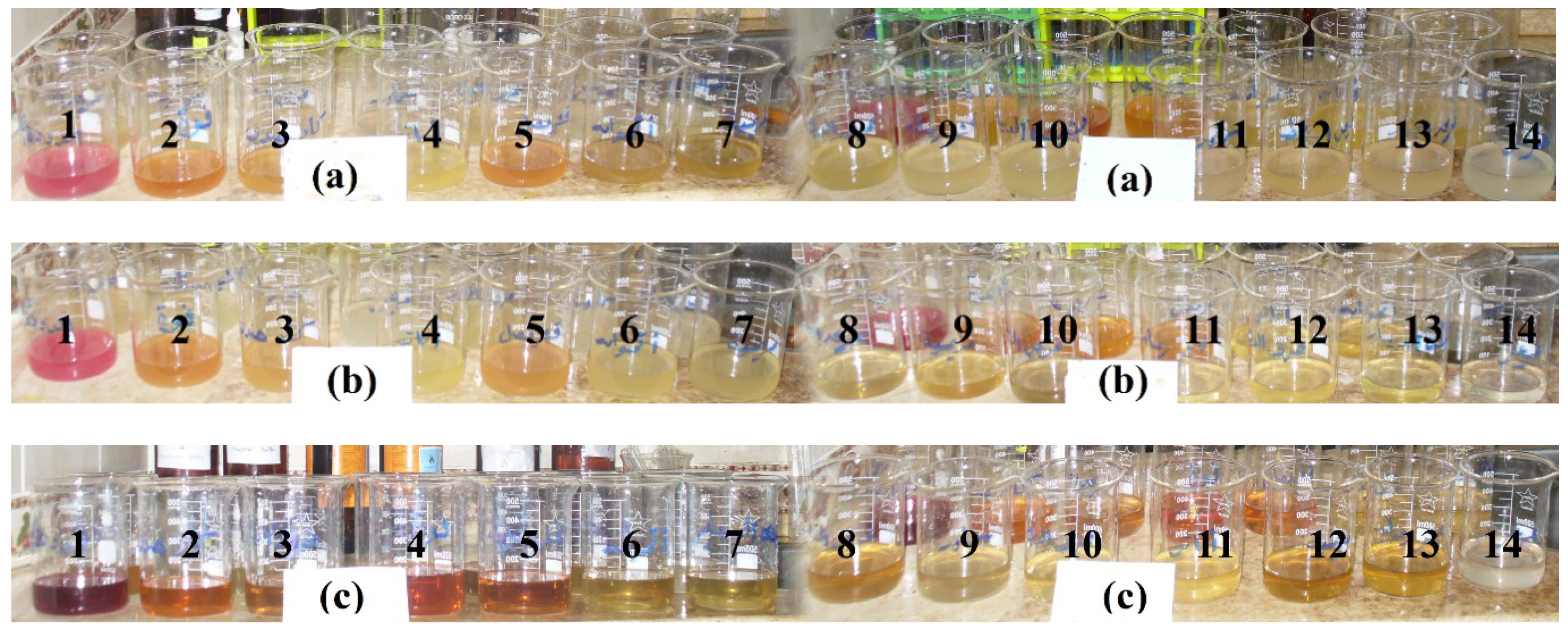


| Fiber | Weave Structure | Basic Weight (g/m2) | Wefts | Warps | Tensile Strength (kg/Strength) | Elongation (%) |
|---|---|---|---|---|---|---|
| Linen | Plain weave | 113 | 13 | 17 | 56.5 | 6.9 |
| Cotton | Plain weave | 179 | 25 | 25 | 66.9 | 19.9 |
| Wool | Plain weave | 148 | 23 | 23 | 13.9 | 12.6 |
| Silk | Plain weave | 143 | 24 | 25 | 33.7 | 14.9 |
| Source of Variance | DF | Sums of Squares | Mean Square | F Value | Pr > F |
| A | 2 | 82.11419 | 41.05710 | 3.28 | 0.0390 |
| B | 13 | 90,562.169 | 6966.32 | 555.94 | <0.0001 |
| C | 3 | 24,577.007 | 8192.335 | 653.78 | <0.0001 |
| A × B | 26 | 1387.205 | 53.354 | 4.26 | <0.0001 |
| A × C | 6 | 90.785 | 15.131 | 1.21 | 0.3018 |
| B × C | 39 | 34,035.76 | 872.711 | 69.65 | <0.0001 |
| A × B × C | 78 | 2308.23 | 29.592 | 2.36 | <0.0001 |
| Error | 336 | 4210.29 | 12.531 | ||
| Corrected Total | 503 | 157,253.569 |
| Enzyme Solution | Dye | Relative Color Strength (%) | Total Color Difference (ΔE) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Linen | Cotton | Silk | Wool | Linen | Cotton | Silk | Wool | ||
| Buffer solution | Annatto | 118.56 ± 4.31 | 117.20 ± 4.41 | 110.60 ± 2.26 | 103.33 ± 2.51 | 3.03 ± 0.05 | 2.76 ± 0.32 | 1.16 ± 0.31 | 1.13 ± 0.15 |
| Bloodroot | 103.36 ± 6.29 | 110.60 ± 1.73 | 125.56 ± 6.83 | 118.76 ± 5.41 | 2.96 ± 0.41 | 3.36 ± 0.66 | 2.83 ± 0.21 | 2.3 ± 0.20 | |
| Catechu tree | 91.80 ± 5.54 | 109.03 ± 4.75 | 103.56 ± 2.43 | 128.73 ± 4.97 | 6.83 ± 0.73 | 5.33 ± 0.25 | 2.36 ± 0.40 | 5.40 ± 0.36 | |
| Cochineal | 74.13 ± 4.39 | 83.16 ± 0.85 | 93.90 ± 1.81 | 100.06 ± 0.95 | 4.00 ± 0.10 | 4.83 ± 0.15 | 3.73 ± 0.15 | 2.86 ± 0.15 | |
| Common juniper | 85.53 ± 4.80 | 89.90 ± 0.00 | 71.60 ± 0.00 | 109.90 ± 2.76 | 2.16 ± 0.37 | 2.16 ± 0.35 | 3.06 ± 0.21 | 0.96 ± 0.15 | |
| Common sage | 68.86 ± 0.21 | 84.43 ± 1.13 | 83.06 ± 2.90 | 120.63 ± 2.04 | 3.30 ± 0.2 | 2.46 ± 0.15 | 1.66 ± 0.15 | 1.20 ±0.1 | |
| Common hops | 92.46 ± 4.98 | 89.70 ± 4.61 | 74.3 ± 1.65 | 101.16 ± 3.00 | 3.60 ±0.45 | 2.86 ±0.05 | 4.86 ±0.56 | 1.26 ± 0.15 | |
| Dog’s fennel | 79.50 ± 2.61 | 68.63 ± 4.53 | 70.53 ± 1.40 | 89.16 ± 3.01 | 3.00 ± 0.10 | 2.90 ± 0.10 | 2.60 ± 0.20 | 0.67 ± 0.04 | |
| Egyptian acacia | 120.46 ± 1.51 | 117.23 ± 3.42 | 122.66 ± 6.95 | 119.53 ± 2.11 | 1.76 ± 0.15 | 2.16 ± 0.25 | 1.53 ± 0.15 | 2.36 ± 0.56 | |
| Goldenrod | 77.83 ± 3.61 | 80.26 ± 1.90 | 84.86 ± 1.26 | 110.86 ± 2.07 | 2.46 ± 0.31 | 2.33 ± 0.25 | 2.03 ± 0.15 | 1.00 ± 0.10 | |
| Madder | 86.16 ± 5.57 | 98.53 ± 1.33 | 83.70 ± 3.04 | 99.20 ± 1.05 | 1.70 ± 0.26 | 1.23 ± 0.05 | 2.23 ± 0.11 | 2.96 ± 0.81 | |
| Onion skin | 68.40 ± 1.76 | 74.80 ± 4.51 | 96.46 ± 3.28 | 89.76 ± 1.88 | 4.76 ± 0.65 | 4.33 ± 0.55 | 5.93 ± 0.81 | 2.36 ± 0.15 | |
| Pomegranate skin | 111.03 ± 2.43 | 107.66 ± 2.87 | 110.26 ± 3.05 | 129.13 ± 3.33 | 1.73 ± 0.25 | 1.05 ± 0.17 | 1.06 ± 0.15 | 2.50 ± 0.10 | |
| Sumac | 109.86 ± 1.55 | 115.03 ± 1.65 | 109.26 ± 2.65 | 126.53 ± 1.98 | 2.26 ± 0.41 | 1.36 ± 0.31 | 2.70 ± 0.10 | 3.00 ± 0.20 | |
| Enzyme concentration A | Annatto | 109.16 ± 8.06 | 112.66 ± 1.98 | 112.66 ± 7.03 | 96.83 ± 6.03 | 3.30 ± 0.55 | 2.90 ± 0.36 | 1.36 ± 0.21 | 1.06 ± 0.15 |
| Bloodroot | 99.20 ± 4.64 | 111.83 ± 4.14 | 129.73 ± 9.37 | 119.60 ± 2.04 | 2.83 ± 0.15 | 2.90 ± 0.40 | 3.20 ± 0.10 | 2.170 ± 0.24 | |
| Catechu tree | 86.93 ± 2.28 | 111.53 ± 2.47 | 107.16 ± 2.44 | 117.30 ± 3.57 | 5.70 ± 0.26 | 4.53 ± 0.37 | 1.90 ± 0.20 | 4.63 ± 0.37 | |
| Cochineal | 81.66 ± 5.15 | 86.13 ± 2.62 | 93.83 ± 4.14 | 102.16 ± 3.35 | 3.23 ± 0.73 | 3.93 ± 0.15 | 3.40 ± 0.43 | 1.94 ± 0.05 | |
| Common juniper | 93.60 ± 3.67 | 90.96 ± 1.51 | 66.56 ± 4.16 | 112.86 ± 1.81 | 1.93 ± 0.35 | 1.70 ± 0.20 | 3.46 ± 0.41 | 1.15 ± 0.18 | |
| Common sage | 70.40 ± 0.65 | 77.40 ± 4.01 | 80.30 ± 0.81 | 118.76 ± 1.81 | 2.80 ± 0.26 | 2.63 ± 0.21 | 2.23 ± 0.15 | 1.20 ± 0.20 | |
| Common hops | 89.00 ± 2.91 | 85.73 ± 1.19 | 72.90 ± 2.70 | 101.63 ± 4.23 | 3.93 ± 0.56 | 2.80 ± 0.30 | 4.20 ± 0.79 | 1.73 ± 0.35 | |
| Dog’s fennel | 84.43 ± 5.35 | 73.56 ± 1.55 | 75.60 ± 3.99 | 91.50 ± 2.16 | 2.56 ± 0.15 | 2.73 ± 0.15 | 2.39 ± 0.10 | 0.61 ± 0.10 | |
| Egyptian acacia | 123.33 ± 2.00 | 107.63 ± 1.96 | 116.33 ± 1.75 | 114.03 ± 2.21 | 1.90 ± 0.20 | 1.93 ± 0.15 | 1.33 ± 0.15 | 1.53 ± 0.31 | |
| Goldenrod | 80.16 ± 1.05 | 79.16 ± 1.85 | 86.83 ± 2.57 | 117.93 ± 2.43 | 1.83 ± 0.21 | 2.36 ± 0.15 | 1.63 ± 0.21 | 1.03 ± 0.15 | |
| Madder | 98.43 ± 3.68 | 101.03 ± 2.21 | 82.30 ± 5.08 | 96.20 ± 5.12 | 1.33 ± 0.05 | 1.26 ± 0.05 | 2.25 ± 0.13 | 3.13 ± 0.60 | |
| Onion skin | 72.20 ± 2.12 | 80.20 ± 5.75 | 104.60 ± 6.13 | 94.73 ± 1.76 | 4.06 ± 0.15 | 4.10 ± 0.10 | 5.66 ± 0.32 | 2.20 ± 0.36 | |
| Pomegranate skin | 117.76 ± 1.89 | 117.23 ± 3.67 | 110.36 ± 5.71 | 137.23 ± 3.74 | 1.93 ± 0.15 | 1.08 ± 0.12 | 1.66 ± 0.66 | 3.00 ± 0.10 | |
| Sumac | 110.4 ± 3.70 | 119.26 ± 3.05 | 107.53 ± 3.05 | 128.00 ± 2.16 | 1.83 ± 0.25 | 1.03 ± 0.15 | 2.56 ± 0.05 | 3.40 ± 0.26 | |
| Enzyme concentration B | Annatto | 114.86 ± 4.41 | 109.60 ± 5.40 | 107.23 ± 4.17 | 109.86 ± 4.2 | 2.86 ± 0.25 | 1.96 ± 0.81 | 1.30 ± 0.40 | 1.43 ± 0.32 |
| Bloodroot | 99.80 ± 1.37 | 106.73 ± 3.45 | 132.20 ± 3.65 | 115.53 ± 2.20 | 3.33 ± 0.21 | 2.90 ± 0.40 | 2.90 ± 0.20 | 2.00 ± 0.36 | |
| Catechu tree | 88.5 ± 4.20 | 107.03 ± 2.71 | 104.16 ± 2.95 | 126.5 ± 4.30 | 6.30 ± 0.26 | 4.53 ± 0.55 | 1.90 ± 0.45 | 4.93 ± 0.25 | |
| Cochineal | 76.00 ± 1.37 | 80.73 ± 0.45 | 87.03 ± 5.56 | 94.20 ± 4.84 | 3.56 ± 0.15 | 4.46 ± 0.25 | 4.23 ± 0.51 | 2.30 ± 0.26 | |
| Common juniper | 88.33 ± 2.45 | 89.70 ± 0.95 | 68.73 ± 3.76 | 111.96 ± 4.21 | 2.10 ± 0.30 | 1.66 ± 0.31 | 3.23 ± 0.25 | 0.90 ± 0.10 | |
| Common sage | 69.16 ± 1.00 | 87.33 ± 4.21 | 77.00 ± 3.08 | 114.43 ± 2.32 | 3.43 ± 0.25 | 2.26 ± 0.21 | 1.36 ± 0.15 | 0.95 ± 0.13 | |
| Common hops | 90.93 ± 1.61 | 80.96 ± 3.02 | 70.03 ± 3.86 | 106.30 ± 3.80 | 3.46 ± 0.50 | 3.13 ± 0.15 | 4.56 ± 0.45 | 1.33 ± 0.15 | |
| Dog’s fennel | 78.430 ± 2.92 | 75.20 ± 3.48 | 72.16 ± 1.76 | 92.56 ± 2.89 | 2.73 ± 0.21 | 2.53 ± 0.25 | 2.50 ± 0.10 | 0.68 ± 0.02 | |
| Egyptian acacia | 116.63 ± 2.46 | 114.16 ± 2.43 | 121.06 ± 4.80 | 120.20 ± 0.55 | 1.76 ± 0.25 | 1.63 ± 0.21 | 1.36 ± 0.21 | 1.80 ± 0.26 | |
| Goldenrod | 79.50 ± 0.87 | 80.46 ± 1.22 | 86.90 ± 1.24 | 108.3 ± 4.71 | 2.76 ± 0.25 | 2.26 ± 0.31 | 1.93 ± 0.21 | 0.96 ± 0.21 | |
| Madder | 94.33 ± 4.11 | 107.13 ± 3.01 | 87.53 ± 2.37 | 100.40 ± 2.91 | 1.30 ± 0.10 | 1.10 ± 0.10 | 1.82 ± 0.91 | 2.26 ± 0.15 | |
| Onion skin | 69.70 ± 2.26 | 73.03 ± 3.05 | 106.50 ± 3.36 | 94.50 ± 4.23 | 4.03 ± 0.21 | 4.23 ± 0.31 | 5.96 ± 0.81 | 2.03 ± 0.25 | |
| Pomegranate skin | 112.56 ± 1.41 | 110.30 ± 5.52 | 109.90 ± 5.18 | 129.30 ± 5.51 | 1.43 ± 0.21 | 1.05 ± 0.21 | 0.95 ± 0.35 | 2.50 ± 0.36 | |
| Sumac | 106.20 ± 4.37 | 114.83 ± 3.90 | 110.43 ± 1.79 | 129.16 ± 1.05 | 1.99 ± 0.17 | 0.96 ± 0.16 | 2.50 ± 0.26 | 3.50 ± 0.45 | |
| Compound | mg/kg Extract | |
|---|---|---|
| Madder | Cochineal | |
| Pyrogallol | - | - |
| Quinol | - | - |
| Gallic acid | 35.12 | - |
| Catechol | 35.96 | - |
| p-Hydroxy benzoic acid | 30.14 | 110.94 |
| Caffeine | 41.17 | - |
| Chlorgenic acid | 45.72 | 5.56 |
| Vanillic acid | 187.11 | 9.71 |
| Caffeic acid | 3.19 | 9.79 |
| Syringic acid | 5.65 | - |
| Vanillin | 31.46 | 40.42 |
| p-Coumaric acid | 2.53 | - |
| Ferulic acid | - | 19.04 |
| Benzoic acid | 582.68 | 49.92 |
| Rutin | 88.78 | 37,732 |
| Ellagic acid | 784.86 | - |
| o-Coumaric acid | 8.87 | - |
| Salicylic acid | 1758.91 | 193.87 |
| Cinnamic acid | 9.39 | - |
| Myricetin | 41.51 | 809.97 |
| Quercetin | 844.23 | 496.76 |
| rosemarinic | 143.57 | - |
| Naringenin | 122.09 | - |
| Kampherol | 232.91 | 915.98 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Salem, M.Z.M.; Ibrahim, I.H.M.; Ali, H.M.; Helmy, H.M. Assessment of the Use of Natural Extracted Dyes and Pancreatin Enzyme for Dyeing of Four Natural Textiles: HPLC Analysis of Phytochemicals. Processes 2020, 8, 59. https://doi.org/10.3390/pr8010059
Salem MZM, Ibrahim IHM, Ali HM, Helmy HM. Assessment of the Use of Natural Extracted Dyes and Pancreatin Enzyme for Dyeing of Four Natural Textiles: HPLC Analysis of Phytochemicals. Processes. 2020; 8(1):59. https://doi.org/10.3390/pr8010059
Chicago/Turabian StyleSalem, Mohamed Z. M., Ibrahim H. M. Ibrahim, Hayssam M. Ali, and Hany M. Helmy. 2020. "Assessment of the Use of Natural Extracted Dyes and Pancreatin Enzyme for Dyeing of Four Natural Textiles: HPLC Analysis of Phytochemicals" Processes 8, no. 1: 59. https://doi.org/10.3390/pr8010059
APA StyleSalem, M. Z. M., Ibrahim, I. H. M., Ali, H. M., & Helmy, H. M. (2020). Assessment of the Use of Natural Extracted Dyes and Pancreatin Enzyme for Dyeing of Four Natural Textiles: HPLC Analysis of Phytochemicals. Processes, 8(1), 59. https://doi.org/10.3390/pr8010059






