Abstract
Recent in vitro experiments have demonstrated the ability of the pathogen Clostridium difficile and commensal gut bacteria to form biofilms on surfaces, and biofilm development in vivo is likely. Various studies have reported that 3%–15% of healthy adults are asymptomatically colonized with C. difficile, with commensal species providing resistance against C. difficile pathogenic colonization. C. difficile infection (CDI) is observed at a higher rate in immunocompromised patients previously treated with broad spectrum antibiotics that disrupt the commensal microbiota and reduce competition for available nutrients, resulting in imbalance among commensal species and dysbiosis conducive to C. difficile propagation. To investigate the metabolic interactions of C. difficile with commensal species from the three dominant phyla in the human gut, we developed a multispecies biofilm model by combining genome-scale metabolic reconstructions of C. difficile, Bacteroides thetaiotaomicron from the phylum Bacteroidetes, Faecalibacterium prausnitzii from the phylum Firmicutes, and Escherichia coli from the phylum Proteobacteria. The biofilm model was used to identify gut nutrient conditions that resulted in C. difficile-associated dysbiosis characterized by large increases in C. difficile and E. coli abundances and large decreases in F. prausnitzii abundance. We tuned the model to produce species abundances and short-chain fatty acid levels consistent with available data for healthy individuals. The model predicted that experimentally-observed host-microbiota perturbations resulting in decreased carbohydrate/increased amino acid levels and/or increased primary bile acid levels would induce large increases in C. difficile abundance and decreases in F. prausnitzii abundance. By adding the experimentally-observed perturbation of increased host nitrate secretion, the model also was able to predict increased E. coli abundance associated with C. difficile dysbiosis. In addition to rationalizing known connections between nutrient levels and disease progression, the model generated hypotheses for future testing and has the capability to support the development of new treatment strategies for C. difficile gut infections.
1. Introduction
The gut microbiota comprise a complex ecological system that maintains a critical symbiotic relationship with the human host [1,2]. The microbiota provide essential nutrients such as short-chain fatty acids (SCFAs; acetate, butyrate, and propionate), support colonization resistance to pathogens, participate in the degradation of toxic compounds, and regulate the immune responses [3,4,5,6,7]. Bacteroidetes and Firmicutes are the two dominant phyla in the healthy gut, comprising approximately 90% of the community. Other important but less abundant phyla are Proteobacteria, Actinobacteria, Euryarchaeota and Verrucomicrobia, as well as Eukaryota such as fungi [8,9]. The gut microbiota composition can be altered by numerous factors including diet, antibiotic treatment, stress, and lifestyle [10,11]. Dietary components including carbohydrates, protein, fat, and host secretions such as primary bile acids and nitrate play a particularly important role in shaping microbiota abundances [12,13,14,15,16,17]. Unhealthy alterations of the gut microbiota are termed as dysbiosis and represent imbalances in species abundances associated with diseases such as inflammatory bowel diseases, Crohn’s disease, obesity, and diabetes [18,19,20].
The anaerobic bacterium Clostridium difficile is an opportunistic human pathogen responsible for infections in the colon of the human gastrointestinal tract [21]. Various studies have reported that 3%–15% of healthy adults are asymptomatically colonized with C. difficile [22,23,24,25,26,27,28]. Commensal species in healthy gut usually provide resistance against C. difficile pathogenic colonization. C. difficile infection (CDI) is most common in patients previously treated with broad spectrum antibiotics that disrupt the healthy gut microbiota and reduce competition for available nutrients [29], resulting in dysbiosis conducive to C. difficile propagation [30,31,32,33]. CDI symptoms can range from mild diarrhea to severe and life-threatening colitis [21,34]. C. difficile virulence is attributable to the secretion of the high molecular weight toxins A and B that promote epithelial tissue damage and rapid fluid loss. Some C. difficile strains have developed resistance to common antibiotics while also exhibiting more severe pathogenicity [35]. CDI has become particularly common in hospital settings due to the ability of C. difficile to form spores that adhere to surfaces and resist common disinfectant protocols. Studies estimate that almost 500,000 CDI cases occur within the U.S. annually [36], resulting in 29,000 deaths and over $4.8 billion in associated costs in acute care facilities alone [37].
Numerous experimental studies have demonstrated that C. difficile [38,39,40,41] can form biofilms in vitro. The other commensal bacteria [42,43] can form biofilms in vivo, which are well known to exhibit phenotypes distinct from planktonic cultures. For example, bacteria in biofilms can tolerate antimicrobial concentrations 10,000-times higher than the same bacteria grown planktonically, making the development of effective treatment strategies a major challenge [44,45]. This difficulty is partially attributable to the spatially-varying biofilm environment, which has profound effects on biofilm development and function [46,47,48]. Mechanistic understanding of the relationships between biofilm spatial variations, species–species interactions, and host–species interactions remains inadequate to systematically analyze and rationally treat CDI [49]. To address these challenges, we added C. difficile to our previous multispecies biofilm model [50,51] consisting of three representative species from the phyla Bacteroidetes (Bacteroides thetaiotaomicron), Firmicutes (Faecalibacterium prausnitzii), and Proteobacteria (Escherichia coli). Model simulations were performed to connect host-induced nutrient changes in the gut environment with observed alternations of species abundances and SCFA levels [52,53,54] to unravel the metabolic determinants of CDI.
2. Results
2.1. Discovery of Putative Byproduct Cross-Feeding Relationships
Our previous modeling study [50] without C. difficile generated three byproduct cross-feeding relationships that were predicted to be necessary and sufficient for the coexistence of the three species: B. thetaiotaomicron consumption of ethanol secreted by E. coli and F. prausnitzii consumption of acetate and succinate secreted by B. thetaiotaomicron and E. coli. Preliminary flux balance analysis (FBA) with the C. difficile reconstruction showed that acetate, butyrate, and propionate were the major byproducts, and succinate and formate could be uptaken as carbon sources in the presence of glucose. With this knowledge, the four-species biofilm model was analyzed to discover additional cross-feeding relationships that support C. difficile coexistence with the three commensal species. Each species was allowed to consume glucose, the eight amino acids, and any available byproduct (acetate, CO, ethanol, formate, lactate, and succinate), assuming no differences in uptake kinetics across species and byproducts (see Materials and Methods). Simulations with a biofilm thickness of 40 microns and bulk concentrations of 8 mmol/L glucose and 0.5 mmol/L each amino acid at the biofilm-stool interface corresponding to the healthy case (Table 1) were run for 300 h to ensure a steady-state solution consistent with obtaining a mature biofilm. A particular cross-feeding relationship was deemed significant if at least one uptake or secretion flux exceeded 1 mmol/gDW·h.

Table 1.
Nutrient concentrations used for healthy and three dysbiosis simulation cases in mmol/L.
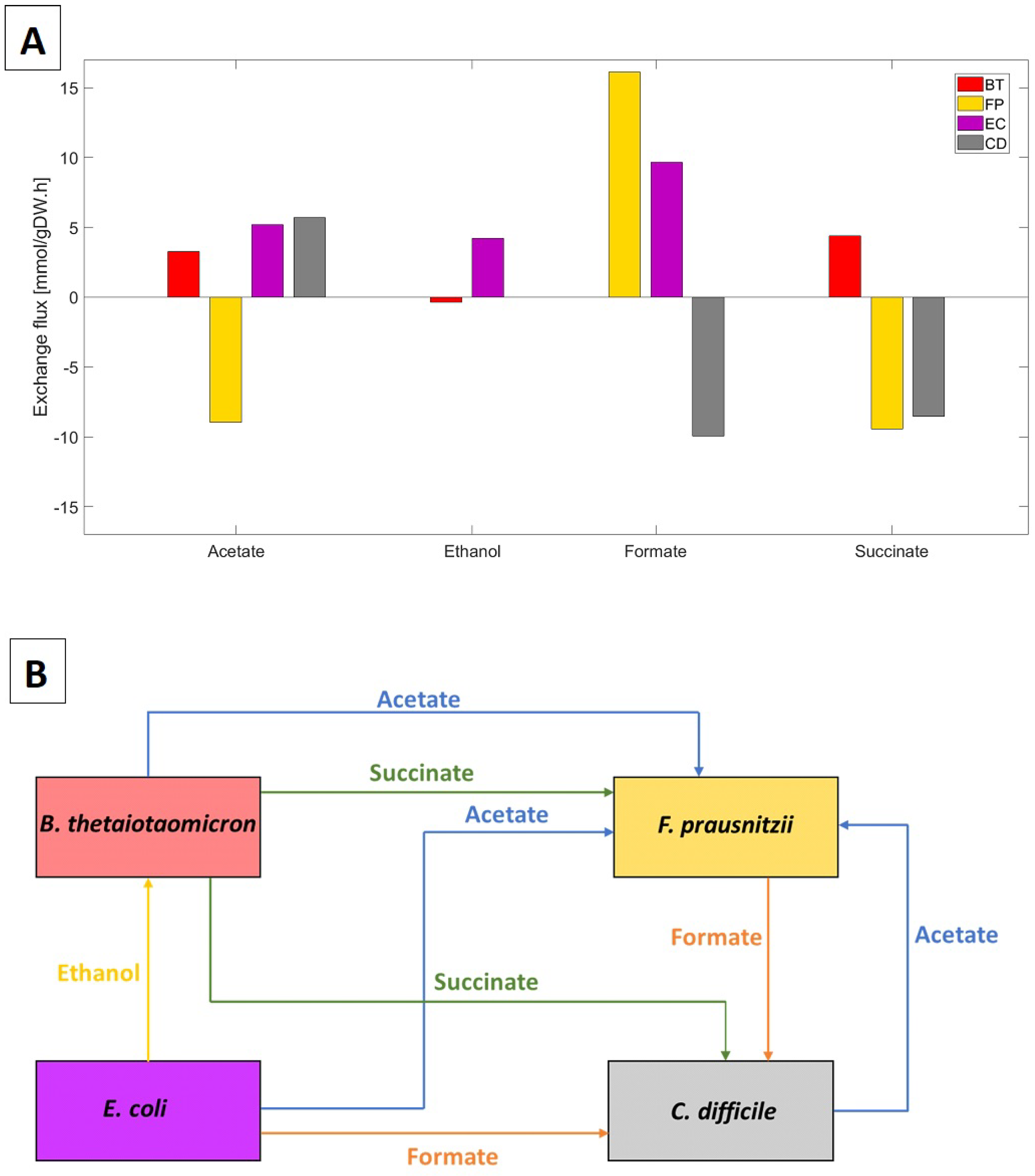
The biofilm model predicted significant cross-feeding of acetate, ethanol, formate, and succinate between the four species (Figure 1A). Lactate and CO cross-feeding were insignificant. Importantly for this study, C. difficile was predicted to: (1) consume formate secreted by F. prausnitzii and E. coli; (2) compete with F. prausnitzii for succinate secreted by B. thetaiotaomicron; and (3) synthesize acetate for consumption by F. prausnitzii (Figure 1B). Experimentally, C. difficile has been shown to uptake succinate and produce butyrate [55] and to produce acetate by consuming formate directly or indirectly by uptaking CO and H [56]. Consequently, we hypothesized that formate and succinate cross-feeding could play a role in C. difficile propagation in vivo.
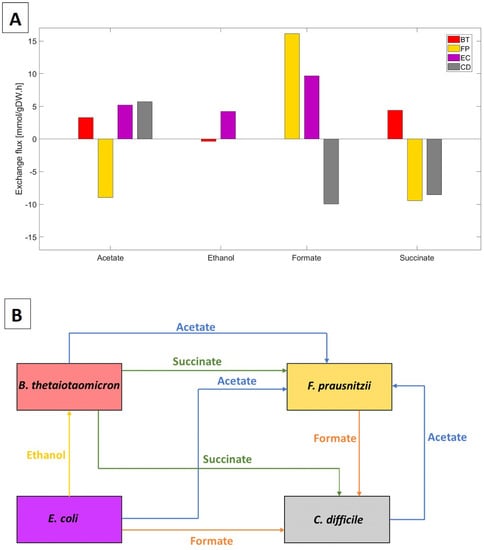
Figure 1.
Predicted cross-feeding of byproducts between the four species. (A) Species exchange rates specified in mmol/gDW/h. Secretion rates are positive, and uptake rates are negative. (B) Byproduct cross-feeding patterns identified from the species uptake and secretion fluxes in (A).
To test community stability and robustness in the absence of C. difficile, the same simulation was performed with the initial C. difficile biomass concentration set to zero. The resulting three-species community remained stable with B. thetaiotaomicron:F. prausnitzii:E. coli abundances of 66%:27%:7%, consistent with a healthy gut community (Supplementary Materials Figure S1). These predictions were aligned with our previous study [50].
2.2. Characterization of Healthy Gut Microbiota
With the putative cross-feeding relationships (Figure 1B) included, the multispecies biofilm model was simulated for a biofilm thickness of 40 microns and the healthy nutrient levels (Table 1). The model was tuned such that the mature biofilm obtained after 300 h of simulation produced B. thetaiotaomicron:F. prausnitzii:E. coli:C. difficile abundances of 71%:21%:7%:1% when averaged across the biofilm (see Materials and Methods). These abundances were consistent with data from in vivo studies [57,58].
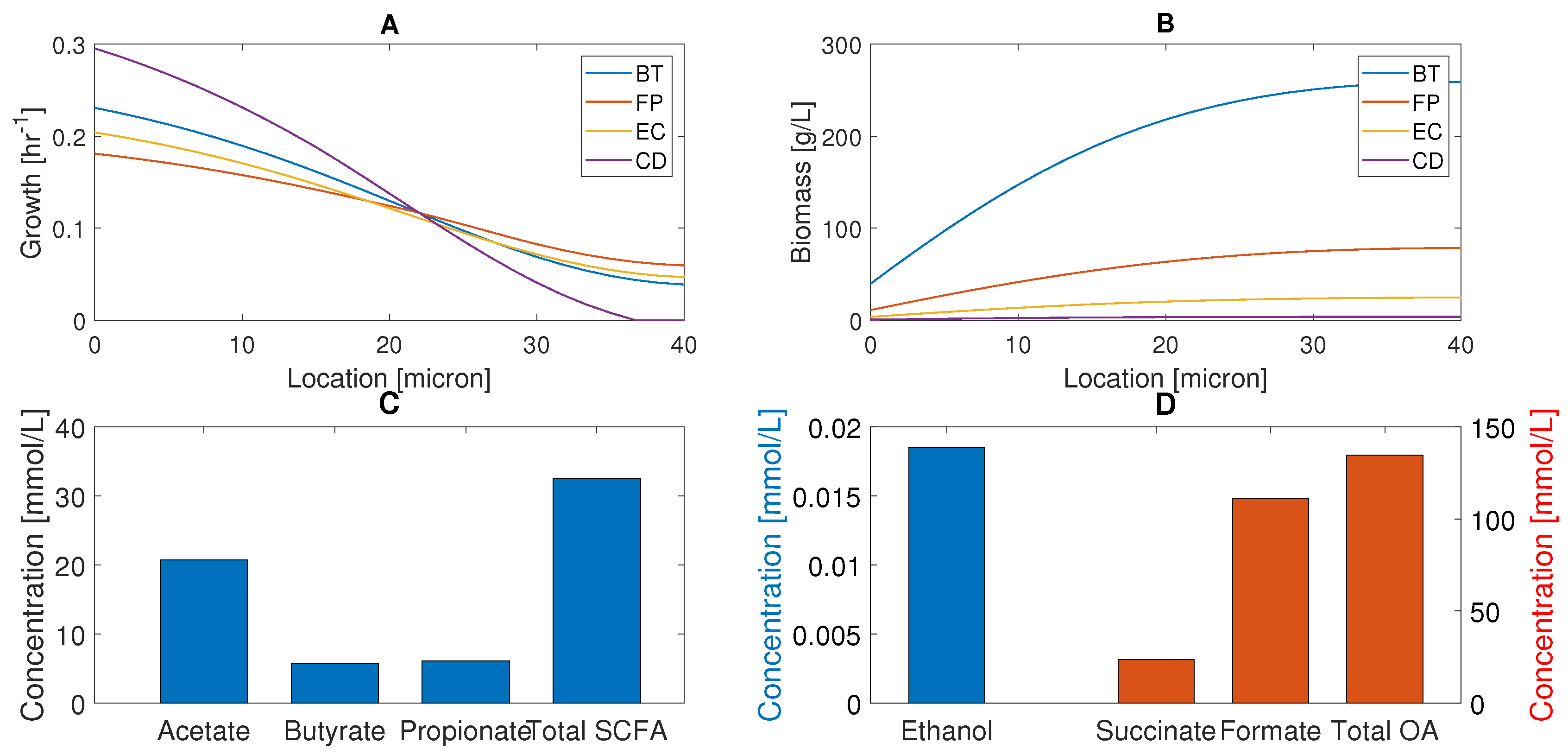
We analyzed species biomass concentrations (Figure 2A) and local growth rates (Figure 2B) with respect to location in the biofilm with nutrients supplied at the biofilm–stool interface (). C. difficile was predicted to have the highest growth rates in the nutrient-rich bottom half of the biofilm, but the lowest growth rates in the nutrient-lean top half. The local growth rates of the three commensal bacteria were comparable across the biofilm, with B. thetaiotaomicron having the highest growth rates in the bottom half and F. prausnitzii having a slight advantage in the top half. Due to its growth advantage in the nutrient-rich bottom half and slow cellular diffusion, B. thetaiotaomicron produced much higher biomass concentrations across the entire biofilm. F. prausnitzii and E. coli established lower biomass concentrations, while C. difficile was present at small concentrations due to its very small growth rate in the nutrient-lean top half. The spatial distributions of supplied nutrients, species biomass, and secreted byproducts were similar to those reported in our previous studies [50,51] and are omitted here. This simulation suggests that the commensal bacteria can sublimate C. difficile propagation through nutrient competition and may help explain how healthy individuals can be asymptomatically colonized.
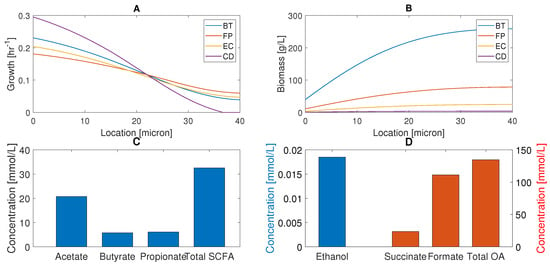
Figure 2.
Predicted multispecies biofilm behavior in the absence of host-microbiota perturbations. (A) Species biomass concentrations across the thickness of the biofilm with nutrients supplied and biomass removed at microns. (B) Local species growth rates across the thickness of the biofilm. (C) Acetate, butyrate, propionate, and total SCFA concentrations averaged across the biofilm. (D) Ethanol, succinate, formate, and total OA levels averaged across the biofilm.
The biofilm model also was tuned for healthy nutrient levels to produce acetate:propionate: butyrate fractions of 60%:20%:20% when averaged across the biofilm to be consistent with in vivo studies [5,59] (see Materials and Methods). The model predicted the total SCFA concentration to be 32.5 mmol/L (Figure 2C), which was in reasonable agreement with an in vivo study with a control diet that yielded 41.1 mmol/L of total SCFAs [60]. One possible explanation for the lower SCFA levels predicted by our model is the simplified diet (glucose, eight amino acids) compared to the control diet used experimentally.
Ethanol was present at a very low level (Figure 2D) due to limited synthesis by the small E. coli population and high consumption by the large B. thetaiotaomicron population. Of the two organic acids (OAs) produced, formate was predicted to be present at a high level because synthesis by F. prausnitzii and E. coli substantially exceeded consumption by C. difficile. Succinate was present at a moderate level since it was consumed by both C. difficile and F. prausnitzii. These predictions suggest that plentiful formate and succinate could be available to promote C. difficile propagation under in vivo perturbations.
2.3. Glucose and Amino Acid Perturbations
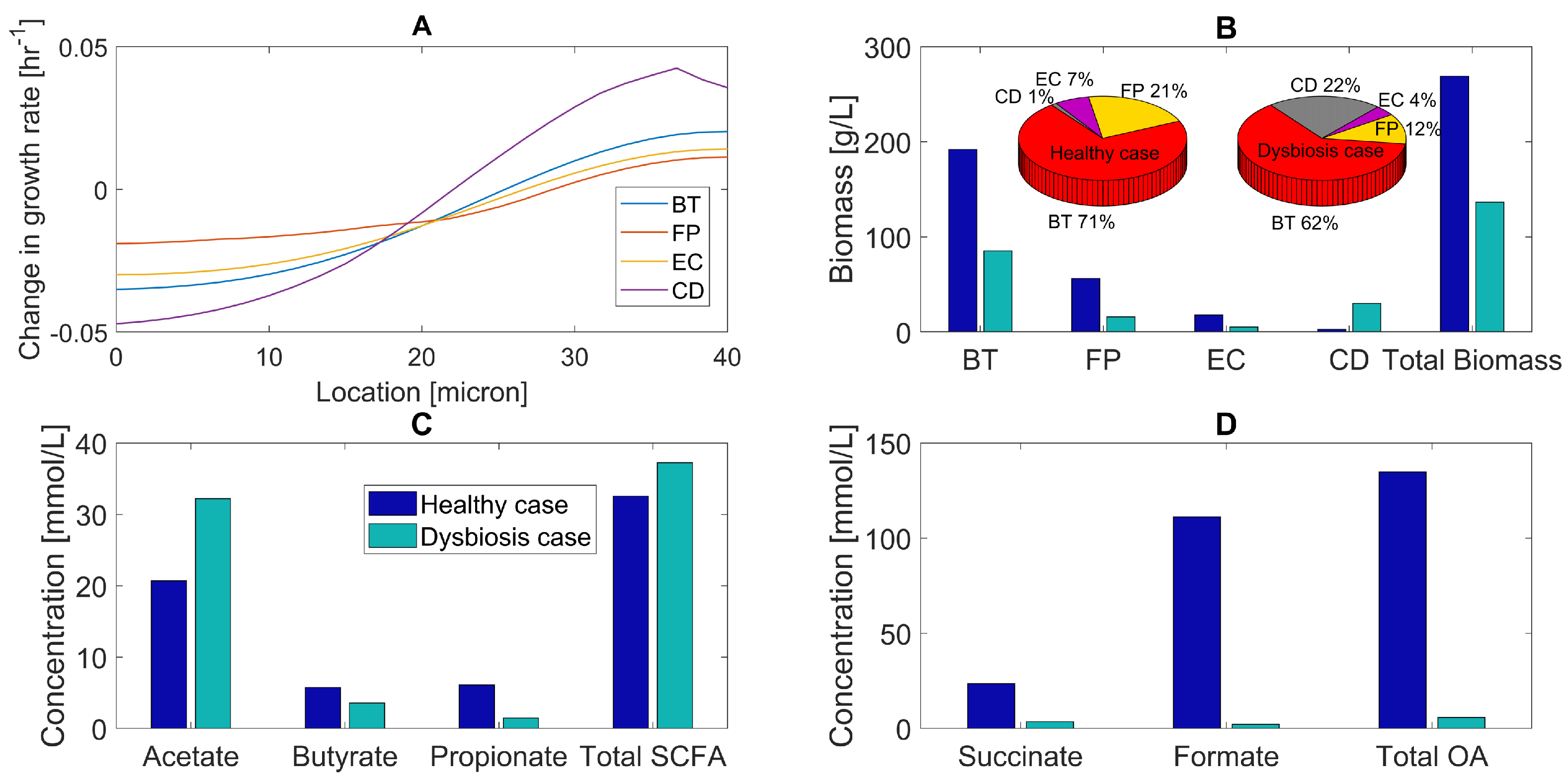
Various in vivo studies have shown that glucose concentration decreases and amino acid concentrations increase in the gut during C. difficile and other types of dysbiosis [12,61,62,63,64]. To investigate the effects of altered nutrient levels associated with host-microbiota perturbations, we performed simulations for a 40-micron biofilm with elevated amino acid and reduced glucose bulk concentrations (Table 1) under the assumption that C. difficile expansion is driven by these experimentally-observed nutrient changes. While in vivo nutrient levels are impacted by diet, host metabolism, and microbiota, this assumption was deemed reasonable given the simplified nature of our model. Given the uncertainty associated with the bulk nutrient concentrations, we performed a sensitivity analysis to explore their effects with respect to the species abundances (Figure S2). This analysis was consistent with the model predictions reported below as long as the glucose to amino acid ratio was sufficiently large. Compared to the healthy case, the local C. difficile growth rate decreased in the bottom half of the biofilm, but increased in the top half (Figure 3A). Similar trends were predicted for the three commensal species, which we attributed to reduced glucose, but increased amino acid penetration into the biofilm. C. difficile is known to grow efficiently on amino acids due to its ability to use amino acid pairs such as leucine and proline to generate ATP via Stickland metabolism [65,66,67].
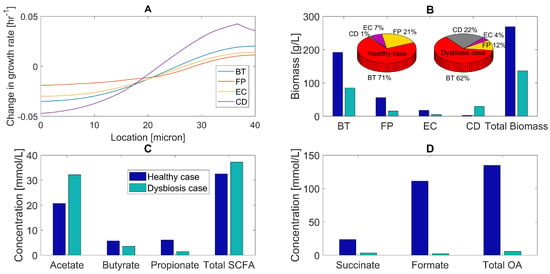
Figure 3.
Predicted multispecies biofilm dysbiosis resulting from host–microbiota perturbations in glucose and amino acid concentrations. (A) Change in species growth rates across the biofilm plotted as the difference between the growth rates for the healthy and dysbiosis cases. (B) Biomass concentrations (bar graphs) and species abundances (pie chart) averaged across the biofilm for healthy and dysbiosis case. (C) Acetate, butyrate, propionate, and total SCFA concentrations averaged across the biofilm. (D) Succinate, formate, and total OA concentrations averaged across the biofilm.
As a result of its enhanced growth in the top half of the biofilm compared to the commensal species, C. difficile increased its average biomass concentration ten-fold and species abundance from 1%–22% compared to the healthy case (Figure 3A). The biomass concentration of each commensal species dropped due to reduced glucose availability. A substantial effect was predicted for F. prausnitzii with its species abundance decreasing from 21%–12%, partially due to increased competition for succinate with C. difficile. These predictions are in agreement with in vivo studies [29,68,69,70], with the exception that dysbiosis during CDI should be accompanied by an increase in E. coli abundance [13,15,71,72,73]. The model predicted reduced total biomass production due to reduced growth of the three commensal species.
Dysbiosis was predicted to result in increased acetate, decreased butyrate and propionate, and lower total SCFA levels compared to the healthy case (Figure 3C). We attributed reduced total SCFA synthesis to lower glucose availability and increased acetate and decreased butyrate levels to a change in the balance of acetate-producing C. difficile and acetate-to-butyrate converting F. prausnitzii. Experimental studies have shown that dysbiosis is associated with reduced butyrate concentrations in the gut [69,74]. The model predicted large changes in organic acid levels, with succinate, formate, and total OA concentrations dropping due to reduced glucose fermentation. These predictions suggest that the combination of decreased carbohydrate and increased amino acid levels could play a role in C. difficile-associated dysbiosis.
2.4. Primary Bile Acid Perturbations
Primary bile acids such as taurocholate are secreted by the liver and transported into the intestines where anaerobic bacteria degrade them into secondary bile acids [75,76,77]. Broad spectrum antibiotics are known to reduce gut microbiota diversity [30,31,32,33,78], including the possible loss of bacterial species from families Lachnospiraceae and Ruminococcaceae responsible for the conversion of primary bile acids. Various in vitro [77,79,80] and in vivo [16,81] studies have shown that C. difficile spores can use primary bile acids for germination. Sodium taurocholate is the typical reagent used to grow C. difficile in vitro [82,83]. We investigated the impact of such perturbations with the multispecies biofilm model by adding taurocholate as a representative primary bile acid (Table 1). While primary bile acids are known to promote C. difficile transition from spores to a vegetative state [79,84], we assumed that C. difficile was already vegetative and investigated the effect of taurocholate on C. difficile growth. Preliminary FBA calculations with the C. difficile metabolic reconstruction showed that taurocholate uptake increased the growth rate, while taurocholate uptake was not possible with the three commensal species reconstructions.
Compared to the healthy case, the introduction of taurocholate was predicted to increase the local C. difficile growth rate across the biofilm (Figure 4A). B. thetaiotaomicron and E. coli growth were largely unaffected, while the F. prausnitzii growth rate decreased due to increased competition for succinate from C. difficile. As a result, the C. difficile abundance increased from 1%–18%, while the F. prausnitzii abundance decreased by 38% (Figure 4B). The B. thetaiotaomicron and E. coli abundances exhibited relatively small decreases, although experimental studies showed that E. coli abundance should increase during dysbiosis [71,73]. The total biomass concentration was predicted to remain almost constant, showing that taurocholate was responsible for changing the species distribution of the biomass.
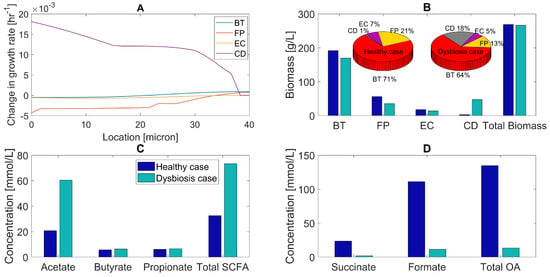
Figure 4.
Predicted multispecies biofilm dysbiosis resulting from host-microbiota perturbations in the concentration of the primary bile acid taurocholate. (A) Change in species growth rates across the biofilm plotted as the difference between the growth rates for the healthy and dysbiosis case. (B) Biomass concentrations (bar graphs) and species abundances (pie charts) averaged across the biofilm for the healthy and dysbiosis case. (C) Acetate, butyrate, propionate, and total SCFA concentrations averaged across the biofilm. (D) Succinate, formate, and total OA concentrations averaged across the biofilm.
The predicted trends for SCFA and OA levels were similar to those observed for the combined glucose/amino acid perturbation. Acetate and total SCFA concentrations increased compared to the healthy case due to increased acetate synthesis by C. difficile and decreased acetate consumption by F. prausnitzii (Figure 4C). The formate concentration decreased because of the same mechanism, while we attributed the reduced succinate concentration to increased succinate consumption by C. difficile (Figure 4D). Butyrate (produced by F. prausnitzii and C. difficile) and propionate (produced by B. thetaiotaomicron and C. difficile) concentrations remained almost constant as C. difficile compensated for reduced SCFA synthesis by the two commensal species. We also simulated a host-microbiota perturbation with decreased glucose/increased amino acids and increased taurocholate to examine the combined effects of these nutrient changes. Compared to either perturbation alone, the model predicted a further increase in C. difficile abundance and a decrease in F. prausnitzii abundance (Figure S3). Overall, these results support the hypothesis that increased primary bile acid levels could contribute to C. difficile propagation in vivo.
2.5. Host-Derived Nitrate Perturbations
The human host is known to secrete nitrate in response to inflammation in the gut [17]. Preliminary FBA calculations showed that nitrate uptake increased the E. coli growth rate, while the other three community members were unable to use nitrate as an electron acceptor. Therefore, we hypothesized that host-derived nitrate would increase E. coli abundance during simulated C. difficile-associated dysbiosis and yield better agreement with experimental studies [71,73]. To quantify the effects of nitrate availability, biofilm simulations were performed with and without nitrate for a dysbiosis case with reduced glucose, increased amino acids, and available taurocholate (Table 1).
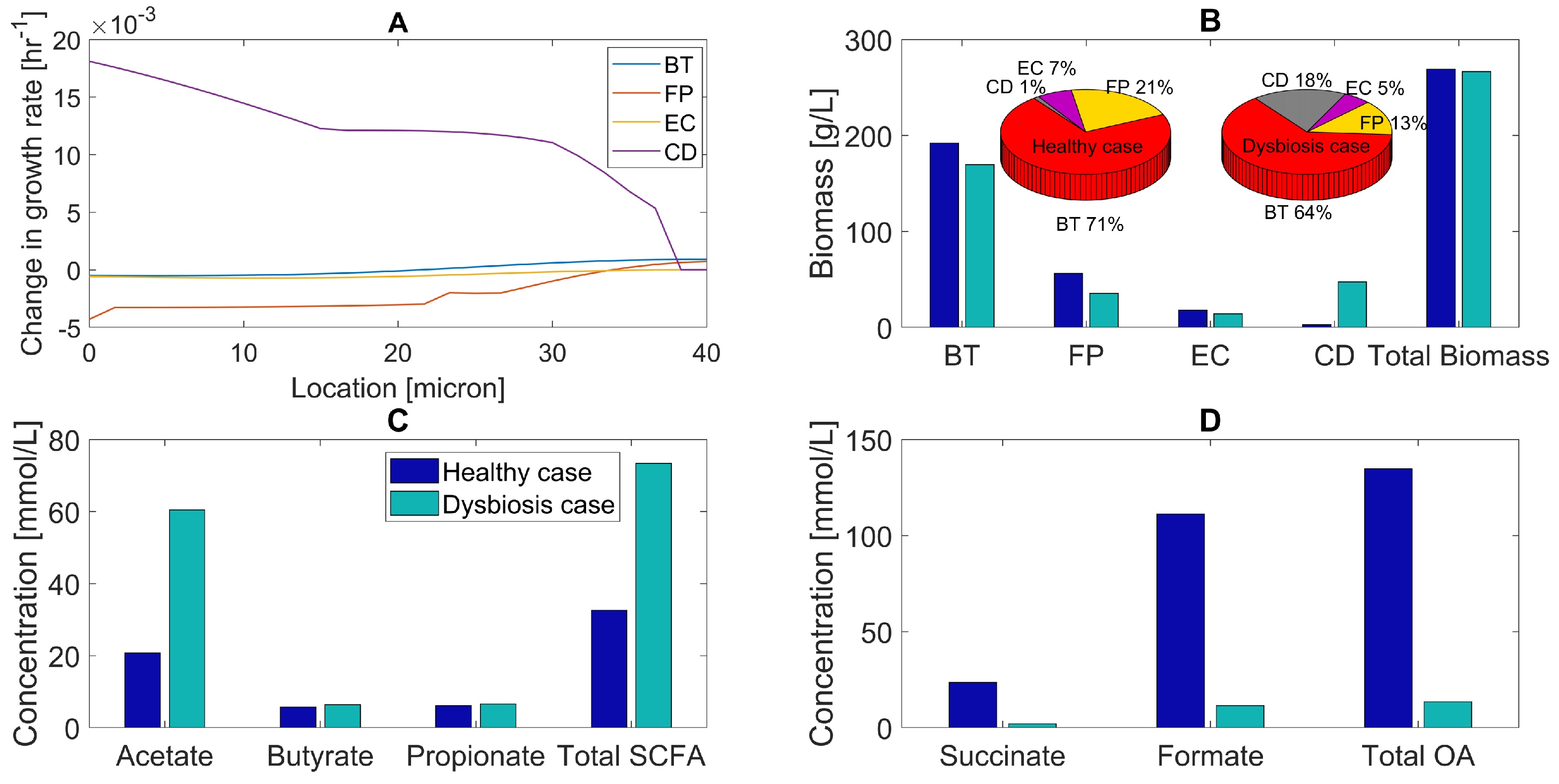
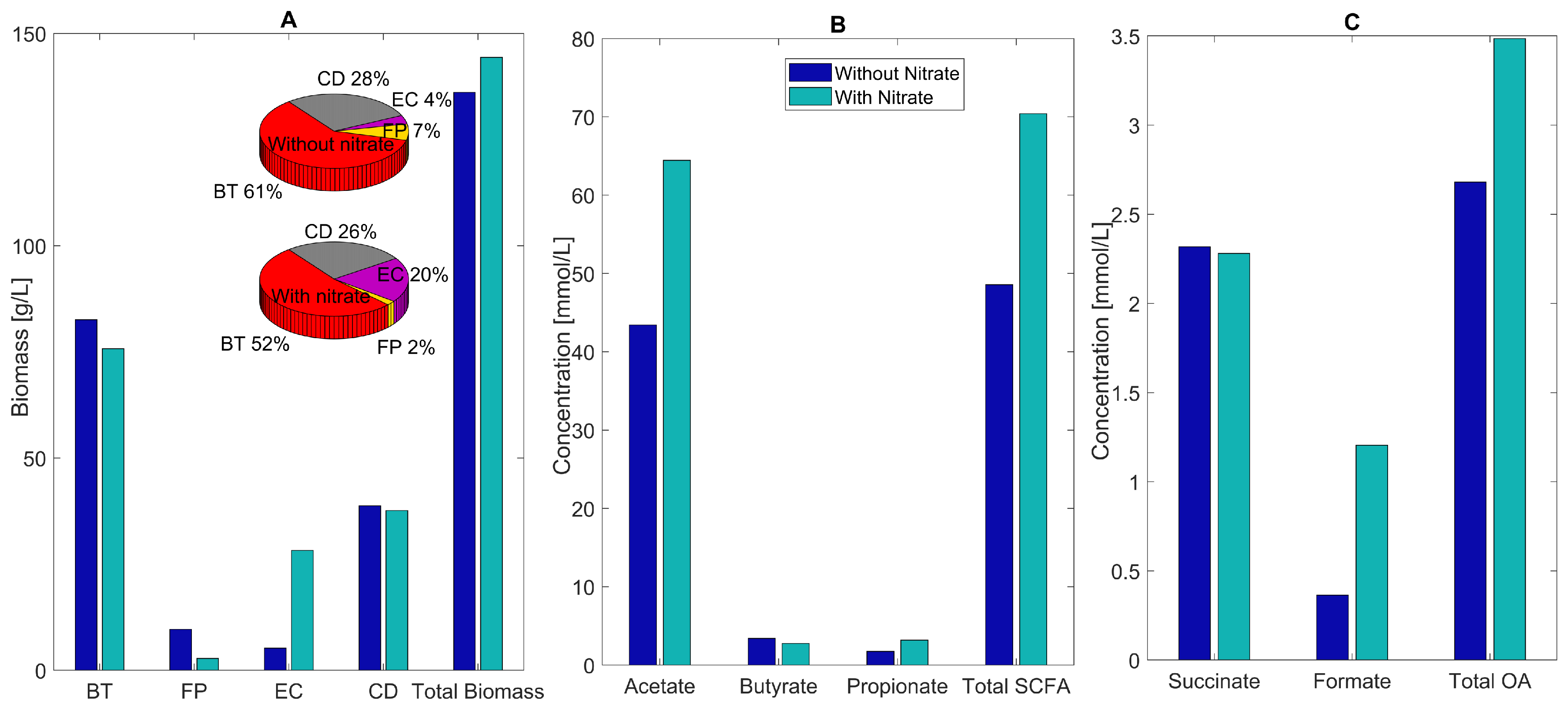
As hypothesized, the main impact of host-derived nitrate was to substantially increase E. coli abundance from 4% without nitrate to 20% with nitrate (Figure 5A). The F. prausnitzii abundance decreased from 7% to 2%, while the abundances of B. thetaiotaomicron and C. difficile decreased modestly to accommodate the increased E. coli. The species abundances predicted with nitrate are in good agreement with experimental studies for C. difficile-associated dysbiosis showing large increases in C. difficile and E. coli, large decreases in F. prausnitzii, and modest changes in B. thetaiotaomicron [85,86,87].
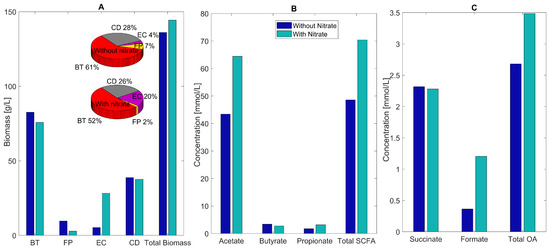
Figure 5.
Predicted multispecies biofilm dysbiosis with and without host-derived nitrate. (A) Biomass concentrations (bar graphs) and species abundances (pie charts) averaged across the biofilm for the healthy and dysbiosis case. (B) Acetate, butyrate, propionate, and total SCFA concentrations (mmol/L) averaged across the biofilm. (C) Succinate, formate, and total OA concentrations averaged across the biofilm.
Nitrate availability was predicted to increase the acetate and total SCFA concentrations substantially due to large changes in E. coli and F. prausnitzii abundances (Figure 5B). Decreased succinate consumption by F. prausnitzii and increased formate synthesis by E. coli results in increased levels of individual and total OAs (Figure 5C). These predictions implicate a role for host-derived nitrate in C. difficile-associated dysbiosis.
We investigated the robustness of the four-species community during dysbiosis with available nitrate by removing selected cross-feeding relationships and varying the biofilm thickness from the nominal value of 40 microns. When C. difficile uptake of formate or succinate was eliminated, the C. difficile abundance dropped substantially (Figure S4), further suggesting that these cross-feeding relationships could be important for C. difficile propagation in vivo. Consistent with our previous study [50], cross-feeding of ethanol was important for B. thetaiotaomicron growth, and cross-feeding of both acetate and succinate was necessary for F. prausnitzii co-existence. For biofilm thicknesses of 30–60 microns, the species abundances were predicted to vary substantially with the most important trend being that thinner biofilms enhanced C. difficile growth (Figures S5 and S6). The growth rate profiles for 60 microns (Figure S7) suggested that C. difficile spores might be formed in the upper half of the biofilm where C. difficile was unable to sustain vegetative growth. Since such spores could be activated by favorable nutrient conditions, the incorporation of C. difficile spore formation and activation could be an interesting direction for future research. Overall, our results could help explain the role of broad spectrum antibiotics during CDI, as antibiotics could be expected to reduce the diversity and density of commensal bacteria that protect the gut from C. difficile expansion.
To gain insights into the internal pathway fluxes associated with the healthy and dysbiosis states, we determined for each species the eight internal fluxes that varied the most between the healthy (Figure 2) and C. difficile dysbiosis (Figure 5) states and identified the internal pathways associated with each of these fluxes. Using simulation data from the stool-biofilm interface at 300 h, the most variable fluxes were determined by computing for each individual flux the difference between the healthy and dysbiosis values and scaling the result by the healthy value (Figure S8). Pathways associated with amino acid metabolism were upregulated in B. thetaiotaomicron and C. difficile, demonstrating the ability of these two species to take advantage of increased amino acid availability. Similarly, the internal flux through the cysteine metabolism pathway was predicted to increase in E. coli. Most internal pathway fluxes in F. prausnitzii were predicted to decrease, suggesting that the dysbiosis environment was unfavorable for its growth, resulting in decreased abundance.
3. Discussion
The gut microbiota serve a broad array of important functions for the human host, including providing colonization resistance to opportunistic pathogens. Unhealthy changes in the microbiota composition, commonly termed dysbiosis, have been correlated to a wide variety of gut and metabolic diseases including inflammatory bowel disease, Crohn’s disease, obesity, diabetes, and chronic gut infections. The opportunistic gut pathogen Clostridium difficile has been estimated to asymptomatically colonize 3%–15% of healthy adults [28]. A common cause of symptomatic C. difficile infection (CDI) is the use of broad spectrum antibiotics, which induce dysbiosis by reducing the diversity and density of gut commensal bacteria that provide resistance to C. difficile expansion [30,31,32,33,78]. Improved understanding of the complex interactions between commensal species, C. difficile, the gut environment, and the human host are needed to treat CDI more rationally.
To help unravel the metabolic determinants of C. difficile-associated dysbiosis, we developed a multispecies biofilm model by combining genome-scale metabolic reconstruction of C. difficile [88] and commensal species representing the three dominant phyla in the gut: Bacteroides thetaiotaomicron (Bacteroidetes) [89], Faecalibacterium prausnitzii (Firmicutes) [90], and Escherichia coli (Proteobacteria) [91]. The chosen species are well-studied representatives of the most dominant phyla in the human gut microbiome, and curated metabolic reconstructions of these species were available. While our four-species model represented a substantial reduction in complexity compared to the actual gut microbiota, the number of species and extracellular metabolites included were limited by computational considerations. Community models with substantially more species and cross-fed metabolites can be formulated and solved by neglecting spatial and temporal variations, as shown in our recent study of the gut microbiota [92]. However, these assumptions are not appropriate for biofilm simulations. Furthermore, our four-species model could be useful for designing in vitro systems for experimentally testing model predictions.
While specific spatial organization of gut microbes is currently unknown, the structure likely includes biofilm growth associated with host mucosa and epithelial tissue [93]. The literature provides significant evidence to support the hypothesis that some gut microbes develop spatially-structured multispecies biofilms [40,43]. We sought to understand how the commensal species could sublimate C. difficile expansion and under what gut conditions colonization resistance could become compromised. The biofilm model was tuned to represent a healthy state with species abundances and concentrations of short-chain fatty acids (SCFAs; acetate, butyrate, propionate) consistent with experimental studies for healthy individuals [5,57,59]. Because our model lacked an explicit description of the human host, we mimicked host-microbiota perturbations associated with CDI by varying nutrient levels guided by experimental observations. More specifically, dysbiosis states were modeled through changes in the concentrations of available glucose, amino acids [12,61,62,63,64], primary bile acids [16,77,81], and nitrate [17].
Our model predicted that cross-feeding of secreted byproducts plays an important role in C. difficile sublimation and expansion. C. difficile consumed formate synthesized by F. prausnitzii and E. coli and succinate synthesized by B. thetaiotaomicron and F. prausnitzii. The existence of both cross-feeding relationships is supported by the experimental literature [55,56]. In silico removal of either cross-feeding relationship was predicted to provide C. difficile colonization resistance, demonstrating the complexity and importance of cross-feeding networks even in this simplified four-species community. These predictions could be tested experimentally through the development of an in vitro model system of the four species. More importantly, these results suggest that therapeutic strategies that target species–species interactions could be promising alternatives to conventional antibiotics that target C. difficile directly.
Host–microbiota perturbations modeled as increases in glucose and decreases in amino acid concentrations reproduced several features of C. difficile-associated dysbiosis including substantially reduced F. prausnitzii and increased C. difficile abundances and an imbalance in SCFA synthesis characterized by increased acetate and reduced butyrate levels [94]. The predicted decrease in anti-inflammatory butyrate would be expected to exasperate dysbiosis and accelerate disease progression [69,74]. Similar results were obtained when glucose and amino acid changes were replaced by increases in the primary bile acid taurocholate, which was predicted to be used as an electron acceptor by C. difficile in vivo to provide a growth advantage in the absence of commensal bacteria that degrade primary bile acids to secondary bile acids [61,95,96]. Taurocholate availability was predicted to have less effect on butyrate and propionate synthesis, but the SCFA imbalance remained due to high acetate synthesis. Our model predicted that dysbiosis could be induced with moderate changes in nutrient concentrations, a prediction that could be tested in vitro and suggesting the possible promise of therapeutic strategies that aim to alter the gut nutritional environment.
Despite their many consistencies with experimental studies [12,97,98], our simulations with glucose, amino acids, and taurocholate changes were unable to reproduce the large increase in E. coli abundance observed during CDI [71,73]. The addition of host-derived nitrate [17,99] to the other nutrient changes rectified this inconsistency and reproduced the key microbiota signatures of C. difficile-associated dysbiosis during CDI: large increases in C. difficile and E. coli abundances, large decreases in health-promoting F. prausnitzii abundance, and moderate changes in B. thetaiotaomicron abundance. The model generated high acetate levels associated with dysbiosis states, a prediction that could be tested through in vitro experiments. We believe further development of our multispecies biofilm model could yield a general computational platform for in silico investigation of CDI, other gut infections, and chronic inflammation disorders such as inflammatory bowel and Crohn’s diseases. Some possibilities include the modeling of C. difficile spore formation/germination, the inclusion of more commensal gut species (e.g., [100]) including those from other phyla [101,102,103], the addition of a broader array of gut nutrients including fibers, oligosaccharides, and fats resulting from realistic diets [12,13,14,15,104], and modeling of the human host through incorporation of available metabolic reconstructions such as Recon 2 or Recon 3D [105,106,107]. A possible drawback of our modeling approach is the lack of species-specific parameters for nutrient uptake kinetics and metabolite-dependent mass transfer coefficients.
4. Materials and Methods
4.1. Biofilm Model Formulation and Solution
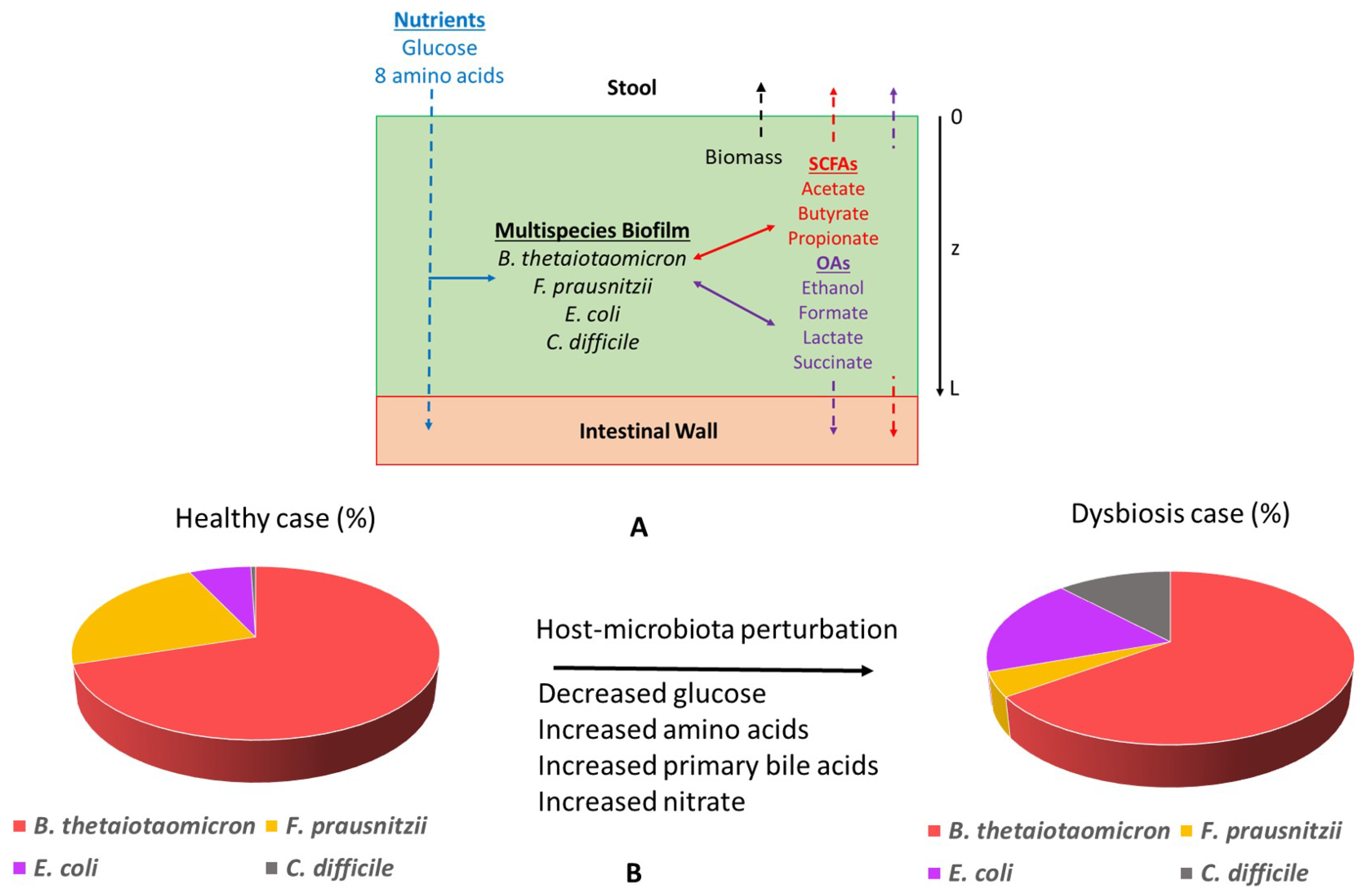
The multispecies biofilm model was constructed by combining genome-scale metabolic reconstructions of C. difficile (Strain 630erm) [88] and three commensal gut species: B. thetaiotaomicron [89], F. prausnitzii (Strain A2-165) [90], and E. coli (Strain K-12 MG1655) [91]. The biofilm was considered to be attached to the colon lining defined as the top of the biofilm (Figure 6A). A minimal defined media (MDM) containing glucose, cysteine, isoleucine, leucine, methionine, proline, serine, tryptophan, and valine along with essential vitamins and minerals was used for all simulations. The amino acids cysteine, isoleucine, leucine, proline, serine, and tryptophan are essential for in vivo C. difficile growth [66,67], while the amino acids methionine, tryptophan, and serine are essential for in vivo F. prausnitzii growth [108]. To simulate various host-microbiota perturbations, the primary bile acid taurocholate and/or the electron acceptor nitrate were added to the media. The diffusion of nutrients, byproducts, and species biomass was assumed to occur only in the axial direction z. Therefore, each variable was considered to be changing with respect to space z and time t over a fixed biofilm thickness L.
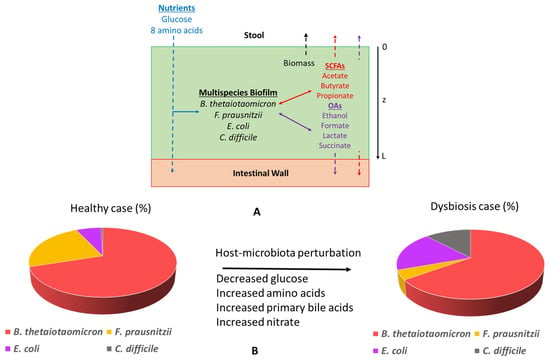
Figure 6.
Schematic representation of the in silico gut community. (A) The model assumed biofilm attachment to the intestinal wall and described diffusion of glucose, amino acids, short-chain fatty acids, organic acids, ethanol, CO, and species biomass in and/or out of the biofilm along the axial direction z. (B) Host-microbiota perturbations were modeled through changes in the bulk concentrations of glucose, amino acids, primary bile acids, and nitrate at the biofilm–stool interface to predict species abundances in healthy and C. difficile-infected guts.
The nutrients were supplied at the top of the biofilm (Figure 6A). SCFAs, ethanol, organic acids, and CO produced by the four species were allowed to diffuse and be removed from both ends of the biofilm. Biomass was assumed to move slowly through the biofilm by diffusion and be removed from the biofilm–stool interface according to a continuous erosion mechanism, as described in our previous publications [50,51,109]. This assumption provided a reasonable mechanism to ensure that biomass generation would be balanced by biomass loss such that a steady-state solution could be obtained. The multispecies biofilm model was tuned with nominal glucose and amino acid concentrations to reproduce species abundances and SCFA levels consistent with experimental studies on healthy individuals [57,58]. This tuned model was referred to as the “healthy case”. Host-microbiota perturbations were simulated by altering glucose/amino acid concentrations and/or by introducing primary bile acids and nitrate as nutrients to predict the resulting species abundances (Figure 6B). These models were collectively referred to as the “dysbiosis case.” In vivo concentrations of glucose and AA in the guts of healthy and C. difficile-infected patients are not commonly available. We have specified the glucose and AA concentrations for the healthy case based on limited experimental data [12,61,62,63,64] and have reduced the glucose concentration and increased AA concentrations for the dysbiosis case consistent with experimental observation [33,110]. We performed a sensitivity analysis of these concentrations to show that a similar behavior (i.e., healthy state) as that reported for the nominal values occurred if the glucose to AA ratio was sufficiently large (Figure S2). By contrast, a CDI dysbiosis-like state was obtained when the glucose to AA ratio was sufficiently small.
Uptake rates of nutrients and byproducts were assumed to follow Michaelis–Menten kinetics. Due to lack of available data, maximum uptake rates and Michaelis–Menten constants were assumed to be independent of species and metabolite. Calculated uptake rates were imposed as lower bounds of the exchange fluxes in the species metabolic reconstructions. The calculated growth rate, uptake fluxes, and secretion fluxes from each reconstruction served as inputs to reaction-diffusion-type equations for the biomass concentration of each species and the molar concentration of each nutrient and byproduct. This formulation yielded a set of 23 partial differential equations (PDEs) in the time and the axial direction z with embedded linear programs (LPs) for species metabolism (see Appendix S1). Following our previous methodology [50,51], lexicographic optimization with growth rate maximization as the primary objective was used to avoid alternative optima that would render the biofilm model non-smooth. This approach yielded a total of 71 LPs.
The biofilm model equations were solved by spatially discretizing the PDEs into a large set of ordinary differential equations (ODEs) [111,112]. We used 25 spatial node points to achieve a suitable compromise between solution accuracy and computational efficiency, which produced a discretized model with 575 ODEs and 1775 LPs that was solved with the MATLAB code DFBAlab [113]. We used Gurobi 6.5.2 for the LP solution, the stiff MATLAB solver ode15s for ODE integration, and DFBAlab running in MATLAB 9.0 (R2016a). Although not explored here, our biofilm modeling method can be extended to more species and extracellular metabolites. For N spatial discretization points, the addition of each new extracellular metabolite would generate N additional ODEs. For m total extracellular metabolites, the addition of each new species would generates N additional ODEs and LPs. Because the LP solution scales more favorably than the ODE solution, we anticipated that models with approximately 1000 ODEs and 7500 LPs would remain computationally viable on a typical desktop computer. These equation numbers translate into approximately 10 species and 30 extracellular metabolites.
4.2. Biofilm Model Parameterization and Tuning
Nominal parameter values used in the multispecies biofilm model are shown in Table 2. The parameters were obtained from the experimental literature to the extent possible and from our previous modeling studies [50,51] as necessary. The bulk glucose and amino acid concentrations at the biofilm–stool interface were specified to reflect healthy gut conditions. Due to the lack of species-specific uptake data, we used published kinetic parameters reported for E. coli [114]. Due to the lack of data, all eight byproducts were assumed to have the same uptake parameters as glucose. For simplicity, all eight amino acids were assumed to have the same uptake parameters obtained as the average of amino acid-dependent values reported for E. coli [114].

Table 2.
Nominal parameter values for the multispecies biofilm model.
With all other parameter values fixed, the biofilm model was qualitatively tuned to achieve biomass and SCFA fractions within experimental ranges for a healthy patient. The species abundances were tuned by adjusting the non-growth-associated ATP maintenance (ATPM) values of the four metabolic reconstructions following our previous studies [50,51]. Our justification for tuning these values was the simple nature of the biofilm model, which neglected other phyla (e.g., Actinobacteria), other nutrients (e.g., oligosaccharides, fats), other species interactions (e.g., Actinobacteria cross-feeding of SCFAs and organic acids), as well as host metabolism present in the actual gut environment. These ATPM values listed in Table 2 produced B. thetaiotaomicron:F. prausnitzii:E. coli:C. difficile abundances of 71%:21%:7%:1%, which were deemed reasonable based on published data [57,58]. We found that the coexistence of the four species was achieved over a range of ATPM values (not shown here).
We adjusted the SCFA mass transfer coefficients controlling metabolite removal from the biofilm to tune the acetate, butyrate, and propionate concentrations for the healthy case. Starting with a value of 5 × 10 cm/s, the butyrate and propionate values were decreased until approximate fractions of 60%:20%:20% consistent with published data [5,59] were obtained. We justified the use of SCFA-dependent values by noting that our model neglected host-microbiota interactions, which would be expected to strongly affect SCFA levels in vivo. Biofilm simulations were performed for four combinations of bulk glucose, amino acid, nitrate, and taurocholate concentrations chosen to mimic a healthy gut environment and three unhealthy nutrient environments (high amino acids, high primary bile acids, high nitrate) experimentally correlated with C. difficile-associated dysbiosis (Table 1). We deemed the actual concentrations used to be less important than the concentration trends (e.g., decreasing glucose and increasing amino acids in the high amino acids case) since our goal was to assess qualitatively the effects of nutrient levels on community behavior.
5. Conclusions
Clostridium difficile infection (CDI) is a common problem in hospital settings, with almost 500,000 CDI cases diagnosed within the U.S. annually in acute care facilities alone. CDI involves dysbiosis of the commensal gut microbiota characterized by a significant reduction of butyrate-producing species, e.g., Faecalibacterium prausnitzii, and a large increase in Proteobacteria, e.g., Escherichia coli, along with uncontrolled propagation of C. difficile. Motivated by recent experimental studies demonstrating the ability of C. difficile and commensal gut bacteria to form biofilms, we developed a multispecies biofilm model with a minimal representation of the gut microbiota containing C. difficile and one species each from the three dominant phyla (F. prausnitzii, E. coli, Bacteroides thetaiotaomicron). The model was used to investigate possible metabolic determinants of CDI mediated through host–microbiota perturbations, modeled as decreased carbohydrate levels and increased amino acid, primary bile acid, and nitrate levels compared to the healthy gut. These nutrient perturbations were shown to mimic microbiota changes characteristic of CDI, namely marked increases in C. difficile and E. coli abundances and a sharp decrease in F. prausnitzii abundance. C. difficile propagation was strongly dependent on cross-feeding of formate and succinate secreted by the commensal species, a prediction in agreement with experimental studies and that provides possible targets for the development of novel therapeutic strategies. While our model is a simplified representation of a complex disease process, the results presented emphasized the importance of metabolic interactions between C. difficile and commensal species in CDI progression.
Supplementary Materials
The following are available online at http://www.mdpi.com/2227-9717/7/2/97/s1. Additional File 1. Model equations and description. Figure S1. Predicted cross-feeding of byproducts with C. difficile removed from the community. Figure S2. Predicted species abundances at various nutrient concentrations. Figure S3. Predicted multispecies biofilm dysbiosis resulting from host–microbiota perturbations in the concentrations of amino acids and the primary bile acid taurocholate. Figure S4. Effect of removing individual cross-feeding relationships on predicted species abundances. Figure S5. Effect of the biofilm length on predicted species abundances for the healthy case. Figure S6. Predicted multispecies biofilm behavior under healthy nutrient conditions for a 30 micron-thick biofilm. Figure S7. Predicted multispecies biofilm behavior under healthy nutrient conditions for a 60 micron-thick biofilm. Figure S8. Differences between healthy case and bile acid dysbiosis case internal pathway fluxes.
Author Contributions
P.P. and M.A.H. conceived of the study. P.P. and M.A.H. developed the model and model solution method. P.P. and M.A.H. performed the simulations and analyzed the results. P.P. and M.A.H. prepared the manuscript. All authors read and approved the final manuscript.
Funding
This research was partially funded by NIH (Award U01EB019416). This work was funded in part by a Fellowship from the University of Massachusetts to P.P. as part of an NIH-funded Training Program (National Research Service Award T32 GM108556). Funds from these grants were used to cover the costs to publish in open access.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| ATPM | ATP maintenance |
| BT | Bacteroides thetaiotaomicron |
| CD | Clostridium difficile |
| CDI | Clostridium difficile infection |
| DFBAlab | Dynamic flux balance analysis laboratory |
| EC | Escherichia coli |
| FBA | Flux balance analysis |
| FP | Faecalibacterium prausnitzii |
| IBD | Inflammatory bowel diseases |
| LP | Linear program |
| MDM | Minimal defined media |
| OA | Organic acid |
| ODE | Ordinary differential equation |
| PDE | Partial differential equation |
| SCFA | Short chain fatty acid |
References
- Gill, S.R.; Pop, M.; DeBoy, R.T.; Eckburg, P.B.; Turnbaugh, P.J.; Samuel, B.S.; Gordon, J.I.; Relman, D.A.; Fraser-Liggett, C.M.; Nelson, K.E. Metagenomic analysis of the human distal gut microbiome. Science 2006, 312, 1355–1359. [Google Scholar] [CrossRef] [PubMed]
- Bäckhed, F.; Ley, R.E.; Sonnenburg, J.L.; Peterson, D.A.; Gordon, J.I. Host-bacterial mutualism in the human intestine. Science 2005, 307, 1915–1920. [Google Scholar] [CrossRef] [PubMed]
- Macpherson, A.J.; Harris, N.L. Interactions between commensal intestinal bacteria and the immune system. Nat. Rev. Immunol. 2004, 4, 478–485. [Google Scholar] [CrossRef] [PubMed]
- Bäumler, A.J.; Sperandio, V. Interactions between the microbiota and pathogenic bacteria in the gut. Nature 2016, 535, 85–93. [Google Scholar] [CrossRef] [PubMed]
- Den Besten, G.; van Eunen, K.; Groen, A.K.; Venema, K.; Reijngoud, D.J.; Bakker, B.M. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J. Lipid Res. 2013, 54, 2325–2340. [Google Scholar] [CrossRef] [PubMed]
- Macfarlane, S.; Steed, H.; Macfarlane, G.T. Intestinal bacteria and inflammatory bowel disease. Crit. Rev. Clin. Lab. Sci. 2009, 46, 25–54. [Google Scholar] [CrossRef] [PubMed]
- Morrison, D.J.; Preston, T. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes 2016, 7, 189–200. [Google Scholar] [CrossRef] [PubMed]
- Rajilić-Stojanović, M.; Smidt, H.; De Vos, W.M. Diversity of the human gastrointestinal tract microbiota revisited. Environ. Microbiol. 2007, 9, 2125–2136. [Google Scholar] [CrossRef]
- Zoetendal, E.; Rajilić-Stojanović, M.; De Vos, W. High-throughput diversity and functionality analysis of the gastrointestinal tract microbiota. Gut 2008, 57, 1605–1615. [Google Scholar] [CrossRef]
- Gerritsen, J.; Smidt, H.; Rijkers, G.T.; Vos, W.M. Intestinal microbiota in human health and disease: The impact of probiotics. Genes Nutr. 2011, 6, 209. [Google Scholar] [CrossRef]
- Young, V.B.; Schmidt, T.M. Antibiotic-associated diarrhea accompanied by large-scale alterations in the composition of the fecal microbiota. J. Clin. Microbiol. 2004, 42, 1203–1206. [Google Scholar] [CrossRef] [PubMed]
- Brown, K.; DeCoffe, D.; Molcan, E.; Gibson, D.L. Diet-induced dysbiosis of the intestinal microbiota and the effects on immunity and disease. Nutrients 2012, 4, 1095–1119. [Google Scholar] [CrossRef] [PubMed]
- Anitha, M.; Reichardt, F.; Tabatabavakili, S.; Nezami, B.G.; Chassaing, B.; Mwangi, S.; Vijay-Kumar, M.; Gewirtz, A.; Srinivasan, S. Intestinal dysbiosis contributes to the delayed gastrointestinal transit in high-fat diet fed mice. CMGH Cell. Mol. Gastroenterol. Hepatol. 2016, 2, 328–339. [Google Scholar] [CrossRef] [PubMed]
- Alou, M.T.; Lagier, J.C.; Raoult, D. Diet influence on the gut microbiota and dysbiosis related to nutritional disorders. Hum. Microb. J. 2016, 1, 3–11. [Google Scholar] [CrossRef]
- Agus, A.; Denizot, J.; Thévenot, J.; Martinez-Medina, M.; Massier, S.; Sauvanet, P.; Bernalier-Donadille, A.; Denis, S.; Hofman, P.; Bonnet, R.; et al. Western diet induces a shift in microbiota composition enhancing susceptibility to Adherent-Invasive E. coli infection and intestinal inflammation. Sci. Rep. 2016, 6, 19032. [Google Scholar] [CrossRef] [PubMed]
- Theriot, C.M.; Bowman, A.A.; Young, V.B. Antibiotic-induced alterations of the gut microbiota alter secondary bile acid production and allow for Clostridium difficile spore germination and outgrowth in the large intestine. MSphere 2016, 1, e00045-15. [Google Scholar] [CrossRef]
- Winter, S.E.; Winter, M.G.; Xavier, M.N.; Thiennimitr, P.; Poon, V.; Keestra, A.M.; Laughlin, R.C.; Gomez, G.; Wu, J.; Lawhon, S.D.; et al. Host-derived nitrate boosts growth of E. coli in the inflamed gut. Science 2013, 339, 708–711. [Google Scholar] [CrossRef]
- Frank, D.N.; Amand, A.L.S.; Feldman, R.A.; Boedeker, E.C.; Harpaz, N.; Pace, N.R. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. USA 2007, 104, 13780–13785. [Google Scholar] [CrossRef] [PubMed]
- Ley, R.E.; Turnbaugh, P.J.; Klein, S.; Gordon, J.I. Microbial ecology: Human gut microbes associated with obesity. Nature 2006, 444, 1022–1023. [Google Scholar] [CrossRef] [PubMed]
- Carding, S.; Verbeke, K.; Vipond, D.T.; Corfe, B.M.; Owen, L.J. Dysbiosis of the gut microbiota in disease. Microb. Ecol. Health Dis. 2015, 26, 26191. [Google Scholar] [CrossRef]
- Surawicz, C.M.; Brandt, L.J.; Binion, D.G.; Ananthakrishnan, A.N.; Curry, S.R.; Gilligan, P.H.; McFarland, L.V.; Mellow, M.; Zuckerbraun, B.S. Guidelines for diagnosis, treatment, and prevention of Clostridium difficile infections. Am. J. Gastroenterol. 2013, 108, 478. [Google Scholar] [CrossRef] [PubMed]
- Shim, J.K.; Johnson, S.; Samore, M.H.; Bliss, D.Z.; Gerding, D.N. Primary symptomless colonisation by Clostridium difficile and decreased risk of subsequent diarrhoea. Lancet 1998, 351, 633–636. [Google Scholar] [CrossRef]
- Kyne, L.; Warny, M.; Qamar, A.; Kelly, C.P. Asymptomatic carriage of Clostridium difficile and serum levels of IgG antibody against toxin A. N. Engl. J. Med. 2000, 342, 390–397. [Google Scholar] [CrossRef] [PubMed]
- Barbut, F.; Petit, J.C. Epidemiology of Clostridium difficile-associated infections. Clin. Microbiol. Infect. 2001, 7, 405–410. [Google Scholar] [CrossRef]
- Kato, H.; Kita, H.; Karasawa, T.; Maegawa, T.; Koino, Y.; Takakuwa, H.; Saikai, T.; Kobayashi, K.; Yamagishi, T.; Nakamura, S. Colonisation and transmission of Clostridium difficile in healthy individuals examined by PCR ribotyping and pulsed-field gel electrophoresis. J. Med. Microbiol. 2001, 50, 720–727. [Google Scholar] [CrossRef] [PubMed]
- Ozaki, E.; Kato, H.; Kita, H.; Karasawa, T.; Maegawa, T.; Koino, Y.; Matsumoto, K.; Takada, T.; Nomoto, K.; Tanaka, R.; et al. Clostridium difficile colonization in healthy adults: transient colonization and correlation with enterococcal colonization. J. Med. Microbiol. 2004, 53, 167–172. [Google Scholar] [CrossRef]
- Poutanen, S.M.; Simor, A.E. Clostridium difficile-associated diarrhea in adults. Can. Med. Assoc. J. 2004, 171, 51–58. [Google Scholar] [CrossRef]
- Furuya-Kanamori, L.; Marquess, J.; Yakob, L.; Riley, T.V.; Paterson, D.L.; Foster, N.F.; Huber, C.A.; Clements, A.C. Asymptomatic Clostridium difficile colonization: epidemiology and clinical implications. BMC Infect. Dis. 2015, 15, 516. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.Y.; Antonopoulos, D.A.; Kalra, A.; Tonelli, A.; Khalife, W.T.; Schmidt, T.M.; Young, V.B. Decreased diversity of the fecal microbiome in recurrent Clostridium difficile-associated diarrhea. J. Infect. Dis. 2008, 197, 435–438. [Google Scholar] [CrossRef]
- Bartlett, J.G. Clostridium difficile: History of its role as an enteric pathogen and the current state of knowledge about the organism. Clin. Infect. Dis. 1994, 18, S265–S272. [Google Scholar] [CrossRef]
- Pérez-Cobas, A.E.; Moya, A.; Gosalbes, M.J.; Latorre, A. Colonization resistance of the gut microbiota against Clostridium difficile. Antibiotics 2015, 4, 337–357. [Google Scholar] [CrossRef] [PubMed]
- Pothoulakis, C. Pathogenesis of Clostridium difficile-associated diarrhoea. Eur. J. Gastroenterol. Hepatol. 1996, 8, 1041–1047. [Google Scholar] [CrossRef] [PubMed]
- Theriot, C.M.; Young, V.B. Interactions between the gastrointestinal microbiome and Clostridium difficile. Annu. Rev. Microbiol. 2015, 69, 445–461. [Google Scholar] [CrossRef] [PubMed]
- Mylonakis, E.; Ryan, E.T.; Calderwood, S.B. Clostridium difficile-associated diarrhea: A review. Arch. Intern. Med. 2001, 161, 525–533. [Google Scholar] [CrossRef] [PubMed]
- Jarrad, A.M.; Karoli, T.; Blaskovich, M.A.; Lyras, D.; Cooper, M.A. Clostridium difficile drug pipeline: Challenges in discovery and development of new agents. J. Med. Chem. 2015, 58, 5164–5185. [Google Scholar] [CrossRef] [PubMed]
- Lessa, F.C.; Mu, Y.; Bamberg, W.M.; Beldavs, Z.G.; Dumyati, G.K.; Dunn, J.R.; Farley, M.M.; Holzbauer, S.M.; Meek, J.I.; Phipps, E.C.; et al. Burden of Clostridium difficile infection in the United States. N. Engl. J. Med. 2015, 372, 825–834. [Google Scholar] [CrossRef]
- Dubberke, E.R.; Olsen, M.A. Burden of Clostridium difficile on the healthcare system. Clin. Infect. Dis. 2012, 55, S88–S92. [Google Scholar] [CrossRef]
- Dawson, L.F.; Valiente, E.; Faulds-Pain, A.; Donahue, E.H.; Wren, B.W. Characterisation of Clostridium difficile biofilm formation, a role for Spo0A. PLoS ONE 2012, 7, e50527. [Google Scholar] [CrossRef]
- Dhapa, T.; Leuzzi, R.; Ng, Y.K.; Baban, S.T.; Adamo, R.; Kuehne, S.A.; Scarselli, M.; Minton, N.P.; Serruto, D.; Unnikrishnan, M. Multiple factors modulate biofilm formation by the anaerobic pathogen Clostridium difficile. J. Bacteriol. 2013, 195, 545–555. [Google Scholar]
- Donelli, G.; Vuotto, C.; Cardines, R.; Mastrantonio, P. Biofilm-growing intestinal anaerobic bacteria. FEMS Immunol. Med. Microbiol. 2012, 65, 318–325. [Google Scholar] [CrossRef]
- Semenyuk, E.G.; Laning, M.L.; Foley, J.; Johnston, P.F.; Knight, K.L.; Gerding, D.N.; Driks, A. Spore formation and toxin production in Clostridium difficile biofilms. PLoS ONE 2014, 9, e87757. [Google Scholar] [CrossRef] [PubMed]
- Swidsinski, A.; Weber, J.; Loening-Baucke, V.; Hale, L.P.; Lochs, H. Spatial organization and composition of the mucosal flora in patients with inflammatory bowel disease. J. Clin. Microbiol. 2005, 43, 3380–3389. [Google Scholar] [CrossRef] [PubMed]
- Macfarlane, S.; Dillon, J. Microbial biofilms in the human gastrointestinal tract. J. Appl. Microbiol. 2007, 102, 1187–1196. [Google Scholar] [CrossRef] [PubMed]
- Costerton, J.W.; Stewart, P.S.; Greenberg, E.P. Bacterial biofilms: A common cause of persistent infections. Science 1999, 284, 1318–1322. [Google Scholar] [CrossRef] [PubMed]
- Anderl, J.N.; Franklin, M.J.; Stewart, P.S. Role of antibiotic penetration limitation in Klebsiella pneumoniae biofilm resistance to ampicillin and ciprofloxacin. Antimicrob. Agents Chemother. 2000, 44, 1818–1824. [Google Scholar] [CrossRef] [PubMed]
- Stewart, P.S.; Costerton, J.W. Antibiotic resistance of bacteria in biofilms. Lancet 2001, 358, 135–138. [Google Scholar] [CrossRef]
- Stewart, P.S. Mechanisms of antibiotic resistance in bacterial biofilms. Int. J. Med. Microbiol. 2002, 292, 107–113. [Google Scholar] [CrossRef] [PubMed]
- Zuroff, T.R.; Bernstein, H.; Lloyd-Randolfi, J.; Jimenez-Taracido, L.; Stewart, P.S.; Carlson, R.P. Robustness analysis of culturing perturbations on Escherichia coli colony biofilm beta-lactam and aminoglycoside antibiotic tolerance. BMC Microbiol. 2010, 10, 185. [Google Scholar] [CrossRef] [PubMed]
- Shreiner, A.B.; Kao, J.Y.; Young, V.B. The gut microbiome in health and in disease. Curr. Opin. Gastroenterol. 2015, 31, 69. [Google Scholar] [CrossRef] [PubMed]
- Henson, M.A.; Phalak, P. Byproduct Cross Feeding and Community Stability in an In Silico Biofilm Model of the Gut Microbiome. Processes 2017, 5, 13. [Google Scholar] [CrossRef]
- Henson, M.A.; Phalak, P. Microbiota dysbiosis in inflammatory bowel diseases: in silico investigation of the oxygen hypothesis. BMC Syst. Biol. 2017, 11, 145. [Google Scholar] [CrossRef] [PubMed]
- Rivière, A.; Selak, M.; Lantin, D.; Leroy, F.; De Vuyst, L. Bifidobacteria and butyrate-producing colon bacteria: Importance and strategies for their stimulation in the human gut. Front. Microbiol. 2016, 7, 979. [Google Scholar] [CrossRef] [PubMed]
- Ríos-Covián, D.; Ruas-Madiedo, P.; Margolles, A.; Gueimonde, M.; de los Reyes-Gavilán, C.G.; Salazar, N. Intestinal short chain fatty acids and their link with diet and human health. Front. Microbiol. 2016, 7, 185. [Google Scholar] [CrossRef] [PubMed]
- Jakobsdottir, G.; Xu, J.; Molin, G.; Ahrne, S.; Nyman, M. High-fat diet reduces the formation of butyrate, but increases succinate, inflammation, liver fat and cholesterol in rats, while dietary fibre counteracts these effects. PLoS ONE 2013, 8, e80476. [Google Scholar] [CrossRef] [PubMed]
- Ferreyra, J.A.; Wu, K.J.; Hryckowian, A.J.; Bouley, D.M.; Weimer, B.C.; Sonnenburg, J.L. Gut microbiota- produced succinate promotes C. difficile infection after antibiotic treatment or motility disturbance. Cell Host Microbe 2014, 16, 770–777. [Google Scholar] [CrossRef] [PubMed]
- Köpke, M.; Straub, M.; Dürre, P. Clostridium difficile is an autotrophic bacterial pathogen. PLoS ONE 2013, 8, e62157. [Google Scholar] [CrossRef] [PubMed]
- Spor, A.; Koren, O.; Ley, R. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat. Rev. Microbiol. 2011, 9, 279–290. [Google Scholar] [CrossRef]
- De Filippo, C.; Cavalieri, D.; Di Paola, M.; Ramazzotti, M.; Poullet, J.B.; Massart, S.; Collini, S.; Pieraccini, G.; Lionetti, P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl. Acad. Sci. USA 2010, 107, 14691–14696. [Google Scholar] [CrossRef]
- Byrne, C.; Chambers, E.; Morrison, D.; Frost, G. The role of short chain fatty acids in appetite regulation and energy homeostasis. Int. J. Obes. 2015, 39, 1331–1338. [Google Scholar] [CrossRef]
- Campbell, J.M.; Fahey, G.C.; Wolf, B.W. Selected indigestible oligosaccharides affect large bowel mass, cecal and fecal short-chain fatty acids, pH and microflora in rats. J. Nutr. 1997, 127, 130–136. [Google Scholar] [CrossRef]
- Theriot, C.M.; Koenigsknecht, M.J.; Carlson Jr, P.E.; Hatton, G.E.; Nelson, A.M.; Li, B.; Huffnagle, G.B.; Li, J.Z.; Young, V.B. Antibiotic-induced shifts in the mouse gut microbiome and metabolome increase susceptibility to Clostridium difficile infection. Nat. Commun. 2014, 5, 3114. [Google Scholar] [CrossRef] [PubMed]
- Utzschneider, K.M.; Kratz, M.; Damman, C.J.; Hullarg, M. Mechanisms linking the gut microbiome and glucose metabolism. J. Clin. Endocrinol. Metab. 2016, 101, 1445–1454. [Google Scholar] [CrossRef] [PubMed]
- Carmody, R.N.; Gerber, G.K.; Luevano, J.M.; Gatti, D.M.; Somes, L.; Svenson, K.L.; Turnbaugh, P.J. Diet dominates host genotype in shaping the murine gut microbiota. Cell Host Microbe 2015, 17, 72–84. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Shen, D.; Fang, Z.; Jie, Z.; Qiu, X.; Zhang, C.; Chen, Y.; Ji, L. Human gut microbiota changes reveal the progression of glucose intolerance. PLoS ONE 2013, 8, e71108. [Google Scholar] [CrossRef] [PubMed]
- Jackson, S.; Calos, M.; Myers, A.; Self, W.T. Analysis of proline reduction in the nosocomial pathogen Clostridium difficile. J. Bacteriol. 2006, 188, 8487–8495. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, S.; Burman, L.G.; Åkerlund, T. Suppression of toxin production in Clostridium difficile VPI 10463 by amino acids. Microbiology 1999, 145, 1683–1693. [Google Scholar] [CrossRef] [PubMed]
- Neumann-Schaal, M.; Hofmann, J.D.; Will, S.E.; Schomburg, D. Time-resolved amino acid uptake of Clostridium difficile 630Δerm and concomitant fermentation product and toxin formation. BMC Microbiol. 2015, 15, 281. [Google Scholar] [CrossRef] [PubMed]
- Antonopoulos, D.A.; Huse, S.M.; Morrison, H.G.; Schmidt, T.M.; Sogin, M.L.; Young, V.B. Reproducible community dynamics of the gastrointestinal microbiota following antibiotic perturbation. Infect. Immunity 2009, 77, 2367–2375. [Google Scholar] [CrossRef] [PubMed]
- Antharam, V.C.; Li, E.C.; Ishmael, A.; Sharma, A.; Mai, V.; Rand, K.H.; Wang, G.P. Intestinal dysbiosis and depletion of butyrogenic bacteria in Clostridium difficile infection and nosocomial diarrhea. J. Clin. Microbiol. 2013, 51, 2884–2892. [Google Scholar] [CrossRef]
- Vincent, C.; Manges, A. Antimicrobial use, human gut microbiota and Clostridium difficile colonization and infection. Antibiotics 2015, 4, 230–253. [Google Scholar] [CrossRef]
- Schippa, S.; Conte, M.P. Dysbiotic events in gut microbiota: Impact on human health. Nutrients 2014, 6, 5786–5805. [Google Scholar] [CrossRef]
- Tamboli, C.; Neut, C.; Desreumaux, P.; Colombel, J. Dysbiosis in inflammatory bowel disease. Gut 2004, 53, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Honda, K.; Littman, D.R. The microbiome in infectious disease and inflammation. Annu. Rev. Immunol. 2012, 30, 759–795. [Google Scholar] [CrossRef] [PubMed]
- Ling, Z.; Liu, X.; Jia, X.; Cheng, Y.; Luo, Y.; Yuan, L.; Wang, Y.; Zhao, C.; Guo, S.; Li, L.; et al. Impacts of infection with different toxigenic Clostridium difficile strains on faecal microbiota in children. Sci. Rep. 2014, 4, 7485. [Google Scholar] [CrossRef] [PubMed]
- Ridlon, J.M.; Kang, D.J.; Hylemon, P.B. Bile salt biotransformations by human intestinal bacteria. J. Lipid Res. 2006, 47, 241–259. [Google Scholar] [CrossRef]
- Setchell, K.; Lawson, A.; Tanida, N.; Sjövall, J. General methods for the analysis of metabolic profiles of bile acids and related compounds in feces. J. Lipid Res. 1983, 24, 1085–1100. [Google Scholar] [PubMed]
- Winston, J.A.; Theriot, C.M. Impact of microbial derived secondary bile acids on colonization resistance against Clostridium difficile in the gastrointestinal tract. Anaerobe 2016, 41, 44–50. [Google Scholar] [CrossRef] [PubMed]
- Rea, M.C.; Dobson, A.; O’Sullivan, O.; Crispie, F.; Fouhy, F.; Cotter, P.D.; Shanahan, F.; Kiely, B.; Hill, C.; Ross, R.P. Effect of broad-and narrow-spectrum antimicrobials on Clostridium difficile and microbial diversity in a model of the distal colon. Proc. Natl. Acad. Sci. USA 2011, 108, 4639–4644. [Google Scholar] [CrossRef]
- Sorg, J.A.; Sonenshein, A.L. Bile salts and glycine as cogerminants for Clostridium difficile spores. J. Bacteriol. 2008, 190, 2505–2512. [Google Scholar] [CrossRef] [PubMed]
- Sorg, J.A.; Sonenshein, A.L. Inhibiting the initiation of Clostridium difficile spore germination using analogs of chenodeoxycholic acid, a bile acid. J. Bacteriol. 2010, 192, 4983–4990. [Google Scholar] [CrossRef] [PubMed]
- Allegretti, J.R.; Kearney, S.; Li, N.; Bogart, E.; Bullock, K.; Gerber, G.K.; Bry, L.; Clish, C.B.; Alm, E.; Korzenik, J. Recurrent Clostridium difficile infection associates with distinct bile acid and microbiome profiles. Aliment. Pharmacol. Ther. 2016, 43, 1142–1153. [Google Scholar] [CrossRef] [PubMed]
- Wilson, K.H.; Kennedy, M.J.; Fekety, F.R. Use of sodium taurocholate to enhance spore recovery on a medium selective for Clostridium difficile. J. Clin. Microbiol. 1982, 15, 443–446. [Google Scholar] [PubMed]
- Buggy, B.; Hawkins, C.; Fekety, R. Effect of adding sodium taurocholate to selective media on the recovery of Clostridium difficile from environmental surfaces. J. Clin. Microbiol. 1985, 21, 636–637. [Google Scholar] [PubMed]
- Wilson, K.H. Efficiency of various bile salt preparations for stimulation of Clostridium difficile spore germination. J. Clin. Microbiol. 1983, 18, 1017–1019. [Google Scholar] [PubMed]
- Seekatz, A.M.; Young, V.B. Clostridium difficile and the microbiota. J. Clin. Investig. 2014, 124, 4182–4189. [Google Scholar] [CrossRef]
- Thursby, E.; Juge, N. Introduction to the human gut microbiota. Biochem. J. 2017, 474, 1823–1836. [Google Scholar] [CrossRef]
- Engevik, M.A.; Engevik, K.A.; Yacyshyn, M.B.; Wang, J.; Hassett, D.J.; Darien, B.; Yacyshyn, B.R.; Worrell, R.T. Human Clostridium difficile infection: Inhibition of NHE3 and microbiota profile. Am. J. Physiol. Gastrointest. Liver Physiol. 2015, 308, G497–G509. [Google Scholar] [CrossRef]
- Dannheim, H.; Will, S.E.; Schomburg, D.; Neumann-Schaal, M. Clostridioides difficile 630Δerm in silico and in vivo–quantitative growth and extensive polysaccharide secretion. FEBS Open Biol. 2017, 7, 602–615. [Google Scholar] [CrossRef]
- Heinken, A.; Sahoo, S.; Fleming, R.M.; Thiele, I. Systems-level characterization of a host-microbe metabolic symbiosis in the mammalian gut. Gut Microbes 2013, 4, 28–40. [Google Scholar] [CrossRef]
- Heinken, A.; Khan, M.T.; Paglia, G.; Rodionov, D.A.; Harmsen, H.J.; Thiele, I. Functional metabolic map of Faecalibacterium prausnitzii, a beneficial human gut microbe. J. Bacteriol. 2014, 196, 3289–3302. [Google Scholar] [CrossRef]
- Baumler, D.J.; Peplinski, R.G.; Reed, J.L.; Glasner, J.D.; Perna, N.T. The evolution of metabolic networks of E. coli. BMC Syst. Biol. 2011, 5, 182. [Google Scholar] [CrossRef] [PubMed]
- Henson, M.A.; Phalak, P. Suboptimal community growth mediated through metabolite cross-feeding promotes species diversity in the gut microbiota. PLoS Comput. Biol. 2018, 14, e1006558. [Google Scholar] [CrossRef] [PubMed]
- Ouwerkerk, J.P.; de Vos, W.M.; Belzer, C. Glycobiome: Bacteria and mucus at the epithelial interface. Best Pract. Res. Clin. Gastroenterol. 2013, 27, 25–38. [Google Scholar] [CrossRef] [PubMed]
- Ross, C.L.; Spinler, J.K.; Savidge, T.C. Structural and functional changes within the gut microbiota and susceptibility to Clostridium difficile infection. Anaerobe 2016, 41, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Browne, H.P.; Forster, S.C.; Anonye, B.O.; Kumar, N.; Neville, B.A.; Stares, M.D.; Goulding, D.; Lawley, T.D. Culturing of ‘unculturable’human microbiota reveals novel taxa and extensive sporulation. Nature 2016, 533, 543–546. [Google Scholar] [CrossRef] [PubMed]
- Staley, C.; Weingarden, A.R.; Khoruts, A.; Sadowsky, M.J. Interaction of gut microbiota with bile acid metabolism and its influence on disease states. Appl. Microbiol. Biotechnol. 2017, 101, 47–64. [Google Scholar] [CrossRef] [PubMed]
- Myers, S.P. The causes of intestinal dysbiosis: A review. Altern. Med. Rev. 2004, 9, 180–197. [Google Scholar]
- Singh, R.K.; Chang, H.W.; Yan, D.; Lee, K.M.; Ucmak, D.; Wong, K.; Abrouk, M.; Farahnik, B.; Nakamura, M.; Zhu, T.H.; et al. Influence of diet on the gut microbiome and implications for human health. J. Transl. Med. 2017, 15, 73. [Google Scholar] [CrossRef]
- Kamada, N.; Chen, G.Y.; Inohara, N.; Núñez, G. Control of pathogens and pathobionts by the gut microbiota. Nat. Immunol. 2013, 14, 685–690. [Google Scholar] [CrossRef]
- Weingarden, A.R.; Chen, C.; Bobr, A.; Yao, D.; Lu, Y.; Nelson, V.M.; Sadowsky, M.J.; Khoruts, A. Microbiota transplantation restores normal fecal bile acid composition in recurrent Clostridium difficile infection. Am. J. Physiol. Gastrointest. Liver Physiol. 2013, 306, G310–G319. [Google Scholar] [CrossRef]
- Heinken, A.; Thiele, I. Systematic prediction of health-relevant human-microbial co-metabolism through a computational framework. Gut Microbes 2015, 6, 120–130. [Google Scholar] [CrossRef] [PubMed]
- Shoaie, S.; Nielsen, J. Elucidating the interactions between the human gut microbiota and its host through metabolic modeling. Front. Genet. 2014, 5, 86. [Google Scholar] [CrossRef]
- Thiele, I.; Heinken, A.; Fleming, R.M. A systems biology approach to studying the role of microbes in human health. Curr. Opin. Biotechnol. 2013, 24, 4–12. [Google Scholar] [CrossRef] [PubMed]
- Glick-Bauer, M.; Yeh, M.C. The health advantage of a vegan diet: Exploring the gut microbiota connection. Nutrients 2014, 6, 4822–4838. [Google Scholar] [CrossRef] [PubMed]
- Swainston, N.; Mendes, P.; Kell, D.B. An analysis of a ‘community-driven’reconstruction of the human metabolic network. Metabolomics 2013, 9, 757–764. [Google Scholar] [CrossRef] [PubMed]
- Thiele, I.; Swainston, N.; Fleming, R.M.; Hoppe, A.; Sahoo, S.; Aurich, M.K.; Haraldsdottir, H.; Mo, M.L.; Rolfsson, O.; Stobbe, M.D.; et al. A community-driven global reconstruction of human metabolism. Nat. Biotechnol. 2013, 31, 419. [Google Scholar] [CrossRef] [PubMed]
- Thiele, I.; Sahoo, S.; Heinken, A.; Heirendt, L.; Aurich, M.K.; Noronha, A.; Fleming, R.M. When metabolism meets physiology: Harvey and Harvetta. bioRxiv 2018. [Google Scholar] [CrossRef]
- Dai, Z.L.; Wu, G.; Zhu, W.Y. Amino acid metabolism in intestinal bacteria: Links between gut ecology and host health. Front. Biosci. 2011, 16, 1768–1786. [Google Scholar] [CrossRef]
- Horn, H.; Lackner, S. Modeling of biofilm systems: A review. In Productive Biofilms; Springer: Cham, Swizterland, 2014; pp. 53–76. [Google Scholar]
- Lawley, T.D.; Clare, S.; Walker, A.W.; Stares, M.D.; Connor, T.R.; Raisen, C.; Goulding, D.; Rad, R.; Schreiber, F.; Brandt, C.; et al. Targeted restoration of the intestinal microbiota with a simple, defined bacteriotherapy resolves relapsing Clostridium difficile disease in mice. PLoS Pathog. 2012, 8, e1002995. [Google Scholar] [CrossRef]
- Chen, J.; Gomez, J.A.; Höffner, K.; Phalak, P.; Barton, P.I.; Henson, M.A. Spatiotemporal modeling of microbial metabolism. BMC Syst. Biol. 2016, 10, 21. [Google Scholar] [CrossRef]
- Phalak, P.; Chen, J.; Carlson, R.P.; Henson, M.A. Metabolic modeling of a chronic wound biofilm consortium predicts spatial partitioning of bacterial species. BMC Syst. Biol. 2016, 10, 90. [Google Scholar] [CrossRef] [PubMed]
- Gomez, J.A.; Hoffner, K.; Barton, P.I. DFBAlab: A fast and reliable MATLAB code for dynamic flux balance analysis. BMC Bioinform. 2014, 15, 409. [Google Scholar] [CrossRef] [PubMed]
- Meadows, A.L.; Karnik, R.; Lam, H.; Forestell, S.; Snedecor, B. Application of dynamic flux balance analysis to an industrial Escherichia coli fermentation. Metab. Eng. 2010, 12, 150–160. [Google Scholar] [CrossRef] [PubMed]
- Stewart, P.S. A review of experimental measurements of effective diffusive permeabilities and effective diffusion coefficients in biofilms. Biotechnol. Bioeng. 1998, 59, 261–272. [Google Scholar] [CrossRef]
- Stewart, P.S. Diffusion in biofilms. J. Bacteriol. 2003, 185, 1485–1491. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).