Metabolic and Evolutionary Engineering of Food Yeasts
Abstract
1. Introduction
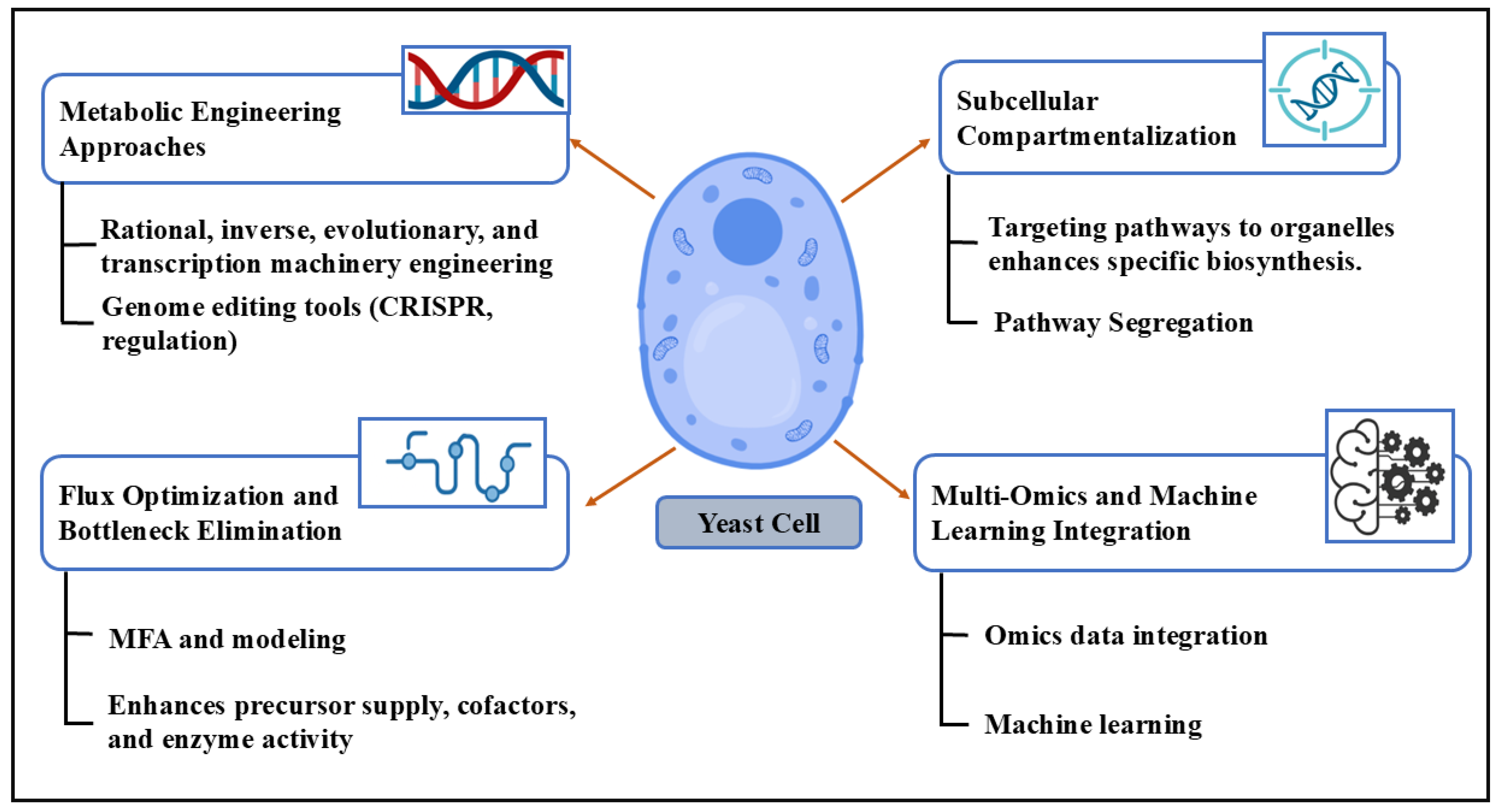
2. Metabolic Engineering
2.1. Gene Editing
| Strategy | Description | Applications | References |
|---|---|---|---|
| Gene Overexpression | Amplifying rate-limiting enzymes to increase metabolite flux | Enhanced ester or flavor compound production in S. cerevisiae | [10,12] |
| Gene Deletion/Knockout | Removing genes that divert or degrade target compounds | Deleting competitive pathways to improve ethanol yield | [11] |
| Heterologous Pathway Introduction | Incorporating metabolic pathways from other organisms | Producing novel flavors or bioactive compounds | [12] |
| CRISPR-Cas9 Gene Editing | Using RNA-guided endonucleases for precise genome modifications | Enhancing stress tolerance, flavor production, and substrate utilization | [13,16] |
| Pathway Compartmentalization | Targeting pathways to specific organelles to boost efficiency | Directing ester synthesis to mitochondria or peroxisomes | [22] |
| Adaptive Laboratory Evolution | Evolving strains under selective pressures to improve traits | Developing yeast strains tolerating high ethanol or temperature | [21] |
2.2. Pathway Optimization
3. Evolutionary Engineering of Food Yeast
3.1. Adaptive Evolution
3.2. Genome-Wide Mutations
4. Different Approaches
4.1. Targeted Genetic Modifications
4.2. Directed Evolution
4.3. Synthetic Biology
5. Current State-of-the-Art Technologies and Advancements
5.1. CRISPR-Cas9 Technology
5.2. Machine Learning
5.3. Metabolic Modeling
5.4. Non-Conventional Yeasts
| Application Area | Specific Traits Enhanced | Approach | References |
|---|---|---|---|
| Flavor Compound Production | Increased ester, acid, and antioxidant production | Overexpressing flavor biosynthesis genes, CRISPR-Cas9 modifications | [1,19] |
| Substrate Utilization | Ability to ferment complex carbohydrates like starch and xylose | Introducing amylolytic or xylose metabolism genes | [17] |
| Stress Tolerance | Tolerance to heat, ethanol, osmotic, and lignocellulosic inhibitors | Enhancing heat shock protein genes, modulating glycerol synthesis | [18] |
| Cell Wall Engineering | Improved flocculation, stress resistance, and enzyme accessibility | Modifying glycosylation patterns via CRISPR | [13] |
| Strain Hybridization | Combining desirable traits of S. cerevisiae and non-Saccharomyces yeasts | Intergeneric genome shuffling and adaptive evolution | [20,21] |
| Synthetic Genome Construction | Creating yeast strains with synthetic DNA for tailored traits | Developing strains with >50% synthetic genome | [19] |
6. Technological Challenges
6.1. Genetic Complexity
6.2. Strain Stability
6.3. Regulatory Hurdles
7. Future Prospects
7.1. Sustainable Production
7.2. Food Innovation
7.3. Pharmaceuticals
7.4. Emerging Directions and Technologies
8. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Chai, K.F.; Ng, K.R.; Samarasiri, M.; Chen, W.N. Precision fermentation to advance fungal food fermentations. Curr. Opin. Food Sci. 2022, 47, 100881. [Google Scholar] [CrossRef]
- Graham, A.E.; Ledesma-Amaro, R. The microbial food revolution. Nat. Commun. 2023, 14, 2231. [Google Scholar] [CrossRef]
- Teng, T.S.; Chin, Y.L.; Chai, K.F.; Chen, W.N. Fermentation for future food systems: Precision fermentation can complement the scope and applications of traditional fermentation. EMBO Rep. 2021, 22, e52680. [Google Scholar] [CrossRef] [PubMed]
- Tekarslan-Sahin, S.H. Metabolic Engineering of Saccharomyces cerevisiae for Industrial Biotechnology; IntechOpen: London, UK, 2021. [Google Scholar]
- Yao, Z.; Wang, Q.; Dai, Z.J. Recent advances in directed yeast genome evolution. J. Fungi 2022, 8, 635. [Google Scholar] [CrossRef]
- Chen, Y.; Li, F.; Nielsen, J. Genome-scale modeling of yeast metabolism: Retrospectives and perspectives. FEMS Yeast Res. 2022, 22, foac003. [Google Scholar] [CrossRef]
- Cao, C.; Gao, J.; Zhu, B.; Zhou, Y.J. Engineering yeast for bio-production of food ingredients. Syst. Microbiol. Biomanuf. 2023, 3, 2–11. [Google Scholar] [CrossRef]
- Gong, G.; Wu, B.; Liu, L.; Li, J.; He, M. Engineering oleaginous red yeasts as versatile chassis for the production of olechemicals and valuable compounds: Current advances and perspectives. Biotechnol. Adv. 2024, 76, 108432. [Google Scholar] [CrossRef]
- Patané, A.; Jansen, G.; Conca, P.; Carapezza, G.; Costanza, J.; Nicosia, G. Multi-objective optimization of genome-scale metabolic models: The case of ethanol production. Ann. Oper. Res. 2019, 276, 211–227. [Google Scholar] [CrossRef]
- Hilgendorf, K.; Wang, Y.; Miller, M.J.; Jin, Y.-S. Precision fermentation for improving the quality, flavor, safety, and sustainability of foods. Curr. Opin. Biotechnol. 2024, 86, 103084. [Google Scholar] [CrossRef]
- Seo, S.-O.; Jin, Y.-S. Next-generation genetic and fermentation technologies for safe and sustainable production of food ingredients: Colors and flavorings. Annu. Rev. Food Sci. Technol. 2022, 13, 463–488. [Google Scholar] [CrossRef]
- Kallscheuer, N. Engineered microorganisms for the production of food additives approved by the European Union—A systematic analysis. Front. Microbiol. 2018, 9, 1746. [Google Scholar] [CrossRef]
- Pan, M.; Barrangou, R.J. Combining omics technologies with CRISPR-based genome editing to study food microbes. Curr. Opin. Biotechnol. 2020, 61, 198–208. [Google Scholar] [CrossRef] [PubMed]
- Patra, P.; Disha, B.; Kundu, P.; Das, M.; Ghosh, A. Recent advances in machine learning applications in metabolic engineering. Biotechnol. Adv. 2023, 62, 108069. [Google Scholar] [CrossRef] [PubMed]
- Steensels, J.; Snoek, T.; Meersman, E.; Nicolino, M.P.; Voordeckers, K.; Verstrepen, K.J. Improving industrial yeast strains: Exploiting natural and artificial diversity. FEMS Microbiol. Rev. 2014, 38, 947–995. [Google Scholar] [CrossRef]
- Costa, J.M.R.; Rodrigues, J.L.; Rodrigues, L.R. CRISPR-Cas9: A powerful tool to efficiently engineer Saccharomyces cerevisiae. Life 2020, 11, 13. [Google Scholar] [CrossRef]
- Ansori, A.N.; Antonius, Y.; Susilo, R.J.; Hayaza, S.; Kharisma, V.D.; Parikesit, A.A.; Zainul, R.; Jakhmola, V.; Saklani, T.; Rebezov, M.J.N.J. Application of CRISPR-Cas9 genome editing technology in various fields: A review. Narra J. 2023, 3, e184. [Google Scholar] [CrossRef] [PubMed]
- Pilap, W.; Thanonkeo, S.; Klanrit, P.; Thanonkeo, P. The potential of multistress tolerant yeast, Saccharomycodes ludwigii, for second-generation bioethanol production. Sci. Rep. 2022, 12, 22062. [Google Scholar] [CrossRef]
- Bourzac, K. Engineered yeast breaks new record: A genome with over 50% synthetic DNA. Nature, 2023; online ahead of print. [Google Scholar]
- Cernak, P.; Estrela, R.; Poddar, S.; Skerker, J.M.; Cheng, Y.-F.; Carlson, A.K.; Chen, B.; Glynn, V.M.; Furlan, M.; Ryan, O.W. Engineering Kluyveromyces marxianus as a robust synthetic biology platform host. mBio 2018, 9, e01410-18. [Google Scholar] [CrossRef]
- Lin, Y.; Cai, Y.; Guo, Y.; Li, X.; Qi, X.; Qi, Q.; Wang, Q. Development and genomic elucidation of hybrid yeast with improved glucose-xylose co-fermentation at high temperature. FEMS Yeast Res. 2019, 19, foz015. [Google Scholar] [CrossRef]
- Choi, B.H.; Kang, H.J.; Kim, S.C.; Lee, P.C. Organelle engineering in yeast: Enhanced production of protopanaxadiol through manipulation of peroxisome proliferation in Saccharomyces cerevisiae. Microorganisms 2022, 10, 650. [Google Scholar] [CrossRef]
- Yu, A.-Q.; Juwono, N.K.P.; Foo, J.L.; Leong, S.S.J.; Chang, M.W. Metabolic engineering of Saccharomyces cerevisiae for the overproduction of short branched-chain fatty acids. Metab. Eng. 2016, 34, 36–43. [Google Scholar] [CrossRef] [PubMed]
- Jojima, T.; Inui, M. Engineering the glycolytic pathway: A potential approach for improvement of biocatalyst performance. Bioengineered 2015, 6, 328–334. [Google Scholar] [CrossRef]
- Wu, Z.; Liang, X.; Li, M.; Ma, M.; Zheng, Q.; Li, D.; An, T.; Wang, G. Advances in the optimization of central carbon metabolism in metabolic engineering. Microb. Cell Factories 2023, 22, 76. [Google Scholar] [CrossRef]
- Selma, S.; Ntelkis, N.; Nguyen, T.H.; Goossens, A. Engineering the plant metabolic system by exploiting metabolic regulation. Plant J. 2023, 114, 1149–1163. [Google Scholar] [CrossRef] [PubMed]
- Radivojević, T.; Costello, Z.; Workman, K.; Garcia Martin, H. A machine learning Automated Recommendation Tool for synthetic biology. Nat. Commun. 2020, 11, 4879. [Google Scholar] [CrossRef]
- Yuan, W.; Du, Y.; Yu, K.; Xu, S.; Liu, M.; Wang, S.; Yang, Y.; Zhang, Y.; Sun, J. The production of pyruvate in biological technology: A critical review. Microorganisms 2022, 10, 2454. [Google Scholar] [CrossRef]
- Dakal, T.C.; Xu, C.; Kumar, A. Advanced Computational Tools, Artificial Intelligence and Machine-learning Approaches in Gut Microbiota and Biomarker Identification. Front. Med. Technol. 2025, 6, 1434799. [Google Scholar] [CrossRef]
- Kang, W.; Ma, T.; Liu, M.; Qu, J.; Liu, Z.; Zhang, H.; Shi, B.; Fu, S.; Ma, J.; Lai, L.T.F. Modular enzyme assembly for enhanced cascade biocatalysis and metabolic flux. Nat. Commun. 2019, 10, 4248. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.Y.; Park, J.M.; Kim, T.Y.; Yun, H.; Lee, S.Y. The genome-scale metabolic network analysis of Zymomonas mobilis ZM4 explains physiological features and suggests ethanol and succinic acid production strategies. Microb. Cell Factories 2010, 9, 94. [Google Scholar] [CrossRef]
- Shi, H.; Shimizu, K. On-line metabolic pathway analysis based on metabolic signal flow diagram. Biotechnol. Bioeng. 1998, 58, 139–148. [Google Scholar] [CrossRef]
- Chandrawanshi, V.; Kulkarni, R.; Prabhu, A.; Mehra, S.J. Enhancing titers and productivity of rCHO clones with a combination of an optimized fed-batch process and ER-stress adaptation. J. Biotechnol. 2020, 311, 49–58. [Google Scholar] [CrossRef] [PubMed]
- Metallo, C.M.; Vander Heiden, M.G. Understanding metabolic regulation and its influence on cell physiology. Mol. Cell 2013, 49, 388–398. [Google Scholar] [CrossRef] [PubMed]
- Bro, C.; Regenberg, B.; Förster, J.; Nielsen, J. In silico aided metabolic engineering of Saccharomyces cerevisiae for improved bioethanol production. Metab. Eng. 2006, 8, 102–111. [Google Scholar] [CrossRef]
- Malina, C.; Di Bartolomeo, F.; Kerkhoven, E.J.; Nielsen, J. Constraint-based modeling of yeast mitochondria reveals the dynamics of protein import and iron-sulfur cluster biogenesis. iScience 2021, 24, 103294. [Google Scholar] [CrossRef]
- Guo, W.; Feng, X. OM-FBA: Integrate transcriptomics data with flux balance analysis to decipher the cell metabolism. PLoS ONE 2016, 11, e0154188. [Google Scholar] [CrossRef]
- Milne, C.B.; Kim, P.J.; Eddy, J.A.; Price, N.D. Accomplishments in genome-scale in silico modeling for industrial and medical biotechnology. Biotechnol. J. 2009, 4, 1653–1670. [Google Scholar] [CrossRef] [PubMed]
- Hong, Y.; Nguyen, T.; Arbter, P.; Utesch, T.; Zeng, A.P. Phenotype analysis of cultivation processes via unsupervised machine learning: Demonstration for Clostridium pasteurianum. Eng. Life Sci. 2022, 22, 85–99. [Google Scholar] [CrossRef]
- Lawson, C.E.; Martí, J.M.; Radivojevic, T.; Jonnalagadda, S.V.R.; Gentz, R.; Hillson, N.J.; Peisert, S.; Kim, J.; Simmons, B.A.; Petzold, C.J. Machine learning for metabolic engineering: A review. Metab. Eng. 2021, 63, 34–60. [Google Scholar] [CrossRef]
- Yildiran, H.; Başyiğit Kiliç, G.; Karahan Çakmakçi, A.G. Characterization and comparison of yeasts from different sources for some probiotic properties and exopolysaccharide production. Food Sci. Technol. 2019, 39, 646–653. [Google Scholar] [CrossRef]
- Vanderwaeren, L.; Dok, R.; Voordeckers, K.; Nuyts, S.; Verstrepen, K.J. Saccharomyces cerevisiae as a model system for eukaryotic cell biology, from cell cycle control to DNA damage response. Int. J. Mol. Sci. 2022, 23, 11665. [Google Scholar] [CrossRef]
- Parapouli, M.; Vasileiadis, A.; Afendra, A.-S.; Hatziloukas, E. Saccharomyces cerevisiae and its industrial applications. AIMS Microbiol. 2020, 6, 1. [Google Scholar] [CrossRef] [PubMed]
- Hanlon, P.; Sewalt, V. GEMs: Genetically engineered microorganisms and the regulatory oversight of their uses in modern food production. Crit. Rev. Food Sci. Nutr. 2021, 61, 959–970. [Google Scholar] [CrossRef] [PubMed]
- Chemler, J.A.; Yan, Y.; Koffas, M.A. Biosynthesis of isoprenoids, polyunsaturated fatty acids and flavonoids in Saccharomyces cerevisiae. Microb. Cell Factories 2006, 5, 20. [Google Scholar] [CrossRef]
- Belda, I.; Williams, T.C.; de Celis, M.; Paulsen, I.T.; Pretorius, I.S. Seeding the idea of encapsulating a representative synthetic metagenome in a single yeast cell. Nat. Commun. 2021, 12, 1599. [Google Scholar] [CrossRef]
- Shomar, H.; Bokinsky, G. Towards a synthetic biology toolset for metallocluster enzymes in biosynthetic pathways: What we know and what we need. Molecules 2021, 26, 6930. [Google Scholar] [CrossRef]
- Mezzetti, F.; Fay, J.C.; Giudici, P.; De Vero, L. Genetic variation and expression changes associated with molybdate resistance from a glutathione producing wine strain of Saccharomyces cerevisiae. PLoS ONE 2017, 12, e0180814. [Google Scholar] [CrossRef]
- Giersch, R.M.; Finnigan, G.C. Yeast still a beast: Diverse applications of CRISPR/Cas editing technology in S. cerevisiae. Yale J. Biol. Med. 2017, 90, 643. [Google Scholar]
- Lian, J.; HamediRad, M.; Zhao, H. Advancing metabolic engineering of Saccharomyces cerevisiae using the CRISPR/Cas system. Biotechnol. J. 2018, 13, 1700601. [Google Scholar] [CrossRef]
- Sander, J.D.; Joung, J.K. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat. Biotechnol. 2014, 32, 347–355. [Google Scholar] [CrossRef]
- Zhang, W.; Nielsen, D.R. Synthetic biology applications in industrial microbiology. Front. Microbiol. 2014, 5, 451. [Google Scholar] [CrossRef]
- McDaniel, E.A.; Wahl, S.A.; Ishii, S.i.; Pinto, A.; Ziels, R.; Nielsen, P.H.; McMahon, K.D.; Williams, R.B. Prospects for multi-omics in the microbial ecology of water engineering. Water Res. 2021, 205, 117608. [Google Scholar] [CrossRef] [PubMed]
- Meng, J.; Qiu, Y.; Shi, S. CRISPR/Cas9 systems for the development of Saccharomyces cerevisiae cell factories. Front. Bioeng. Biotechnol. 2020, 8, 594347. [Google Scholar] [CrossRef]
- Jensen, M.K.; Keasling, J.D. Recent applications of synthetic biology tools for yeast metabolic engineering. FEMS Yeast Res. 2015, 15, 1–10. [Google Scholar] [CrossRef]
- Saha, T.R.; Kang, N.K.; Lee, E.Y. Advanced metabolic Engineering strategies for the sustainable production of free fatty acids and their derivatives using yeast. J. Biol. Eng. 2024, 18, 73. [Google Scholar] [CrossRef]
- Jiang, W.; Li, C.; Li, Y.; Peng, H. Metabolic engineering strategies for improved lipid production and cellular physiological responses in yeast Saccharomyces cerevisiae. J. Fungi 2022, 8, 427. [Google Scholar] [CrossRef] [PubMed]
- Rainha, J.; Gomes, D.; Rodrigues, L.R.; Rodrigues, J.L. Synthetic biology approaches to engineer Saccharomyces cerevisiae towards the industrial production of valuable polyphenolic compounds. Life 2020, 10, 56. [Google Scholar] [CrossRef]
- Malcı, K.; Walls, L.E.; Rios-Solis, L. Multiplex genome engineering methods for yeast cell factory development. Front. Bioeng. Biotechnol. 2020, 8, 589468. [Google Scholar] [CrossRef] [PubMed]
- Shi, S.; Chen, Y.; Nielsen, J. Metabolic Engineering of Yeast. Annu. Rev. Biophys. 2025, 54, 101–120. [Google Scholar] [CrossRef]
- Lacerda, M.P.; Oh, E.J.; Eckert, C. The model system Saccharomyces cerevisiae versus emerging non-model yeasts for the production of biofuels. Life 2020, 10, 299. [Google Scholar] [CrossRef]
- Liu, Z.; Moradi, H.; Shi, S.; Darvishi, F. Yeasts as microbial cell factories for sustainable production of biofuels. Renew. Sustain. Energy Rev. 2021, 143, 110907. [Google Scholar] [CrossRef]
- Sahoo, A.; Das, P.K.; Patra, S.; Veeranki, V.D. Engineered yeasts for the production of biofuel and platform chemicals. In Advances in Yeast Biotechnology for Biofuels and Sustainability; Elsevier: Amsterdam, The Netherlands, 2023; pp. 21–46. [Google Scholar]
- Liu, Z.; Wang, J.; Nielsen, J. Yeast synthetic biology advances biofuel production. Curr. Opin. Microbiol. 2022, 65, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Yook, S.; Alper, H.S. Recent advances in genetic engineering and chemical production in yeast species. FEMS Yeast Res. 2025, 25, foaf009. [Google Scholar] [CrossRef] [PubMed]
- Taghon, G.J.; Strychalski, E.A. Rise of synthetic yeast: Charting courses to new applications. Cell Genom. 2023, 3, 100438. [Google Scholar] [CrossRef]
- Schindler, D. Genetic engineering and synthetic genomics in yeast to understand life and boost biotechnology. Bioengineering 2020, 7, 137. [Google Scholar] [CrossRef]
- Liu, Z.; Zhang, Y.; Nielsen, J. Synthetic biology of yeast. Biochemistry 2019, 58, 1511–1520. [Google Scholar] [CrossRef]
- Thak, E.J.; Yoo, S.J.; Moon, H.Y.; Kang, H.A. Yeast synthetic biology for designed cell factories producing secretory recombinant proteins. FEMS Yeast Res. 2020, 20, foaa009. [Google Scholar] [CrossRef]
- Ma, J.; Gu, Y.; Marsafari, M.; Xu, P. Synthetic biology, systems biology, and metabolic engineering of Yarrowia lipolytica toward a sustainable biorefinery platform. J. Ind. Microbiol. Biotechnol. 2020, 47, 845–862. [Google Scholar] [CrossRef]
- Shi, S.; Zhao, H.J. Metabolic engineering of oleaginous yeasts for production of fuels and chemicals. Front. Microbiol. 2017, 8, 2185. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Wu, L.; Guo, S.; Cao, W.; Ma, W.; Wang, X.; Shen, J.; Wang, M.; Zhang, Q.; Huang, M. Metabolic engineering of yeast for the production of carbohydrate-derived foods and chemicals from C1–3 molecules. Nat. Catal. 2024, 7, 21–34. [Google Scholar] [CrossRef]
- Li, T.; Yang, Y.; Qi, H.; Cui, W.; Zhang, L.; Fu, X.; He, X.; Liu, M.; Li, P.-F.; Yu, T.; et al. CRISPR/Cas9 therapeutics: Progress and prospects. Signal Transduct. Target. Ther. 2023, 8, 36. [Google Scholar] [CrossRef]
- Chettri, D.; Verma, A.K.; Verma, A.K. Past, present and future of yeast engineering. In Advances in Yeast Biotechnology for Biofuels and Sustainability; Elsevier: Amsterdam, The Netherlands, 2023; pp. 3–20. [Google Scholar]
- Stovicek, V.; Holkenbrink, C.; Borodina, I. CRISPR/Cas system for yeast genome engineering: Advances and applications. FEMS Yeast Res. 2017, 17, fox030. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Lage, M.; Torres-Ruiz, R.; Rodriguez-Perales, S. CRISPR/Cas9 technology: Applications and human disease modeling. Prog. Mol. Biol. Transl. Sci. 2017, 152, 23–48. [Google Scholar]
- Li, S.; Zhang, C.; Li, J.; Yan, L.; Wang, N.; Xia, L. Present and future prospects for wheat improvement through genome editing and advanced technologies. Plant Commun. 2021, 2, 100211. [Google Scholar] [CrossRef]
- Das, J.; Kumar, S.; Mishra, D.C.; Chaturvedi, K.K.; Paul, R.K.; Kairi, A. Machine learning in the estimation of CRISPR-Cas9 cleavage sites for plant system. Front. Genet. 2023, 13, 1085332. [Google Scholar] [CrossRef]
- Scheben, A.; Wolter, F.; Batley, J.; Puchta, H.; Edwards, D. Towards CRISPR/Cas crops–bringing together genomics and genome editing. New Phytol. 2017, 216, 682–698. [Google Scholar] [CrossRef] [PubMed]
- Topaloğlu, A.; Esen, Ö.; Turanlı-Yıldız, B.; Arslan, M.; Çakar, Z.P. From Saccharomyces cerevisiae to ethanol: Unlocking the power of evolutionary engineering in metabolic engineering applications. J. Fungi 2023, 9, 984. [Google Scholar] [CrossRef] [PubMed]
- Lian, J.; Mishra, S.; Zhao, H.J. Recent advances in metabolic engineering of Saccharomyces cerevisiae: New tools and their applications. Metab. Eng. 2018, 50, 85–108. [Google Scholar] [CrossRef]
- Song, Y.; Prather, K.L. Strategies in engineering sustainable biochemical synthesis through microbial systems. Curr. Opin. Chem. Biol. 2024, 81, 102493. [Google Scholar] [CrossRef]
- Sarker, I.H. Machine learning: Algorithms, real-world applications and research directions. SN Comput. Sci. 2021, 2, 160. [Google Scholar] [CrossRef]
- Cavallaro, C.; Cutello, V.; Pavone, M.; Zito, F. Machine Learning and Genetic Algorithms: A case study on image reconstruction. Knowl.-Based Syst. 2024, 284, 111194. [Google Scholar] [CrossRef]
- Bottou, L.; Curtis, F.E.; Nocedal, J. Optimization methods for large-scale machine learning. SIAM Rev. 2018, 60, 223–311. [Google Scholar] [CrossRef]
- Boob, A.G.; Chen, J.; Zhao, H. Enabling pathway design by multiplex experimentation and machine learning. Metab. Eng. 2024, 81, 70–87. [Google Scholar] [CrossRef]
- Merzbacher, C.; Oyarzún, D.A. Applications of artificial intelligence and machine learning in dynamic pathway engineering. Biochem. Soc. Trans. 2023, 51, 1871–1879. [Google Scholar] [CrossRef]
- Bai, L.; You, Q.; Zhang, C.; Sun, J.; Liu, L.; Lu, H.; Chen, Q. Advances and applications of machine learning and intelligent optimization algorithms in genome-scale metabolic network models. Syst. Microbiol. Biomanuf. 2023, 3, 193–206. [Google Scholar] [CrossRef]
- Lu, R.; Shi, T.-Q.; Lin, L.; Ledesma-Amaro, R.; Ji, X.-J.; Huang, H. Advances in metabolic engineering of yeasts for the production of fatty acid-derived hydrocarbon fuels. Green Chem. Eng. 2022, 3, 289–303. [Google Scholar] [CrossRef]
- Sánchez, B.J.; Zhang, C.; Nilsson, A.; Lahtvee, P.J.; Kerkhoven, E.J.; Nielsen, J. Improving the phenotype predictions of a yeast genome-scale metabolic model by incorporating enzymatic constraints. Mol. Syst. Biol. 2017, 13, 935. [Google Scholar] [CrossRef] [PubMed]
- Timouma, S.; Balarezo-Cisneros, L.N.; Schwartz, J.-M.; Delneri, D. Development of a genome-scale metabolic model for the lager hybrid yeast S. pastorianus to understand the evolution of metabolic pathways in industrial settings. Msystems 2024, 9, e00424–e00429. [Google Scholar] [CrossRef]
- Elsemman, I.E.; Rodriguez Prado, A.; Grigaitis, P.; Garcia Albornoz, M.; Harman, V.; Holman, S.W.; van Heerden, J.; Bruggeman, F.J.; Bisschops, M.M.; Sonnenschein, N. Whole-cell modeling in yeast predicts compartment-specific proteome constraints that drive metabolic strategies. Nat. Commun. 2022, 13, 801. [Google Scholar] [CrossRef]
- Qian, Z.; Song, L.; Liu, Q.; Gong, X.; Kang, Y.; He, Z.; Long, H.; Cai, M. Biosynthesis of natural products by non-conventional yeasts. Sheng Wu Gong Cheng Xue Bao 2023, 39, 2284–2312. [Google Scholar]
- Rodriguez-Ocasio, E.; Khalid, A.; Truka, C.J.; Blenner, M.A.; Jarboe, L.R. Survey of nonconventional yeasts for lipid and hydrocarbon biotechnology. J. Ind. Microbiol. Biotechnol. 2022, 49, kuac010. [Google Scholar] [CrossRef]
- Gu, Y.; Lu, X.; Liu, T.; Song, Y.; Sang, E.; Ding, S.; Liu, L.; Xue, F.; Xu, P. Engineering the oleaginous yeast Yarrowia lipolytica to produce nutraceuticals: From metabolic design to industrial applications. Food Bioeng. 2023, 2, 187–199. [Google Scholar] [CrossRef]
- Solieri, L. The revenge of Zygosaccharomyces yeasts in food biotechnology and applied microbiology. World J. Microbiol. Biotechnol. 2021, 37, 96. [Google Scholar] [CrossRef]
- Binati, R.L.; Salvetti, E.; Bzducha-Wróbel, A.; Bašinskienė, L.; Čižeikienė, D.; Bolzonella, D.; Felis, G.E. Non-conventional yeasts for food and additives production in a circular economy perspective. FEMS Yeast Res. 2021, 21, foab052. [Google Scholar] [CrossRef] [PubMed]
- Sibirny, A.A. Metabolic engineering of non-conventional yeasts for construction of the advanced producers of biofuels and high-value chemicals. BBA Adv. 2023, 3, 100071. [Google Scholar] [CrossRef] [PubMed]
- Geijer, C.; Ledesma-Amaro, R.; Tomás-Pejó, E. Unraveling the potential of non-conventional yeasts in biotechnology. FEMS Yeast Res. 2022, 22, foab071. [Google Scholar] [CrossRef]
- Navarrete, C.; Martínez, J.L. Non-conventional yeasts as superior production platforms for sustainable fermentation based bio-manufacturing processes. AIMS Bioeng. 2020, 7, 289–305. [Google Scholar] [CrossRef]
- Park, J.; Kim, I.J.; Kim, S.R. Nonconventional yeasts engineered using the CRISPR-cas system as emerging microbial cell factories. Fermentation 2022, 8, 656. [Google Scholar] [CrossRef]
- Löbs, A.-K.; Schwartz, C.; Wheeldon, I. Genome and metabolic engineering in non-conventional yeasts: Current advances and applications. Synth. Syst. Biotechnol. 2017, 2, 198–207. [Google Scholar] [CrossRef]
- Saerens, S.M.; Duong, C.T.; Nevoigt, E. Genetic improvement of brewer’s yeast: Current state, perspectives and limits. Appl. Microbiol. Biotechnol. 2010, 86, 1195–1212. [Google Scholar] [CrossRef]
- Bro, C.; Regenberg, B.; Nielsen, J. Yeast functional genomics and metabolic engineering: Past, present and future. In Functional Genetics of Industrial Yeasts; Springer: Berlin/Heidelberg, Germany, 2003; pp. 331–360. [Google Scholar]
- Xia, Y.; Li, Y.; Shen, W.; Yang, H.; Chen, X. CRISPR-Cas technology for bioengineering conventional and non-conventional yeasts: Progress and new challenges. Int. J. Mol. Sci. 2023, 24, 15310. [Google Scholar] [CrossRef]
- Yuan, B.; Wang, W.-B.; Wang, Y.-T.; Zhao, X.-Q. Regulatory mechanisms underlying yeast chemical stress response and development of robust strains for bioproduction. Curr. Opin. Biotechnol. 2024, 86, 103072. [Google Scholar] [CrossRef] [PubMed]
- Peng, B.; Wood, R.J.; Nielsen, L.K.; Vickers, C.E. An expanded heterologous GAL promoter collection for diauxie-inducible expression in Saccharomyces cerevisiae. ACS Synth. Biol. 2018, 7, 748–751. [Google Scholar] [CrossRef]
- D’ambrosio, V.; Dore, E.; Di Blasi, R.; van den Broek, M.; Sudarsan, S.; Ter Horst, J.; Ambri, F.; Sommer, M.O.; Rugbjerg, P.; Keasling, J.D. Regulatory control circuits for stabilizing long-term anabolic product formation in yeast. Metab. Eng. 2020, 61, 369–380. [Google Scholar] [CrossRef]
- Xu, X.; Liu, C.; Niu, C.; Wang, J.; Zheng, F.; Li, Y.; Li, Q. Rationally designed perturbation factor drives evolution in Saccharomyces cerevisiae for industrial application. J. Ind. Microbiol. Biotechnol. 2018, 45, 869–880. [Google Scholar] [CrossRef]
- Gowrishankar, J. Regulation of genetically modified organisms: Has the time come to amend the law? Curr. Sci. 2009, 96, 1574. [Google Scholar]
- Mirsalami, S.M.; Mirsalami, M. Advances in genetically engineered microorganisms: Transforming food production through precision fermentation and synthetic biology. Futur. Foods 2025, 11, 100601. [Google Scholar] [CrossRef]
- Bratlie, S.; Halvorsen, K.; Myskja, B.K.; Mellegård, H.; Bjorvatn, C.; Frost, P.; Heiene, G.; Hofmann, B.; Holst-Jensen, A.; Holst-Larsen, T. A novel governance framework for GMO: A tiered, more flexible regulation for GMO s would help to stimulate innovation and public debate. EMBO Rep. 2019, 20, e47812. [Google Scholar] [CrossRef] [PubMed]
- Peng, B.; Plan, M.R.; Carpenter, A.; Nielsen, L.K.; Vickers, C.E. Coupling gene regulatory patterns to bioprocess conditions to optimize synthetic metabolic modules for improved sesquiterpene production in yeast. Biotechnol. Biofuels 2017, 10, 43. [Google Scholar] [CrossRef] [PubMed]
- Hammer, S.K.; Avalos, J.L. Harnessing yeast organelles for metabolic engineering. Nat. Chem. Biol. 2017, 13, 823–832. [Google Scholar] [CrossRef]
- Mamta, S.; Supriya, A.; Devi, L.; Nagma, K.; Chandana, B.; Verma, S.K. Biosafety Concerns and Regulatory Framework for Transgenics. Res. J. Agric. For. Sci. 2014, 2, 7–13. [Google Scholar]
- Lee, D.; Chen, A.; Nair, R. Genetically engineered crops for biofuel production: Regulatory perspectives. Biotechnol. Genet. Eng. Rev. 2008, 25, 331–362. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Hussain, A.; Manghwar, H.; Xie, K.; Xie, S.; Zhao, S.; Larkin, R.M.; Qing, P.; Jin, S.; Ding, F. Genome editing with the CRISPR-Cas system: An art, ethics and global regulatory perspective. Plant Biotechnol. J. 2020, 18, 1651–1669. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Hoang Nguyen Tran, P.; Lee, S.-M. Current challenges and opportunities in non-native chemical production by engineered yeasts. Front. Bioeng. Biotechnol. 2020, 8, 594061. [Google Scholar] [CrossRef]
- Turanlı-Yıldız, B.; Hacısalihoğlu, B.; Çakar, Z.P. Advances in Metabolic Engineering of Saccharomyces cerevisiae for the Production of Industrially and Clinically Important Chemicals; IntechOpen: London, UK, 2017. [Google Scholar]
- Yang, L.; Liu, H.; Jin, Y.; Liu, J.; Deng, L.; Wang, F. Recent advances in multiple strategies for the synthesis of terpenes by engineered yeast. Fermentation 2022, 8, 615. [Google Scholar] [CrossRef]
- Dixon, T.A.; Walker, R.S.; Pretorius, I.S. Visioning synthetic futures for yeast research within the context of current global techno-political trends. Yeast 2023, 40, 443–456. [Google Scholar] [CrossRef]
- Pretorius, I.S. Synthetic genome engineering forging new frontiers for wine yeast. Crit. Rev. Biotechnol. 2017, 37, 112–136. [Google Scholar] [CrossRef]
- Schindler, D.; Walker, R.S.; Cai, Y. Methodological advances enabled by the construction of a synthetic yeast genome. Cell Rep. Methods 2024, 4, 100761. [Google Scholar] [CrossRef]
- Pretorius, I.S. Visualizing the next frontiers in wine yeast research. FEMS Yeast Res. 2022, 22, foac010. [Google Scholar] [CrossRef]
- Patra, P.; Das, M.; Kundu, P.; Ghosh, A. Recent advances in systems and synthetic biology approaches for developing novel cell-factories in non-conventional yeasts. Biotechnol. Adv. 2021, 47, 107695. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dagariya, S.; Bhatankar, J.; Dakal, T.C.; Gadi, B.R.; Giudici, P. Metabolic and Evolutionary Engineering of Food Yeasts. Processes 2025, 13, 1852. https://doi.org/10.3390/pr13061852
Dagariya S, Bhatankar J, Dakal TC, Gadi BR, Giudici P. Metabolic and Evolutionary Engineering of Food Yeasts. Processes. 2025; 13(6):1852. https://doi.org/10.3390/pr13061852
Chicago/Turabian StyleDagariya, Sakshi, Janvi Bhatankar, Tikam Chand Dakal, Bhana Ram Gadi, and Paolo Giudici. 2025. "Metabolic and Evolutionary Engineering of Food Yeasts" Processes 13, no. 6: 1852. https://doi.org/10.3390/pr13061852
APA StyleDagariya, S., Bhatankar, J., Dakal, T. C., Gadi, B. R., & Giudici, P. (2025). Metabolic and Evolutionary Engineering of Food Yeasts. Processes, 13(6), 1852. https://doi.org/10.3390/pr13061852






