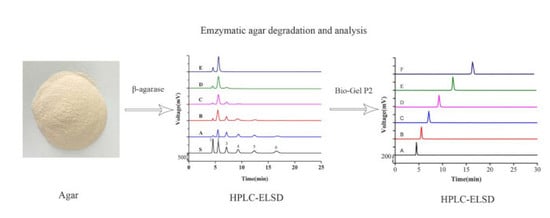
Simple Preparation of Diverse Neoagaro-Oligosaccharides
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains and Reagents
2.2. Optimization of Enzymatic Hydrolysis Condition
2.3. Preparation of NAOS Products with Different Enzymolysis Time
2.4. Analysis of NAOS Products by HPLC-ELSD
2.5. Purification of NAOS
2.6. Identification of NAOS
3. Results and Discussion
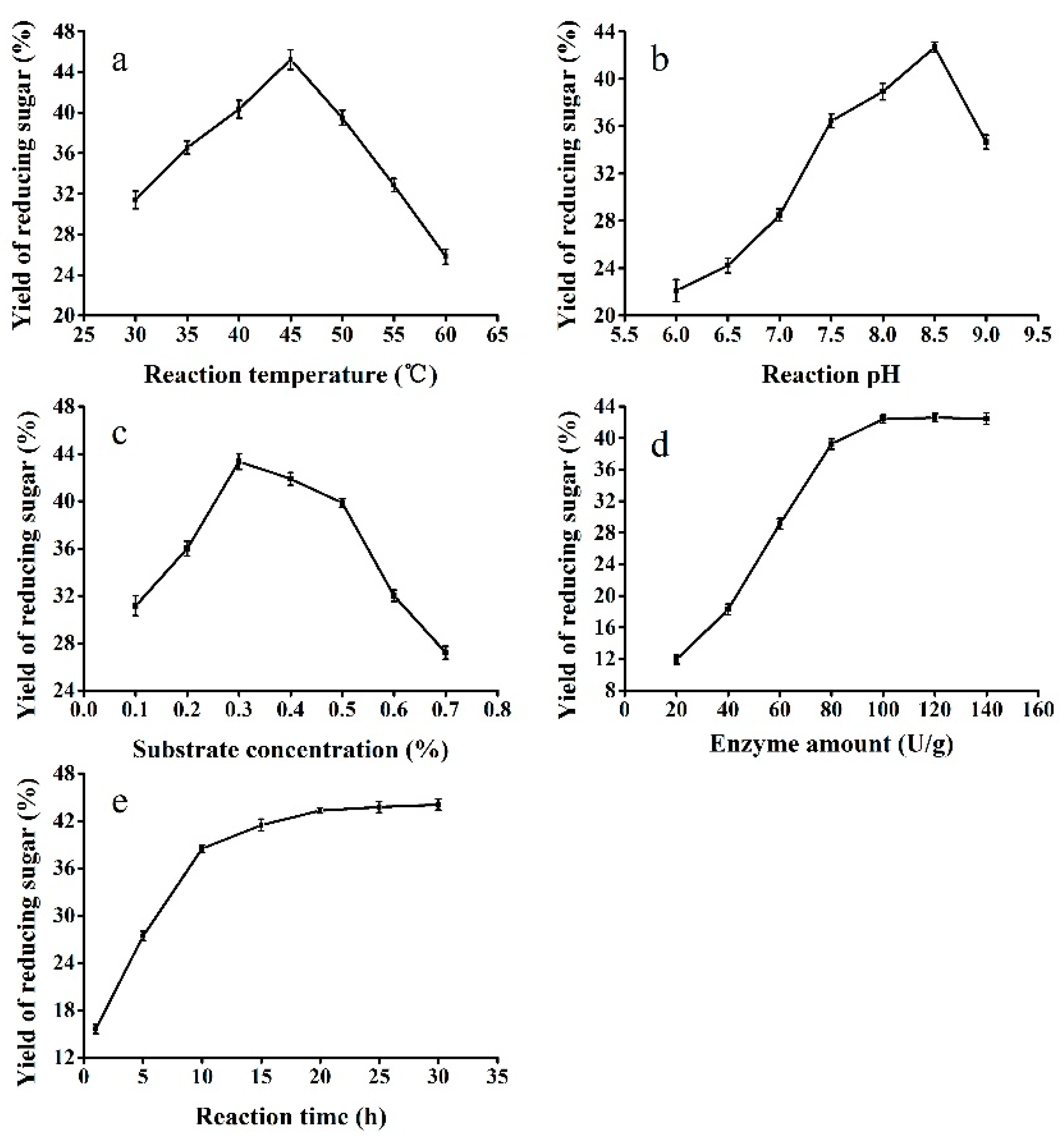
3.1. Optimization of Hydrolysis Parameters by Single Factor Experiments
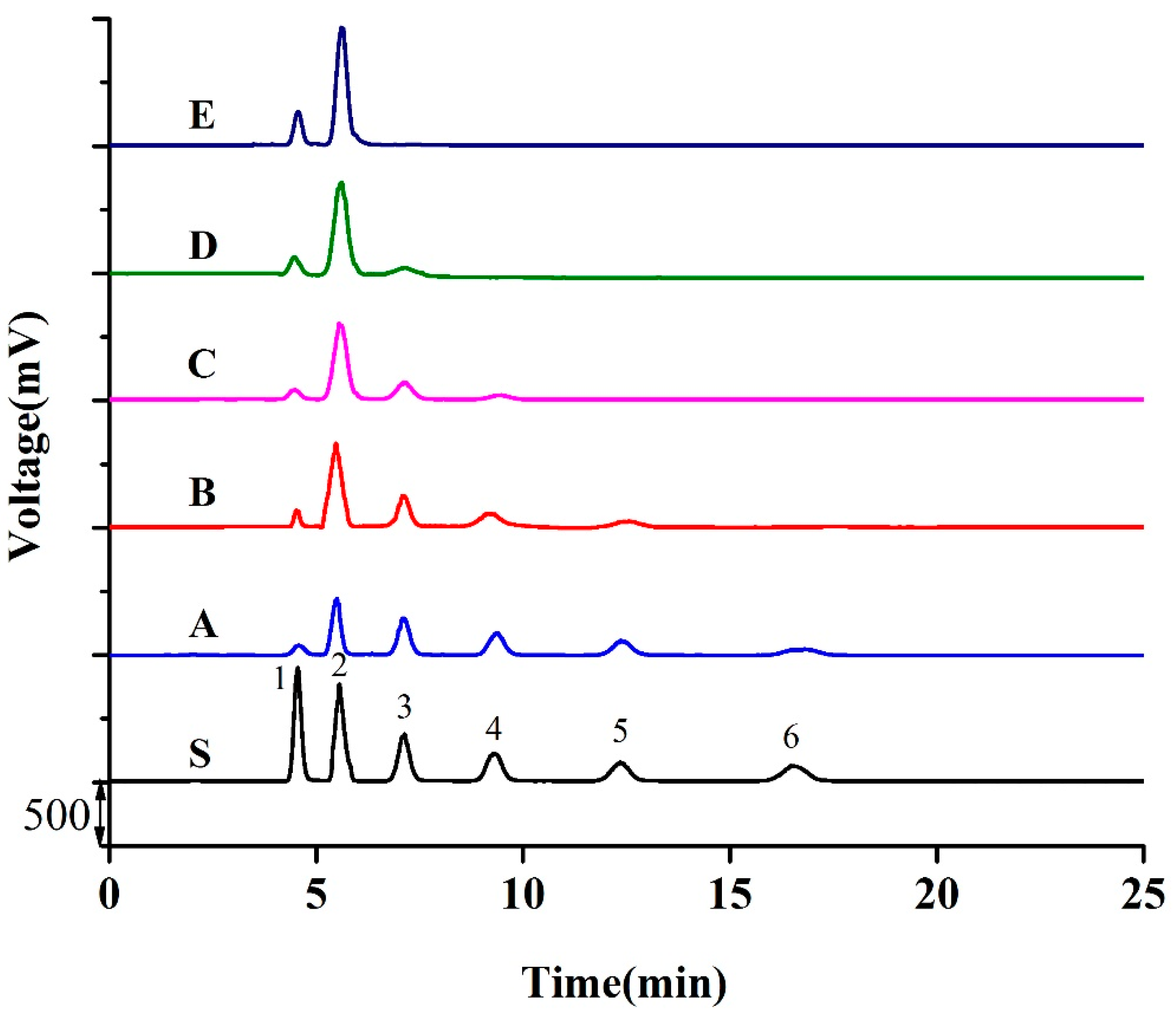
3.2. Preparation of NAOS with Different DP
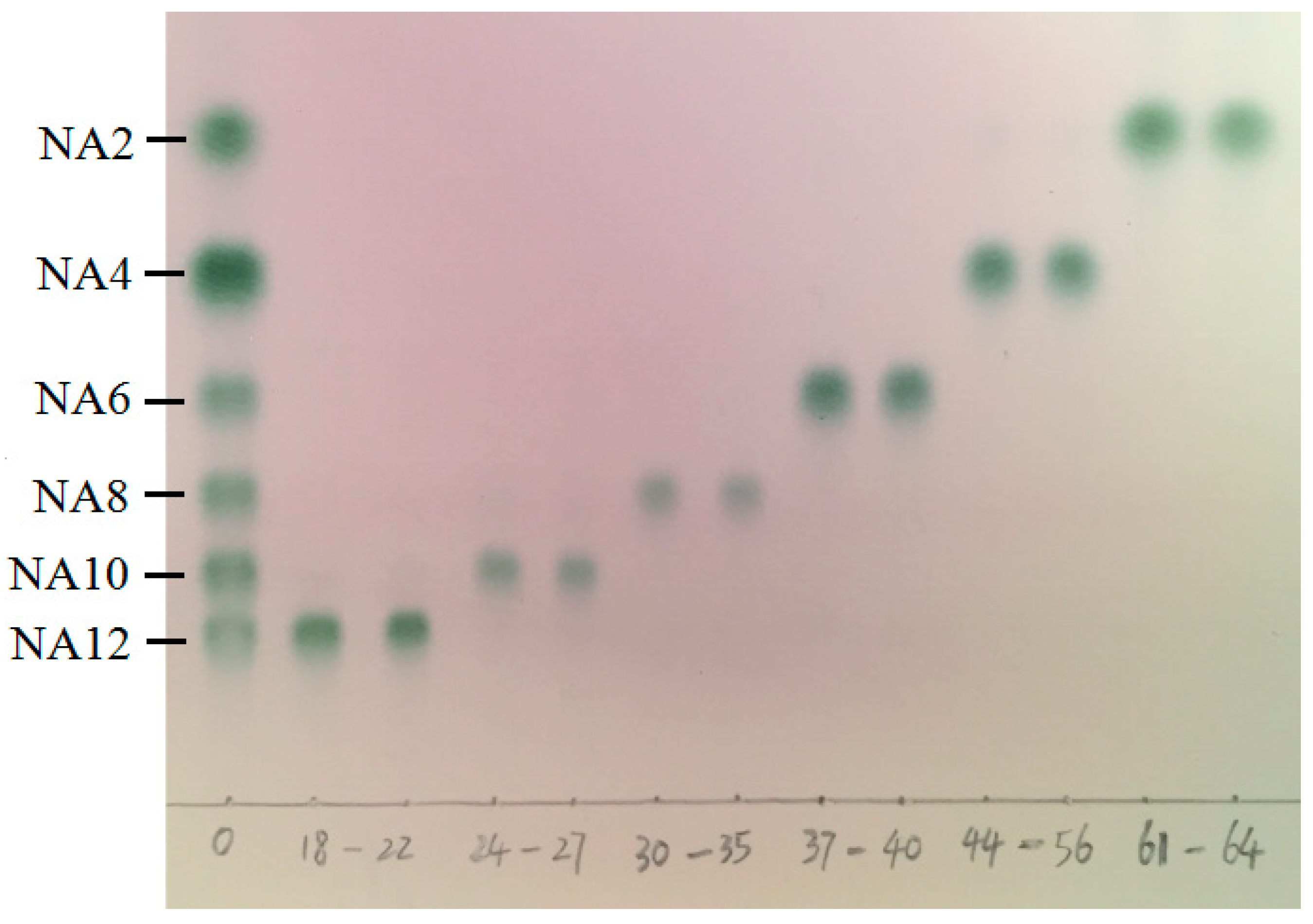
3.3. Separation of NAOS
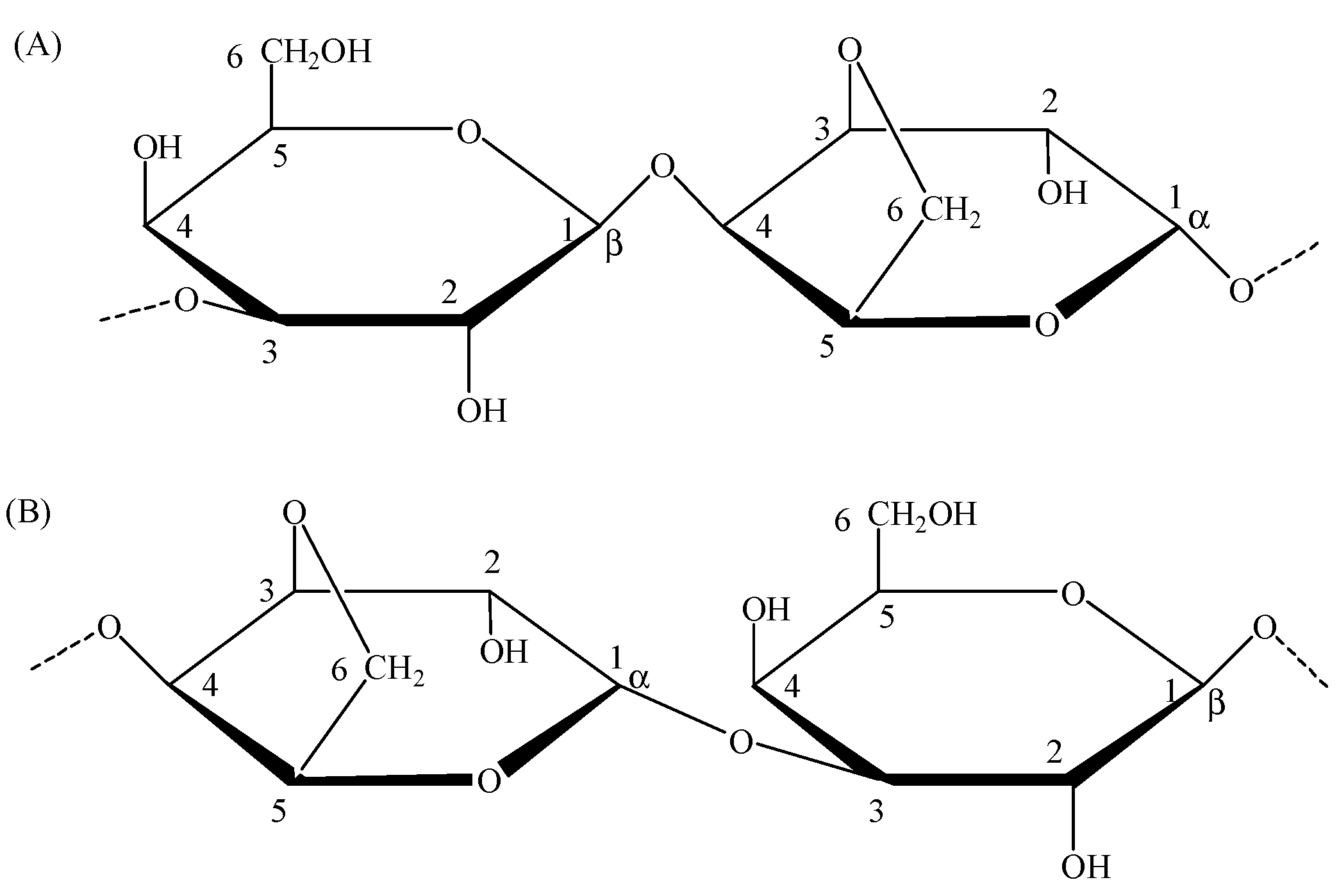
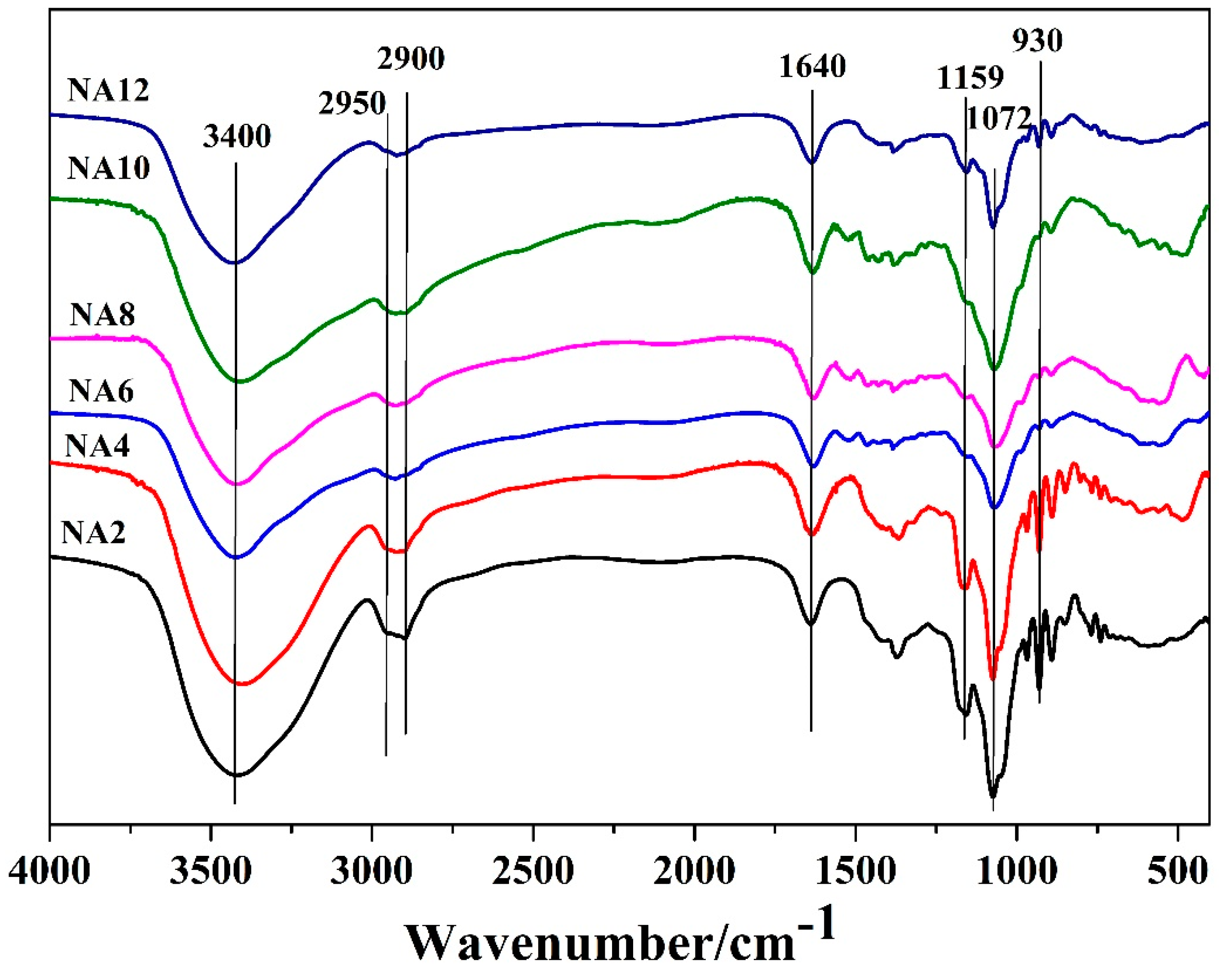
3.4. Characterization of NAOS
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hamer, G.K.; Bhattacharjee, S.S.; Yaphe, W. Analysis of the enzymlc hydmlysls products of agahose by C-NMR. Carbohydr. Res. 1977, 54, C7–C10. [Google Scholar] [CrossRef]
- Jensen, A. Present and future needs for algae and algal products. Hydrobiologia 1993, 260–261, 15–23. [Google Scholar] [CrossRef]
- Chi, W.J.; Chang, Y.K.; Hong, S.K. Agar degradation by microorganisms and agar-degrading enzymes. Appl. Microbiol. Biotechnol. 2012, 94, 917–930. [Google Scholar] [CrossRef]
- Chen, H.M.; Zheng, L.; Yan, X.J. The preparation and bioactivity research of agaro-oligosaccharides. Food Technol. Biotech. 2005, 43, 29–36. [Google Scholar]
- Chen, H.M.; Yan, X.J. Antioxidant activities of agaro-oligosaccharides with different degrees of polymerization in cell-based system. Biochim. Biophys. Acta 2005, 1722, 103–111. [Google Scholar] [CrossRef]
- Wu, S.C.; Wen, T.N.; Pan, C.L. Algal-oligosaccharide-lysates prepared by two bacterial agarases stepwise hydrolyzed and their anti-oxidative properties. Fish. Sci. 2005, 71, 1149–1159. [Google Scholar] [CrossRef]
- Chen, H.; Yan, X.; Zhu, P.; Lin, J. Antioxidant activity and hepatoprotective potential of agaro-oligosaccharides in vitro and in vivo. Nutr. J. 2006, 5, 31–42. [Google Scholar] [CrossRef] [PubMed]
- Kang, O.L.; Ghani, M.; Hassan, O.; Rahmati, S.; Ramli, N. Novel agaro-oligosaccharide production through enzymatic hydrolysis: Physicochemical properties and antioxidant activities. Food Hydrocolloids 2014, 42, 304–308. [Google Scholar] [CrossRef]
- Xu, S.Y.; Huang, X.; Cheong, K.L. Recent advances in marine algae polysaccharides: Isolation, structure, and activities. Mar. Drugs 2017, 15, 388. [Google Scholar] [CrossRef] [PubMed]
- Higashimura, Y.; Naito, Y.; Takagi, T.; Uchiyama, K.; Mizushima, K.; Ushiroda, C.; Ohnogi, H.; Kudo, Y.; Yasui, M.; Inui, S.; et al. Protective effect of agaro-oligosaccharides on gut dysbiosis and colon tumorigenesis in high-fat diet-fed mice. Am. J. Physiol-Gastr. Liver Physiol. 2016, 310, G367–G375. [Google Scholar] [CrossRef]
- Higashimura, Y.; Baba, Y.; Inoue, R.; Takagi, T.; Mizushima, K.; Ohnogi, H.; Honda, A.; Matsuzaki, Y.; Naito, Y. Agaro-oligosaccharides regulate gut microbiota and adipose tissue accumulation in mice. J. Nutr. Sci. Vitaminol. 2017, 63, 269–276. [Google Scholar] [CrossRef] [PubMed]
- Enoki, T.; Tanabe, M.; Shimomura, M.; Ohnogi, H. Induction mechanism of heme oxygenase-1 and anti-inflammatory activity by agaro-oligosaccharides. Nippon Shokuhin Kogyo Gakkaishi 2010, 57, 157–162. [Google Scholar] [CrossRef][Green Version]
- Enoki, T.; Tominaga, T.; Takashima, F.; Ohnogi, H.; Sagawa, H.; Kato, I. Anti-tumor-promoting activities of agaro-oligosaccharides on two-stage mouse skin carcinogenesis. Biol. Pharm. Bull. 2012, 35, 1145–1149. [Google Scholar] [CrossRef] [PubMed]
- Higashimura, Y.; Naito, Y.; Takagi, T.; Tanimura, Y.; Mizushima, K.; Harusato, A.; Fukui, A.; Yoriki, H.; Handa, O.; Ohnogi, H.; et al. Preventive effect of agaro-oligosaccharides on non-steroidal anti-inflammatory drug-induced small intestinal injury in mice. J. Gastroen. Hepatol. 2014, 29, 310–317. [Google Scholar] [CrossRef]
- Kobayashi, R.; Takisada, M.; Suzuki, T.; Kirimuraab, K.; Usamiab, S. Neoagarobiose as a novel moisturizer with whitening effect. Biosci. Biotechnol. Biochem. 1997, 61, 162–163. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Yun, E.J.; Yu, S.; Kim, K.H.; Kang, N.J. Different levels of skin whitening activity among 3,6-Anhydro-l-galactose, Agarooligosaccharides, and Neoagarooligosaccharides. Mar. Drugs 2017, 15, 321. [Google Scholar] [CrossRef] [PubMed]
- Potin, P.; Richard, C.; Rochas, C.; Kloareg, B. Purification and characterization of the α-agarase from Alteromonas agarlyticus (Cataldi) comb. nov., strain GJ1B. Eur. J. Biochem. 1993, 214, 559–607. [Google Scholar] [CrossRef]
- Hassairi, I.; Amar, R.B.; Nonus, M.; Gupta, B.B. Production and separation of α-agarase from Altermonas agarlyticus strain GJ1B. Bioresour. Technol. 2001, 79, 47–51. [Google Scholar] [CrossRef]
- Ohta, Y.; Hatada, Y.; Miyazaki, M.; Nogi, Y.; Ito, S.; Horikoshi, K. Purification and characterization of a novel alpha-agarase from a Thalassomonas sp. Curr. Microbiol. 2005, 50, 212–216. [Google Scholar] [CrossRef] [PubMed]
- Kirimura, K.; Masuda, N.; Iwasaki, Y.; Nakagawa, H.; Kobayashi, R.; Usami, S. Purification and characterization of a novel β-agarase from an alkalophilic bacterium, Alteromonas sp. E-1. J. Biosci. Bioeng. 1999, 87, 436–441. [Google Scholar] [CrossRef]
- Kang, N.Y.; Choi, Y.L.; Cho, Y.S.; Kim, B.K.; Jeon, B.S.; Cha, J.Y.; Kim, C.H.; Lee, Y.C. Cloning, expression and characterization of a β-agarase gene from a marine bacterium, Pseudomonas sp. SK38. Biotechnol. Lett. 2003, 25, 1165–1170. [Google Scholar] [CrossRef] [PubMed]
- Fu, W.; Han, B.; Duan, D.; Liu, W.; Wang, C. Purification and characterization of agarases from a marine bacterium Vibrio sp. F-6. J. Ind. Microbiol. Biotechnol. 2008, 35, 915–922. [Google Scholar] [CrossRef] [PubMed]
- Bhattarai, Y.; Kashyap, P.C. Agaro-oligosaccharides: A new frontier in the fight against colon cancer? Am. J. Physiol-Gastr. Liver Physiol. 2016, 310, G335–G336. [Google Scholar] [CrossRef]
- Xiao, Q.; Liu, C.; Ni, H.; Zhu, Y.; Jiang, Z.; Xiao, A. beta-Agarase immobilized on tannic acid-modified Fe3O4 nanoparticles for efficient preparation of bioactive neoagaro-oligosaccharide. Food Chem. 2019, 272, 586–595. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Gong, Q.; Wang, Y.; Ma, Y.; Li, J.; Yu, W. Prebiotic effects of neoagaro-oligosaccharides prepared by enzymatic hydrolysis of agarose. Anaerobe 2006, 12, 260–266. [Google Scholar] [CrossRef]
- Wang, W.; Liu, P.; Hao, C.; Wu, L.; Wan, W.; Mao, X. Neoagaro-oligosaccharide monomers inhibit inflammation in LPS-stimulated macrophages through suppression of MAPK and NF-kappaB pathways. Sci. Rep. 2017, 7, 44252. [Google Scholar] [CrossRef]
- Xu, X.Q.; Su, B.M.; Xie, J.S.; Li, R.K.; Yang, J.; Lin, J.; Ye, X.Y. Preparation of bioactive neoagaroligosaccharides through hydrolysis of Gracilaria lemaneiformis agar: A comparative study. Food Chem. 2018, 240, 330–337. [Google Scholar] [CrossRef]
- Shi, Y.L.; Lu, X.Z.; Yu, W.G. A new β-agarase from marine bacterium Janthinobacterium sp. SY12. World J. Microbiol. Biotechnol. 2008, 24, 2659–2664. [Google Scholar] [CrossRef]
- Fu, X.T.; Lin, H.; Kim, S.M. Purification and characterization of a novel beta-agarase, AgaA34, from Agarivorans albus YKW-34. Appl. Microbiol. Biotechnol. 2008, 78, 265–273. [Google Scholar] [CrossRef]
- Sugano, Y.; Terada, I.; Arita, M.; Noma, M.; Matsumoto, T. Purification and characterization of a new agarase from a marine bacterium, Vibrio sp. strain JT0107. Appl. Environ. Micro. 1993, 59, 1549–1554. [Google Scholar]
- Su, Q.; Jin, T.; Yu, Y.; Yang, M.; Mou, H.; Li, L. Extracellular expression of a novel beta-agarase from Microbulbifer sp. Q7, isolated from the gut of sea cucumber. AMB Express 2017, 7, 220. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Mou, H.; Jiang, X.; Guan, H. Characterization of a novel beta-agarase from marine Alteromonas sp. SY37-12 and its degrading products. Appl. Microbiol. Biotechnol. 2006, 71, 833–839. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Chu, Y.; Wu, Q.; Gu, Y.; Han, F.; Yu, W. Cloning, expression and characterization of a new agarase-encoding gene from marine Pseudoalteromonas sp. Biotechnol. Lett. 2009, 31, 1565–1570. [Google Scholar] [CrossRef] [PubMed]
- Long, M.; Yu, Z.; Xu, X. A novel beta-agarase with high pH stability from marine Agarivorans sp. LQ48. Mar. Biotechnol. 2010, 12, 62–69. [Google Scholar] [CrossRef] [PubMed]
- Oh, C.; Nikapitiya, C.; Lee, Y.; Whang, I.; Kim, S.J.; Kang, D.H.; Lee, J. Cloning, purification and biochemical characterization of beta agarase from the marine bacterium Pseudoalteromonas sp. AG4. J. Ind. Microbiol. Biotechnol. 2010, 37, 483–494. [Google Scholar] [CrossRef]
- Lee, D.G.; Jang, M.K.; Lee, O.H.; Kim, N.Y.; Ju, S.A.; Lee, S.H. Over-production of a glycoside hydrolase family 50 beta-agarase from Agarivorans sp. JA-1 in Bacillus subtilis and the whitening effect of its product. Biotechnol. Lett. 2008, 30, 911–918. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Zhao, R.; Xiao, A.; Li, L.; Jiang, Z.; Chen, F.; Ni, H. Characterization of an alkaline beta-agarase from Stenotrophomonas sp. NTa and the enzymatic hydrolysates. Int. J. Biol. Macromol. 2016, 86, 525–534. [Google Scholar] [CrossRef]
- Yang, M.; Mao, X.; Liu, N.; Qiu, Y.; Xue, C. Purification and characterization of two agarases from Agarivorans albus OAY02. Process. Biochem. 2014, 49, 905–912. [Google Scholar] [CrossRef]
- Ma, C.; Lu, X.; Shi, C.; Li, J.; Gu, Y.; Ma, Y.; Chu, Y.; Han, F.; Gong, Q.; Yu, W. Molecular cloning and characterization of a novel beta-agarase, AgaB, from marine Pseudoalteromonas sp. CY24. J. Biol. Chem. 2007, 282, 3747–3754. [Google Scholar] [CrossRef]
- Osumi, Y.; Kawai, M.; Amano, H.; Noda, H. Purification and structure of oligosaccharides from porphyran degradated by enzymes from Arthrobacter sp. S-22. Nippon Suisan Gakkaishi 1998, 64, 88–97. [Google Scholar] [CrossRef]
- Chen, H.M.; Zheng, L.; Lin, W.; Yan, X.J. Product monitoring and quantitation of oligosaccharides composition in agar hydrolysates by precolumn labeling HPLC. Talanta 2004, 64, 773–777. [Google Scholar] [CrossRef]
- Li, J.; Han, F.; Lu, X.; Fu, X.; Ma, C.; Chu, Y.; Yu, W. A simple method of preparing diverse neoagaro-oligosaccharides with beta-agarase. Carbohydr. Res. 2007, 342, 1030–1033. [Google Scholar] [CrossRef]
- Morrice, L.M.; McLean, M.W.; Long, W.F.; Williamson, F.B. β-Agarases I and II from Pseudomonas atlantica. Eur. J. Biochem. 1983, 116–117, 576–579. [Google Scholar] [CrossRef]
- Kazlowski, B.; Pan, C.L.; Ko, Y.T. Separation and quantification of neoagaro-and agaro-oligosaccharide products generated from agarose digestion by beta-agarase and HCl in liquid chromatography systems. Carbohydr. Res. 2008, 343, 2443–2450. [Google Scholar] [CrossRef]
- Kazlowski, B.; Pan, C.L.; Ko, Y.T. Monitoring and preparation of neoagaro- and agaro-oligosaccharide products by high performance anion exchange chromatography systems. Carbohydr. Polym. 2015, 122, 351–358. [Google Scholar] [CrossRef]
- Han, J.P.; Huang, Y.Y.; Ye, J.; Xiao, M.T. Screening and identification of a bacterium capable of converting agar to neoagaro oligosaccharides. Acta Microbiol. Sin. 2015, 55, 1126–1132. [Google Scholar]
- Fan, J.M.; Xie, C.Q.; Jia, J.; Yin, X.U.; Zhang, C.X. Opitimization of extraction processing on polysaccharides from Pleurotus eryngii by cellulase enzymolysis. Food. Sci. Tech. 2013, 38, 192–196. [Google Scholar] [CrossRef]
- Liu, J.B.; Zhao, S.N.; Lin, S.Y.; Zhang, Y.; Ren, M. Process optimization on extracting collagen with enzymolysis method from black fungus. Trans. Chin. Soc. Agric. Eng. 2012, 28, 282–286. [Google Scholar] [CrossRef]
- Hong, S.J.; Lee, J.H.; Kim, E.J.; Yang, H.J.; Park, J.S.; Hong, S.K. Toxicological evaluation of neoagarooligosaccharides prepared by enzymatic hydrolysis of agar. Regul. Toxicol. Pharm. 2017, 90, 9–21. [Google Scholar] [CrossRef]
- Mei, J.; Shao, J.; Wang, Q.; Wang, H.; Yi, Y.; Ying, G. Separation and quantification of neoagaro-oligosaccharides. J. Food. Sci. Tech. Mys. 2013, 50, 1217–1221. [Google Scholar] [CrossRef]


| Oligosaccharide | Equation | R2 |
|---|---|---|
| NA2 | y = 6.29305x − 2.21022 | 0.992 |
| NA4 | y = 6.05888x − 3.18131 | 0.996 |
| NA6 | y = 5.54881x − 3.71928 | 0.997 |
| NA8 | y = 4.85490x − 4.42057 | 0.996 |
| NA10 | y = 4.48493x − 3.88547 | 0.994 |
| NA12 | y = 4.25971x − 3.75222 | 0.992 |
| A | B | C | D | E | |
|---|---|---|---|---|---|
| NA2 | 5.0 | 6.0 | 9.3 | 13.8 | 21.6 |
| NA4 | 38.9 | 52.7 | 59.5 | 68.4 | 71.1 |
| NA6 | 18.1 | 15.2 | 13.7 | 9.6 | nd |
| NA8 | 16.8 | 13.7 | 11.7 | nd | nd |
| NA10 | 13.8 | 11.0 | nd | nd | nd |
| NA12 | 2.5 | nd | nd | nd | nd |
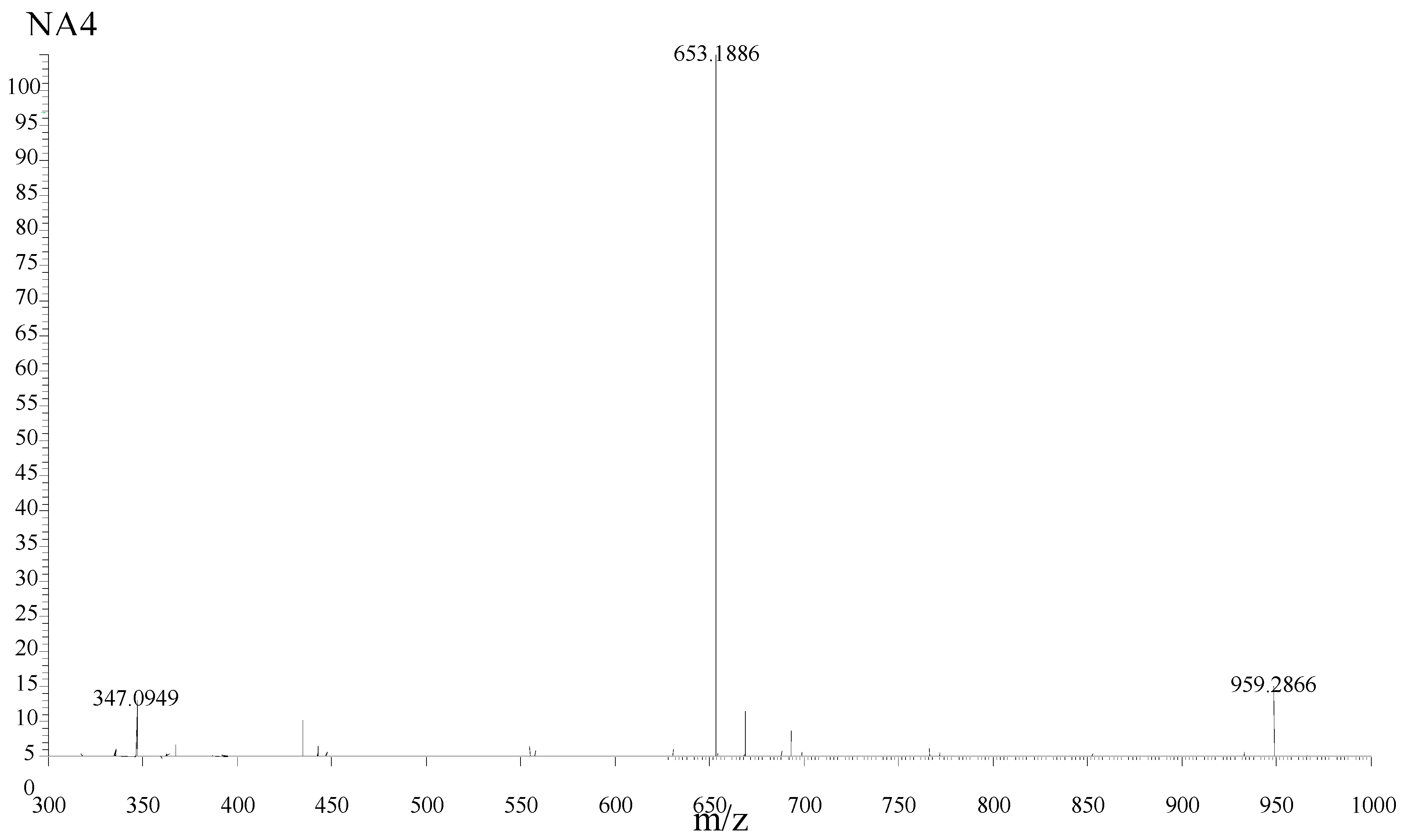
| Purified Products | MS Signals (m/z) [M + Na]+ | Calculated Molecular Weights |
|---|---|---|
| NA2 | 347.0949 | 347.0954 |
| NA4 | 653.1886 | 653.1905 |
| NA6 | 959.2866 | 959.2856 |
| NA8 | 1265.3752 | 1265.3807 |
| NA10 | 1571.4752 | 1571.4757 |
| NA12 | 1877.5751 | 1877.5708 |
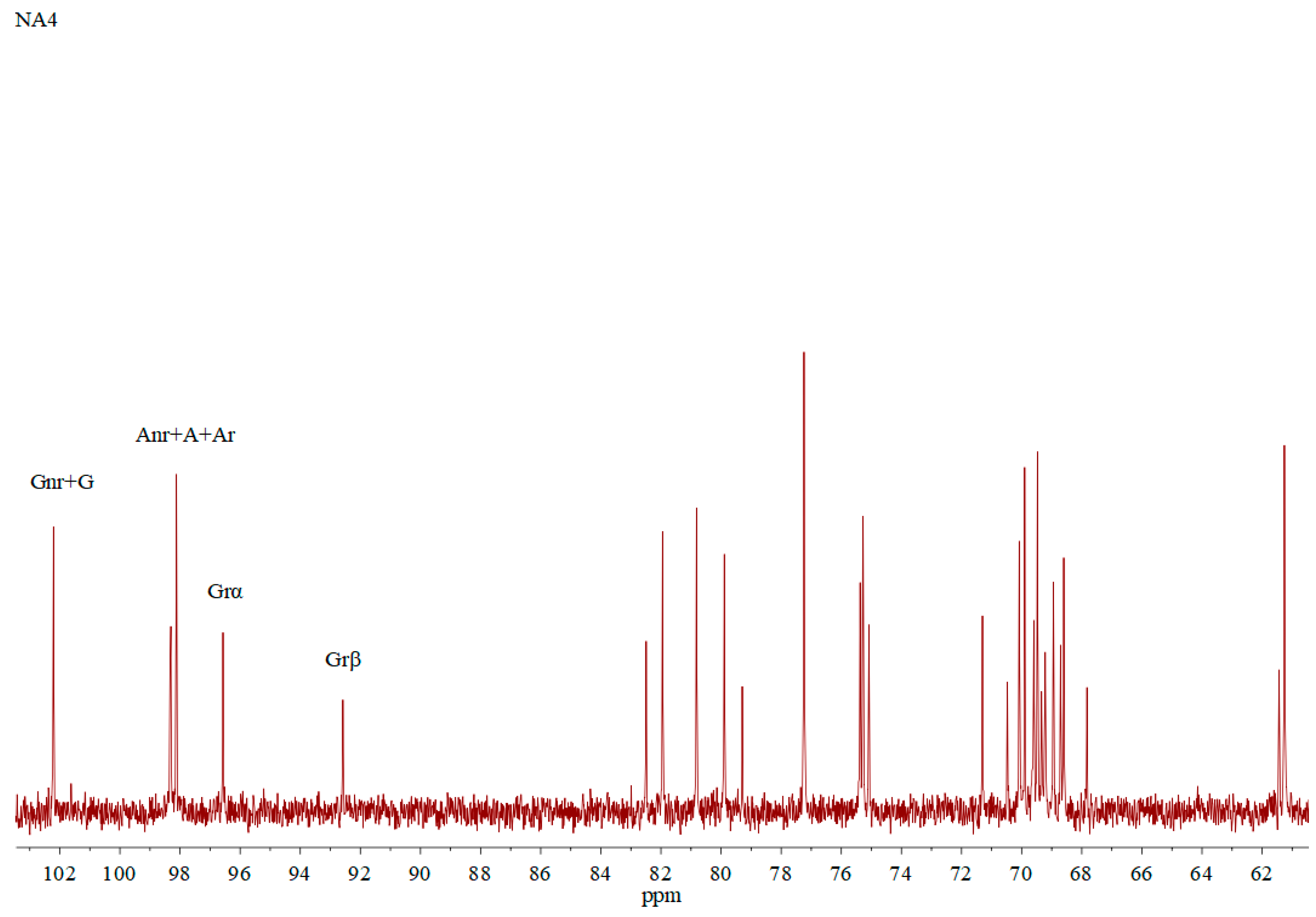
| Unit | C1 | C2 | C3 | C4 | C5 | C6 |
|---|---|---|---|---|---|---|
| Gnr | 102.2 | 69.9 | 82.0 | 67.8 | 75.1 | 61.3 |
| Grβ | 96.6 | 69.9 | 82.5 | 68.6 | 75.1 | 61.3 |
| Grα | 92.6 | 67.8 | 79.9 | 69.2 | 69.9 | 61.4 |
| Anr | 98.3 | 69.4 | 80.8 | 69.6 | 77.3 | 68.7 |
| Arα | 98.1 | 69.5 | 79.9 | 77.3 | 75.3 | 69.0 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, F.; Ye, J.; Huang, Y.; Yang, Y.; Xiao, M. Simple Preparation of Diverse Neoagaro-Oligosaccharides. Processes 2019, 7, 267. https://doi.org/10.3390/pr7050267
Lin F, Ye J, Huang Y, Yang Y, Xiao M. Simple Preparation of Diverse Neoagaro-Oligosaccharides. Processes. 2019; 7(5):267. https://doi.org/10.3390/pr7050267
Chicago/Turabian StyleLin, Fudi, Jing Ye, Yayan Huang, Yucheng Yang, and Meitian Xiao. 2019. "Simple Preparation of Diverse Neoagaro-Oligosaccharides" Processes 7, no. 5: 267. https://doi.org/10.3390/pr7050267
APA StyleLin, F., Ye, J., Huang, Y., Yang, Y., & Xiao, M. (2019). Simple Preparation of Diverse Neoagaro-Oligosaccharides. Processes, 7(5), 267. https://doi.org/10.3390/pr7050267