Prediction of N-linked Glycoform Profiles of Monoclonal Antibody with Extracellular Metabolites and Two-Step Intracellular Models
Abstract
1. Introduction
2. Materials and Methods
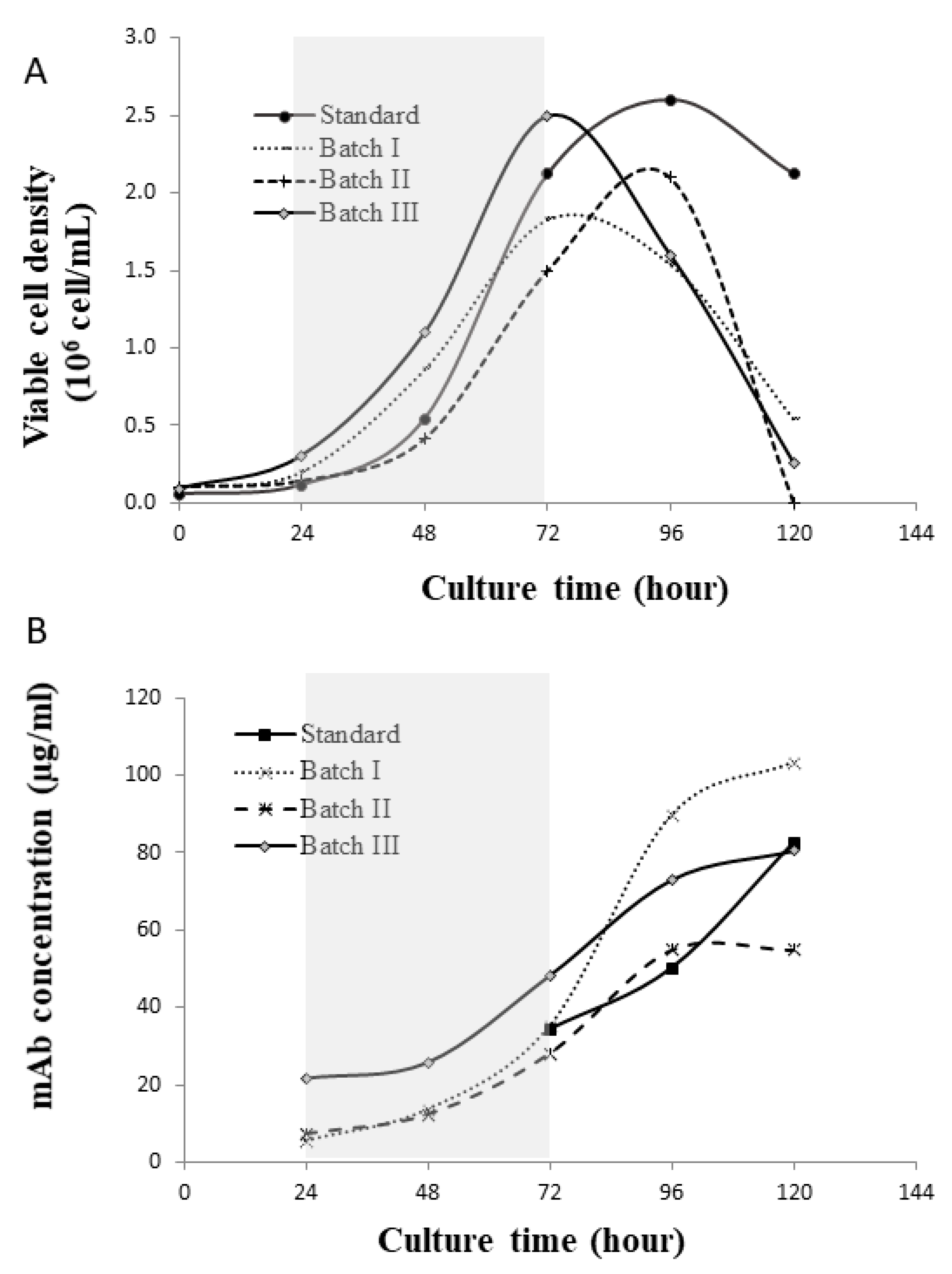
2.1. Bioreactor Process and Analytics
2.2. Rationale of Data Selection from the Previous Study
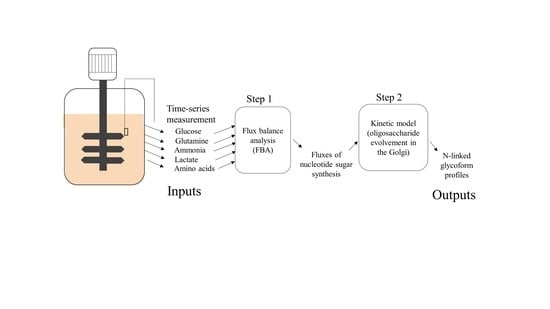
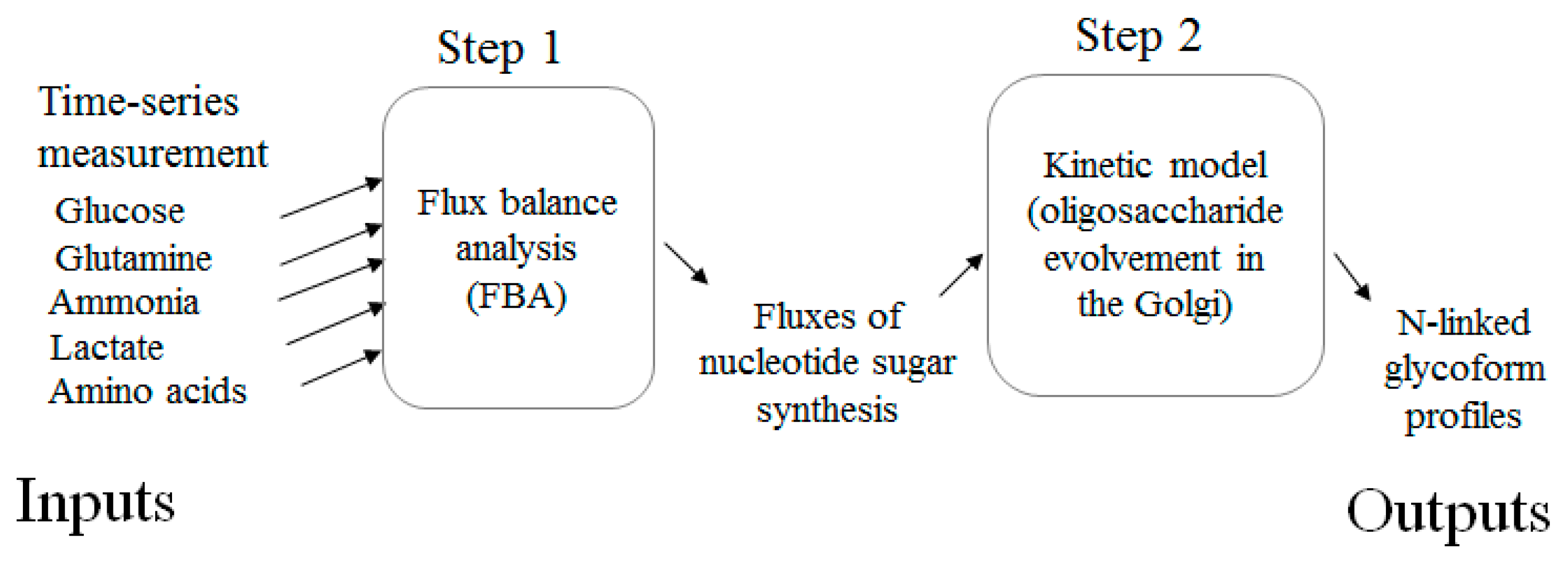
3. Modeling
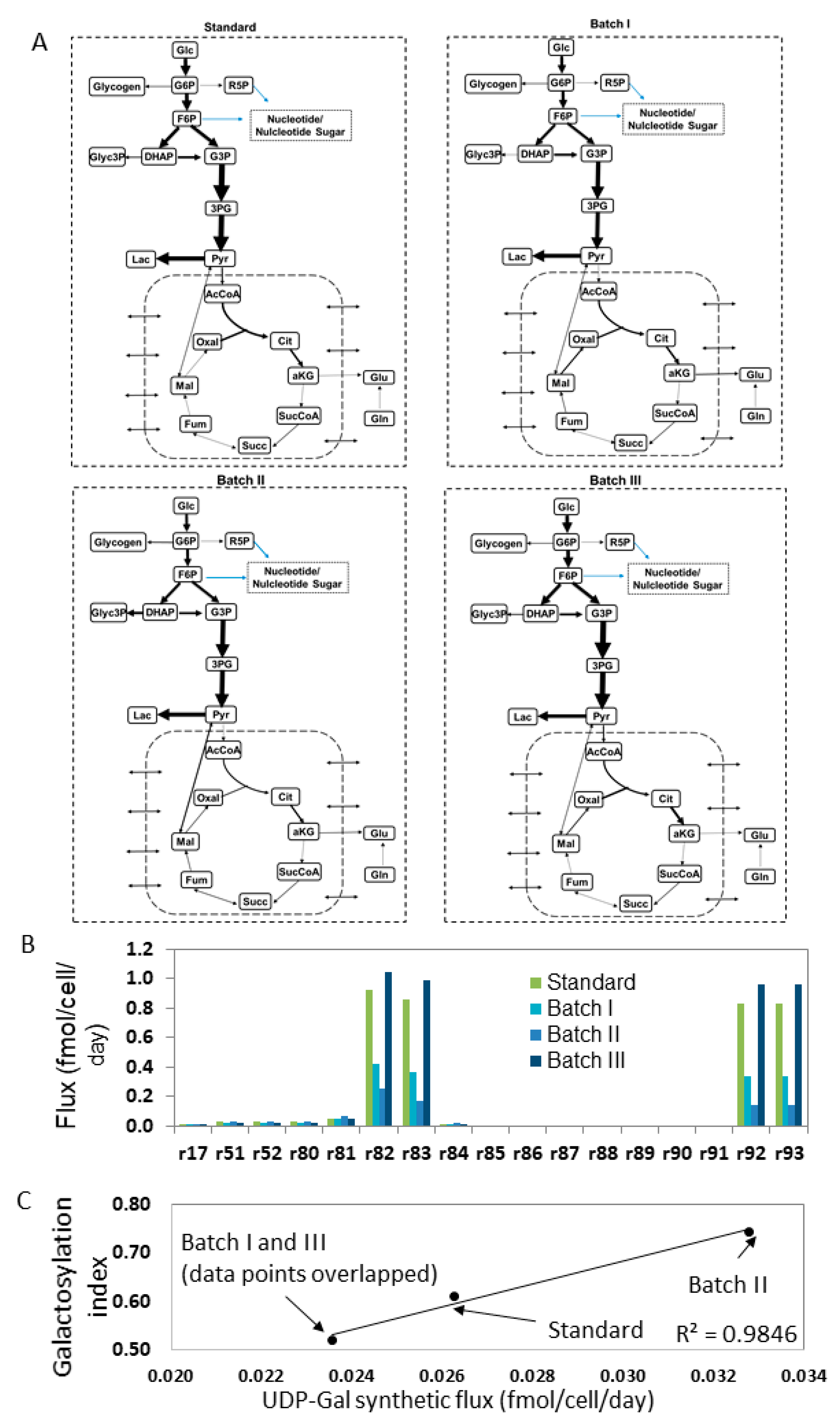
3.1. Estimating Fluxes of Nucleotide Sugar Syntheses from FBA
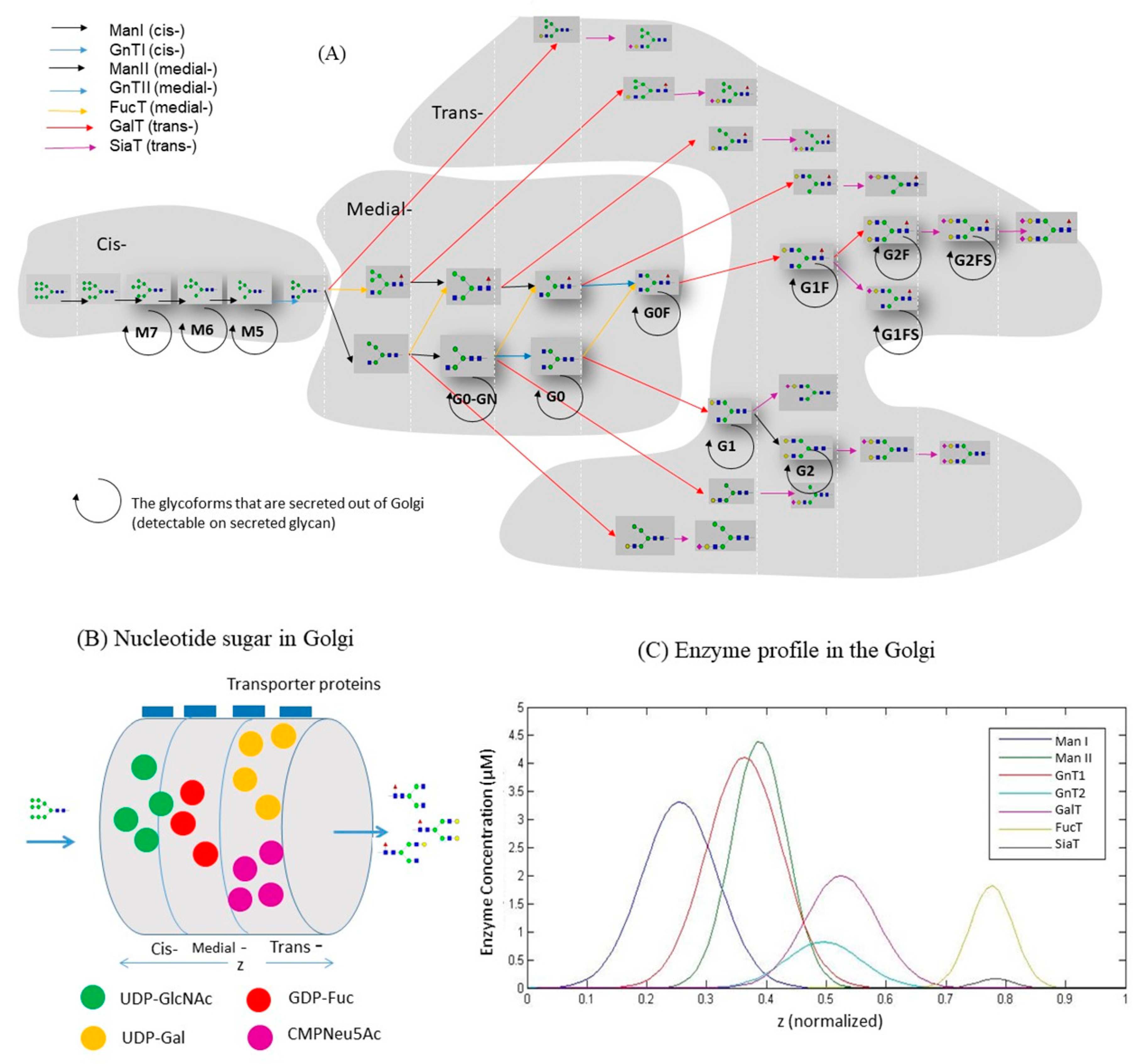
3.2. Construction of Glycosylation Kinetic Model
4. Results
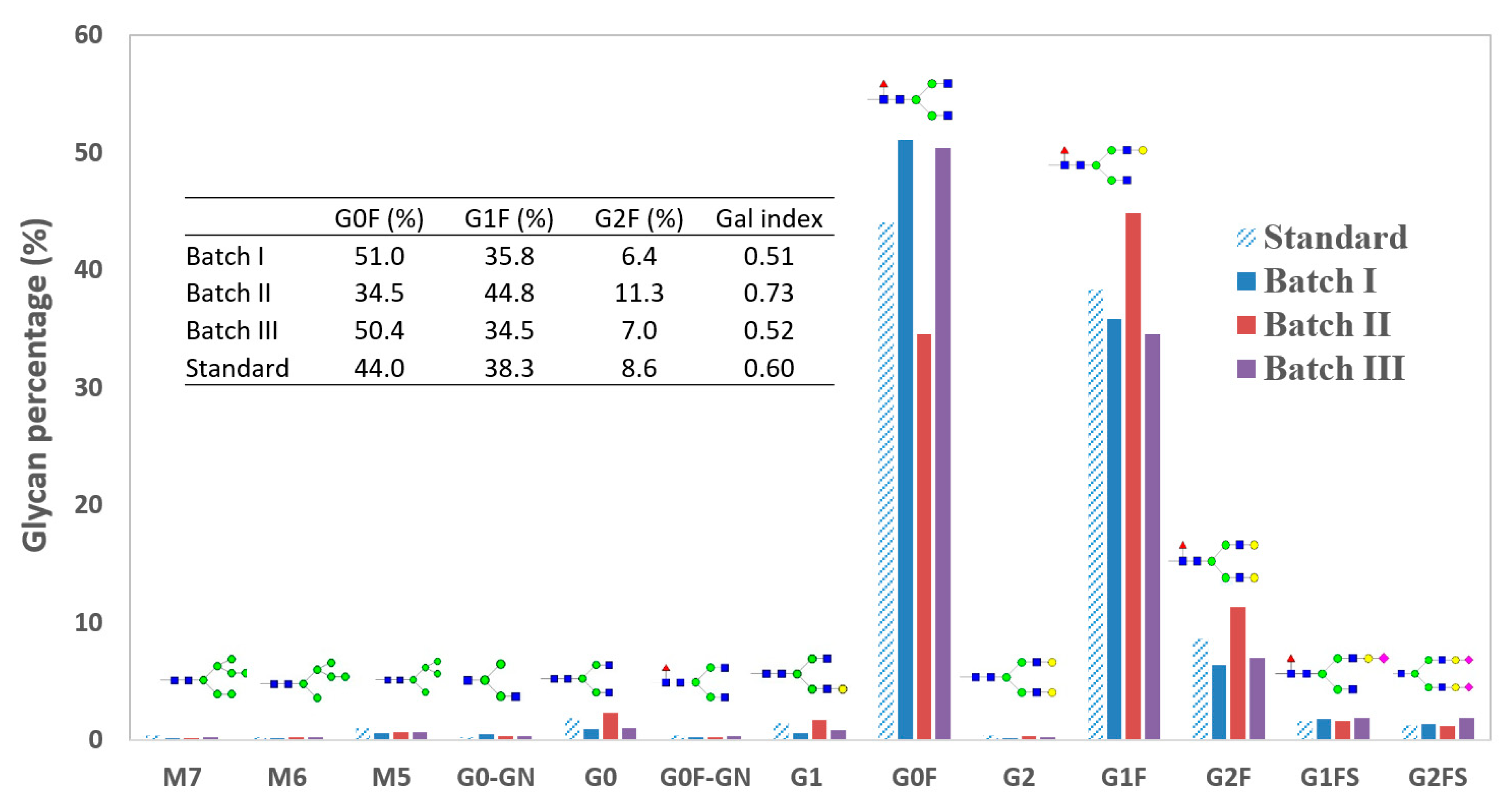
4.1. Glycan Variation across Batches
4.2. Estimating Nucleotide Sugar Fluxes by FBA
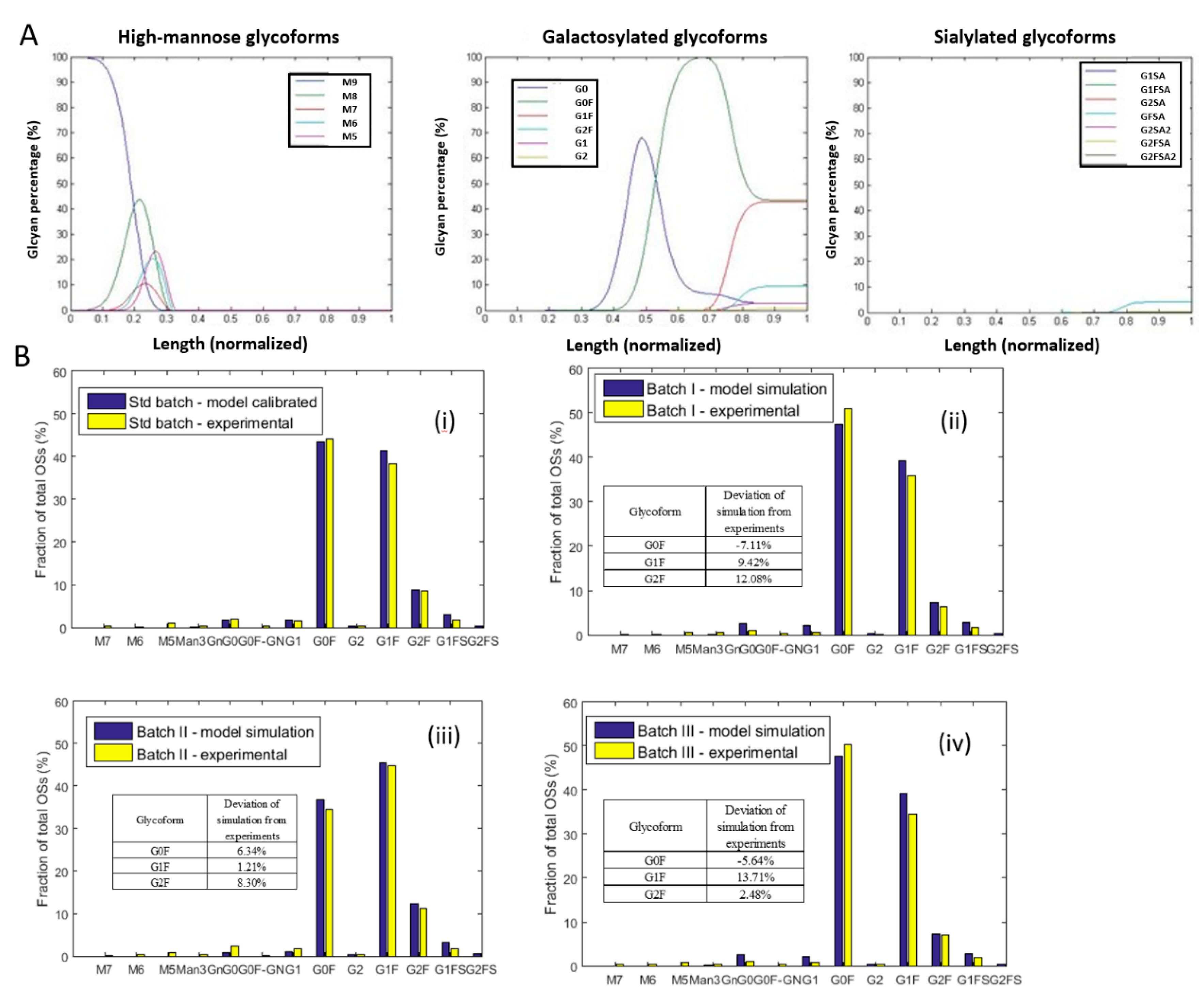
4.3. Model Calibration
4.4. Sensitivity Analysis of the Kinetic Model
4.5. Prediction of Glycoforms in Altered Culture Conditions
5. Discussion
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Aggarwal, S. What’s fueling the biotech engine—2012 to 2013. Nat. Biotechnol. 2014, 32, 32–39. [Google Scholar] [CrossRef]
- Siemiatkoski, J.; Ma, S.; Park, J.; Brorson, K.; Swann, P.; McLeod, L. Glycosylation of therapeutic proteins—Current understanding of structure-function relationships. BioProcess Int. 2011, 9, 48–53. [Google Scholar]
- Sha, S.; Agarabi, C.; Brorson, K.; Lee, D.Y.; Yoon, S. N-Glycosylation Design and Control of Therapeutic Monoclonal Antibodies. Trends Biotechnol. 2016, 34, 835–846. [Google Scholar] [CrossRef] [PubMed]
- Schiestl, M.; Stangler, T.; Torella, C.; Cepeljnik, T.; Toll, H.; Grau, R. Acceptable changes in quality attributes of glycosylated biopharmaceuticals. Nat. Biotechnol. 2011, 29, 310–312. [Google Scholar] [CrossRef]
- Raju, T.S.; Jordan, R.E. Galactosylation variations in marketed therapeutic antibodies. MAbs 2012, 4, 385–391. [Google Scholar] [CrossRef]
- Zupke, C.; Brady, L.J.; Slade, P.G.; Clark, P.; Caspary, R.G.; Livingston, B.; Taylor, L.; Bigham, K.; Morris, A.E.; Bailey, R.W. Real-time product attribute control to manufacture antibodies with defined N-linked glycan levels. Biotechnol. Prog. 2015, 31, 1433–1441. [Google Scholar] [CrossRef] [PubMed]
- Pais, D.A.; Carrondo, M.J.; Alves, P.M.; Teixeira, A.P. Towards real-time monitoring of therapeutic protein quality in mammalian cell processes. Curr. Opin. Biotechnol. 2014, 30, 161–167. [Google Scholar] [CrossRef] [PubMed]
- Read, E.K.; Bradley, S.A.; Smitka, T.A.; Agarabi, C.D.; Lute, S.C.; Brorson, K.A. Fermentanomics Informed Amino Acid Supplementation of an Antibody Producing Mammalian Cell Culture. Biotechnol. Prog. 2012, 29, 745–753. [Google Scholar] [CrossRef]
- Zhang, A.; Tsang, V.L.; Moore, B.; Shen, V.; Huang, Y.M.; Kshirsagar, R.; Ryll, T. Advanced process monitoring and feedback control to enhance cell culture process production and robustness. Biotechnol. Bioeng. 2015, 112, 2495–2504. [Google Scholar] [CrossRef] [PubMed]
- Larson, T.M.; Gawlitzek, M.; Evans, H.; Albers, U.; Cacis, J. Chemometric evaluation of on-line high-pressure liquid chromatography in mammalian cell cultures: Analysis of amino acids and glucose. Biotechnol. Bioeng. 2002, 77, 553–563. [Google Scholar] [CrossRef] [PubMed]
- Bhatia, H.; Mehdizadeh, H.; Drapeau, D.; Yoon, S. In-line monitoring of amino acids in mammalian cell cultures using raman spectroscopy and multivariate chemometrics models. Eng. Life Sci. 2018, 18, 55–61. [Google Scholar] [CrossRef]
- Cha, H.-M.; Lim, J.-H.; Lee, K.-S.; Kim, D.-I. Nucleotide sugar precursor feeding strategy to enhance sialylation of albumin-erythropoietin in CHO cell cultures. Process Biochem. 2018, 66, 197–204. [Google Scholar] [CrossRef]
- Villiger, T.K.; Steinhoff, R.F.; Ivarsson, M.; Solacroup, T.; Stettler, M.; Broly, H.; Krismer, J.; Pabst, M.; Zenobi, R.; Morbidelli, M.; et al. High-throughput profiling of nucleotides and nucleotide sugars to evaluate their impact on antibody N-glycosylation. J. Biotechnol. 2016, 229, 3–12. [Google Scholar] [CrossRef]
- Blondeel, E.J.; Braasch, K.; McGill, T.; Chang, D.; Engel, C.; Spearman, M.; Butler, M.; Aucoin, M.G. Tuning a MAb glycan profile in cell culture: Supplementing N-acetylglucosamine to favour G0 glycans without compromising productivity and cell growth. J. Biotechnol. 2015, 214, 105–112. [Google Scholar] [CrossRef]
- Kochanowski, N.; Blanchard, F.; Cacan, R.; Chirat, F.; Guedon, E.; Marc, A.; Goergen, J.L. Influence of intracellular nucleotide and nucleotide sugar contents on recombinant interferon-gamma glycosylation during batch and fed-batch cultures of CHO cells. Biotechnol. Bioeng. 2008, 100, 721–733. [Google Scholar] [CrossRef]
- Chee Furng Wong, D.; Tin Kam Wong, K.; Tang Goh, L.; Kiat Heng, C.; Gek Sim Yap, M. Impact of dynamic online fed-batch strategies on metabolism, productivity and N-glycosylation quality in CHO cell cultures. Biotechnol. Bioeng. 2005, 89, 164–177. [Google Scholar] [CrossRef] [PubMed]
- Lewis, A.M.; Croughan, W.D.; Aranibar, N.; Lee, A.G.; Warrack, B.; Abu-Absi, N.R.; Patel, R.; Drew, B.; Borys, M.C.; Reily, M.D.; et al. Understanding and Controlling Sialylation in a CHO Fc-Fusion Process. PLoS ONE 2016, 11, e0157111. [Google Scholar] [CrossRef]
- Sou, S.N.; Sellick, C.; lEE, K.; Mason, A.; Kyriakopoulos, S.; Polizzi, K.M.; Kontoravdi, C. How does mild hypothermia affect monoclonal antibody glycosylation. Biotechnol. Bioeng. 2015, 112, 1165–1176. [Google Scholar] [CrossRef]
- St Amand, M.M.; Radhakrishnan, D.; Robinson, A.S.; Ogunnaike, B.A. Identification of manipulated variables for a glycosylation control strategy. Biotechnol. Bioeng. 2014, 111, 1957–1970. [Google Scholar] [CrossRef] [PubMed]
- Sha, S.; Yoon, S. An investigation of nucleotide sugar dynamics under the galactose supplementation in CHO cell culture. Process Biochem. 2019. [Google Scholar] [CrossRef]
- Agarabi, C.D.; Schiel, J.E.; Lute, S.C.; Chavez, B.K.; Boyne, M.T.; Brorson, K.A.; Khan, M.; Read, E.K. Bioreactor process parameter screening utilizing a Plackett–Burman design for a model monoclonal antibody. J. Pharm. Sci. 2015, 104, 1919–1928. [Google Scholar] [CrossRef] [PubMed]
- Ohadi, K.; Aghamohseni, H.; Gädke, J.; Moo-Young, M.; Legge, R.L.; Scharer, J.; Budman, H.M. Novel Dynamic Model to Predict the Glycosylation Pattern of Monoclonal Antibodies from Extracellular Cell Culture Conditions. In Proceedings of the 12th IFAC Symposium on Computer Applications in Biotechnology 2013, Mumbai, India, 16–20 December 2013. [Google Scholar]
- Leighty, R.W.; Antoniewicz, M.R. Dynamic metabolic flux analysis (DMFA): A framework for determining fluxes at metabolic non-steady state. Metab. Eng. 2011, 13, 745–755. [Google Scholar] [CrossRef] [PubMed]
- Selvarasu, S.; Ho, Y.S.; Chong, W.P.; Wong, N.S.; Yusufi, F.N.; Lee, Y.Y.; Yap, M.G.; Lee, D.Y. Combined in silico modeling and metabolomics analysis to characterize fed-batch CHO cell culture. Biotechnol. Bioeng. 2012, 109, 1415–1429. [Google Scholar] [CrossRef]
- Krambeck, F.J.; Betenbaugh, M.J. A mathematical model of N-linked glycosylation. Biotechnol. Bioeng. 2005, 92, 711–728. [Google Scholar] [CrossRef] [PubMed]
- Hossler, P.; Mulukutla, B.C.; Hu, W.-S. Systems analysis of N-Glycan processing in mammalian cells. PLoS ONE 2007, 2, e713. [Google Scholar] [CrossRef]
- Kontoravdi, C.; Asprey, S.P.; Pistikopoulos, E.N.; Mantalaris, A. Development of a dynamic model of monoclonal antibody production and glycosylation for product quality monitoring. Comput. Chem. Eng. 2007, 31, 392–400. [Google Scholar] [CrossRef]
- del Val, I.J.; Nagy, J.M.; Kontoravdi, C. A dynamic mathematical model for monoclonal antibody N-linked glycosylation and nucleotide sugar donor transport within a maturing Golgi apparatus. Biotechnol. Prog. 2011, 27, 1730–1743. [Google Scholar] [CrossRef]
- Jedrzejewski, P.M.; del Val, I.J.; Constantinou, A.; Dell, A.; Haslam, S.M.; Polizzi, K.M.; Kontoravdi, C. Towards controlling the glycoform: A model framework linking extracellular metabolites to antibody glycosylation. Int. J. Mol. Sci. 2014, 15, 4492–4522. [Google Scholar] [CrossRef]
- Kyriakopoulos, S.; Ang, K.S.; Lakshmanan, M.; Huang, Z.; Yoon, S.; Gunawan, R.; Lee, D.Y. Kinetic Modeling of Mammalian Cell Culture Bioprocessing: The Quest to Advance Biomanufacturing. Biotechnol. J. 2017. [Google Scholar] [CrossRef]
- Sha, S.; Huang, Z.; Wang, Z.; Yoon, S. Mechanistic modeling and applications for CHO cell culture development and production. Curr. Opin. Chem. Eng. 2018, 22, 54–61. [Google Scholar] [CrossRef]
- Shi, H.H.; Goudar, C.T. Recent Advances in the Understanding of Biological Implications and Modulation Methodologies of Monoclonal Antibody N-Linked High Mannose Glycans. Biotechnol. Bioeng. 2014, 111, 1907–1919. [Google Scholar] [CrossRef]
- Gramer, M.J.; Eckblad, J.J.; Donahue, R.; Brown, J.; Shultz, C.; Vickerman, K.; Priem, P.; van den Bremer, E.T.; Gerritsen, J.; van Berkel, P.H. Modulation of antibody galactosylation through feeding of uridine, manganese chloride and galactose. Biotechnol. Bioeng. 2011, 108, 1591–1602. [Google Scholar] [CrossRef] [PubMed]
- Wong, N.S.; Wati, L.; Nissom, P.M.; Feng, H.T.; Lee, M.M.; Yap, M.G. An investigation of intracellular glycosylation activities in CHO cells: Effects of nucleotide sugar precursor feeding. Biotechnol. Bioeng. 2010, 107, 321–336. [Google Scholar] [CrossRef]
- Chen, P.; Harcum, S.W. Differntial display identifies genes in CHO cells sensitive to elevated ammonium. Appl. Biochem. Biotechnol. 2007, 141, 349–359. [Google Scholar] [CrossRef]
- Huang, Z.; Lee, D.Y.; Yoon, S. Quantitative intracellular flux modeling and applications in biotherapeutic development and production using CHO cell cultures. Biotechnol. Bioeng. 2017, 114, 2717–2728. [Google Scholar] [CrossRef]

| 0 h | 48–72 h | 72 h | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Batch ID | % DO Set Point | Sparge Rate (SLPH) | Agitation Rate (RPM) | Temperature (°C) | Inoculation Density (Cells/mL) | Nonessential Amino Acids | Fatty Acids | Hydrocortisone | Feeding Strategy | Temperature Shift (at 72 h) |
| Standard batch | 20 | 0.3 | 90 | 37 | 50,000 | Add | Add | Add | Bolus | Shift |
| Batch I | 40 | 0.3 | 170 | 37 | 100,000 | Add | Bolus | No Shift | ||
| Batch II | 40 | 0.3 | 90 | 35.5 | 100,000 | Add | Add | Drip | No Shift | |
| Batch III | 30 | 0.5 | 135 | 37 | 75,000 | Bolus | No Shift | |||
| Standard | Batch I | Batch II | Batch III | |
|---|---|---|---|---|
| UDP-Gal | 0.0262 | 0.0236 | 0.0328 | 0.024 |
| GDP-Man | 0.0394 | 0.0353 | 0.0492 | 0.035 |
| GDP-Fuc | 0.0131 | 0.0118 | 0.0164 | 0.012 |
| UDP-GlcNAc | 0.0656 | 0.0589 | 0.0820 | 0.058 |
| CMP-SA | 0.0262 | 0.0236 | 0.0328 | 0.023 |
| Standard a | Batch I b | Batch II b | Batch III b | |
|---|---|---|---|---|
| UDP-Gal | 9.4 | 8 | 12.6 | 8 |
| UDP-GlcNAc | 1600 | 1436 | 1998 | 1403 |
| GDP-FUC | 576 | 517 | 719 | 517 |
| CMP-SA | 950 | 852 | 1187 | 833 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sha, S.; Huang, Z.; Agarabi, C.D.; Lute, S.C.; Brorson, K.A.; Yoon, S. Prediction of N-linked Glycoform Profiles of Monoclonal Antibody with Extracellular Metabolites and Two-Step Intracellular Models. Processes 2019, 7, 227. https://doi.org/10.3390/pr7040227
Sha S, Huang Z, Agarabi CD, Lute SC, Brorson KA, Yoon S. Prediction of N-linked Glycoform Profiles of Monoclonal Antibody with Extracellular Metabolites and Two-Step Intracellular Models. Processes. 2019; 7(4):227. https://doi.org/10.3390/pr7040227
Chicago/Turabian StyleSha, Sha, Zhuangrong Huang, Cyrus D. Agarabi, Scott C. Lute, Kurt A. Brorson, and Seongkyu Yoon. 2019. "Prediction of N-linked Glycoform Profiles of Monoclonal Antibody with Extracellular Metabolites and Two-Step Intracellular Models" Processes 7, no. 4: 227. https://doi.org/10.3390/pr7040227
APA StyleSha, S., Huang, Z., Agarabi, C. D., Lute, S. C., Brorson, K. A., & Yoon, S. (2019). Prediction of N-linked Glycoform Profiles of Monoclonal Antibody with Extracellular Metabolites and Two-Step Intracellular Models. Processes, 7(4), 227. https://doi.org/10.3390/pr7040227