The Effect of a Photoactivated Ruthenium Nitrocomplex [RuCl(NO2)(dppb)(4,4-2 Mebipy)] on the Viability of Eukaryotic and Prokaryotic Cells, Including Bacterial Biofilms
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Synthesis of the Nitro Complex [RuCl(NO2)(dppb)(4,4-Mebipy)]
2.3. X-Ray Structure Determination
2.4. Continuous Photolysis
2.5. Electrochemical Experiments
2.6. Carboxy-PTIO Reaction to Identify NO
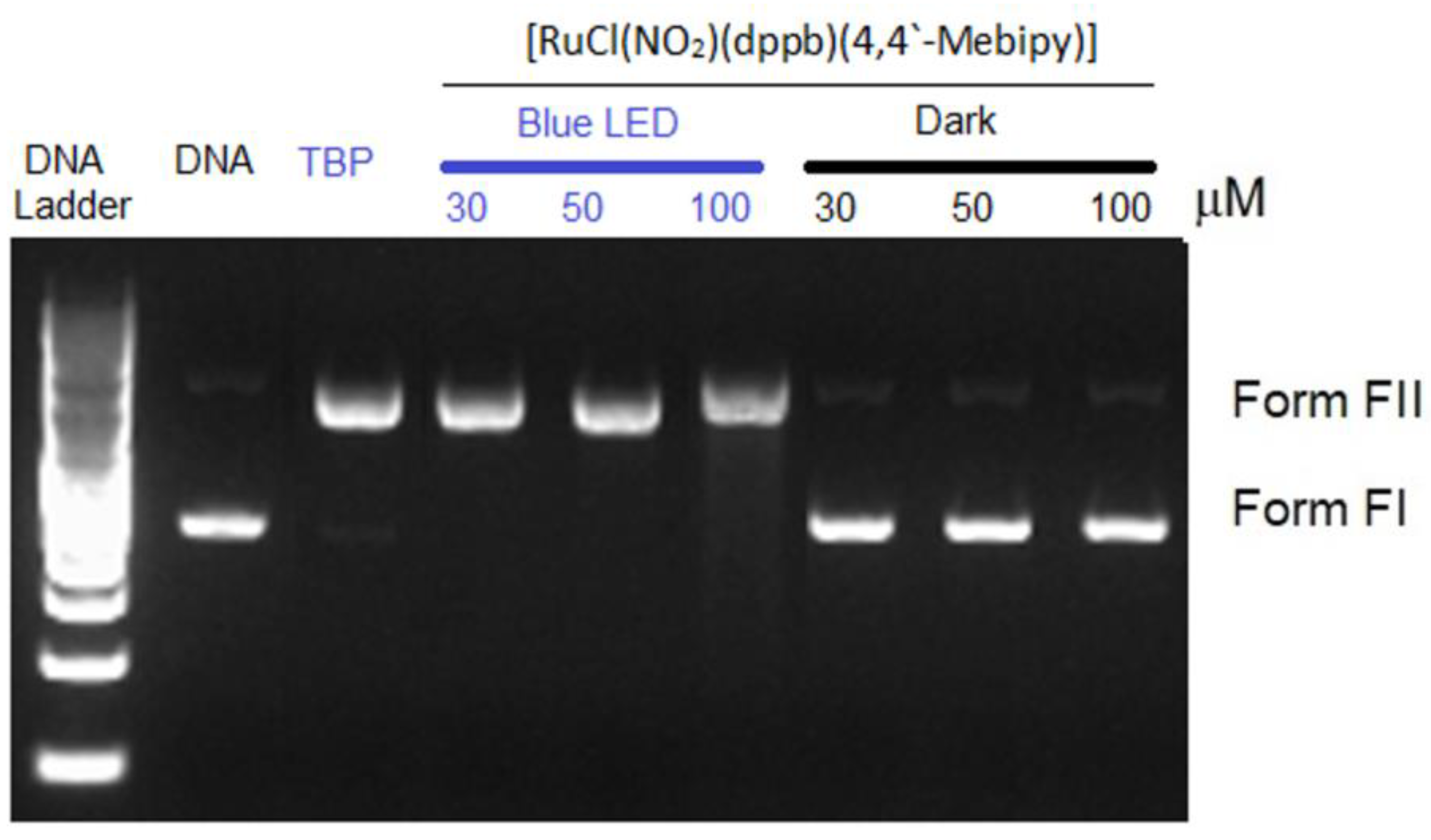
2.7. DNA Photocleavage Assay
2.8. Bacterial Strains and Culture Conditions
2.9. Minimum Inhibitory Concentration (MIC) and Minimum Bactericidal Concentration (MBC) Determination
2.10. Checkerboard Assay
2.11. Biofilm Susceptibility Assay
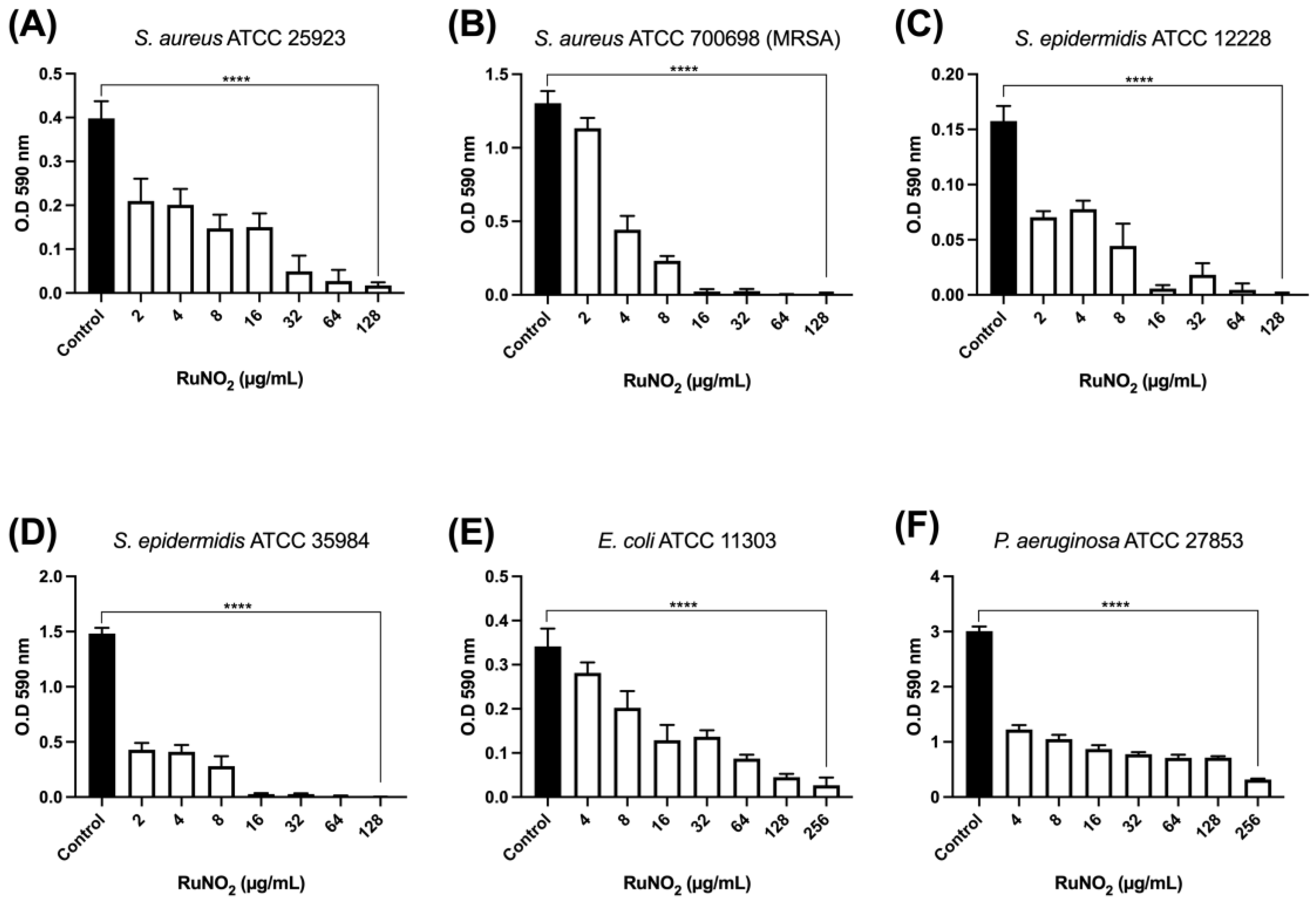
2.11.1. Quantification of Total Biomass
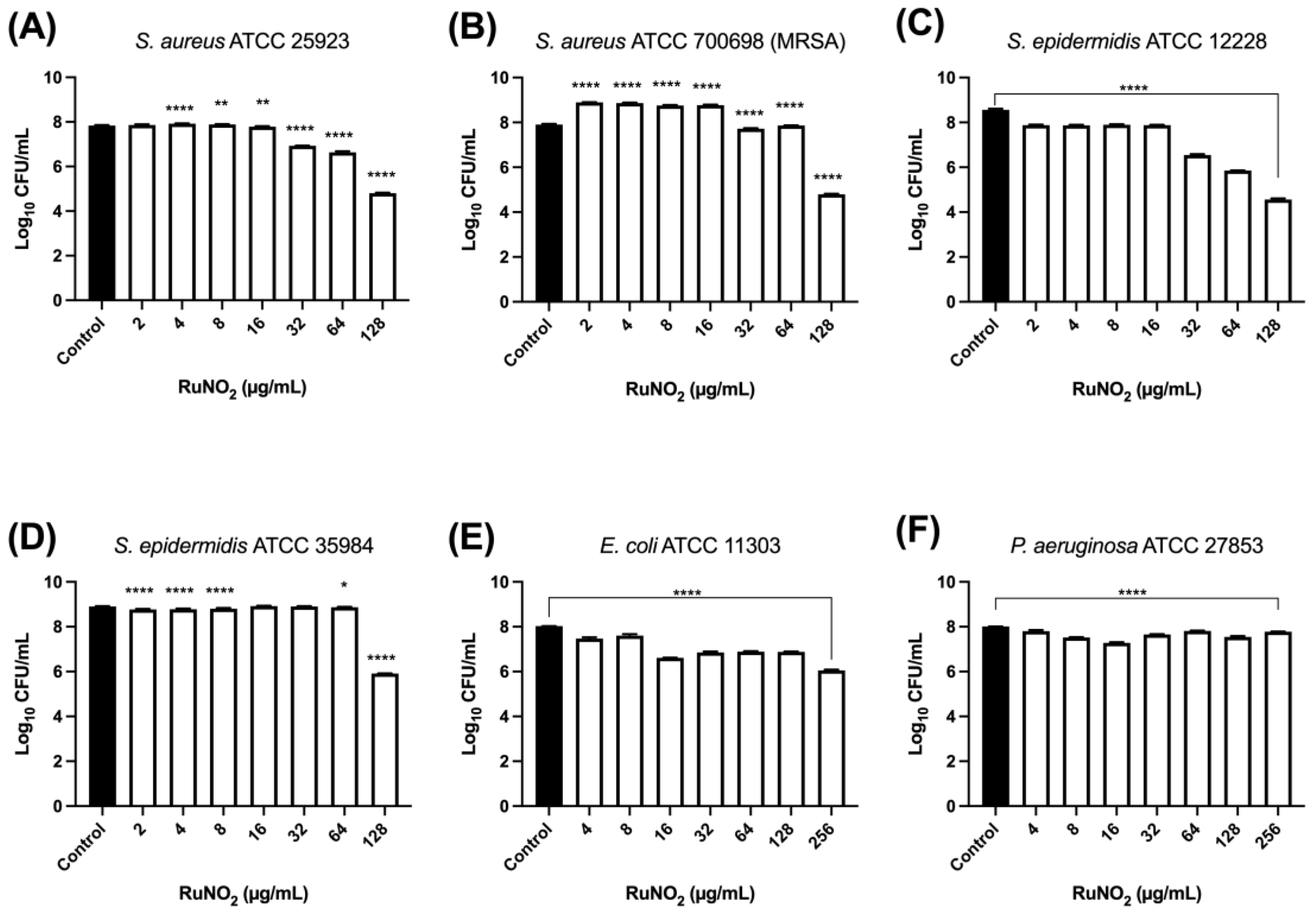
2.11.2. Viable Cell Count
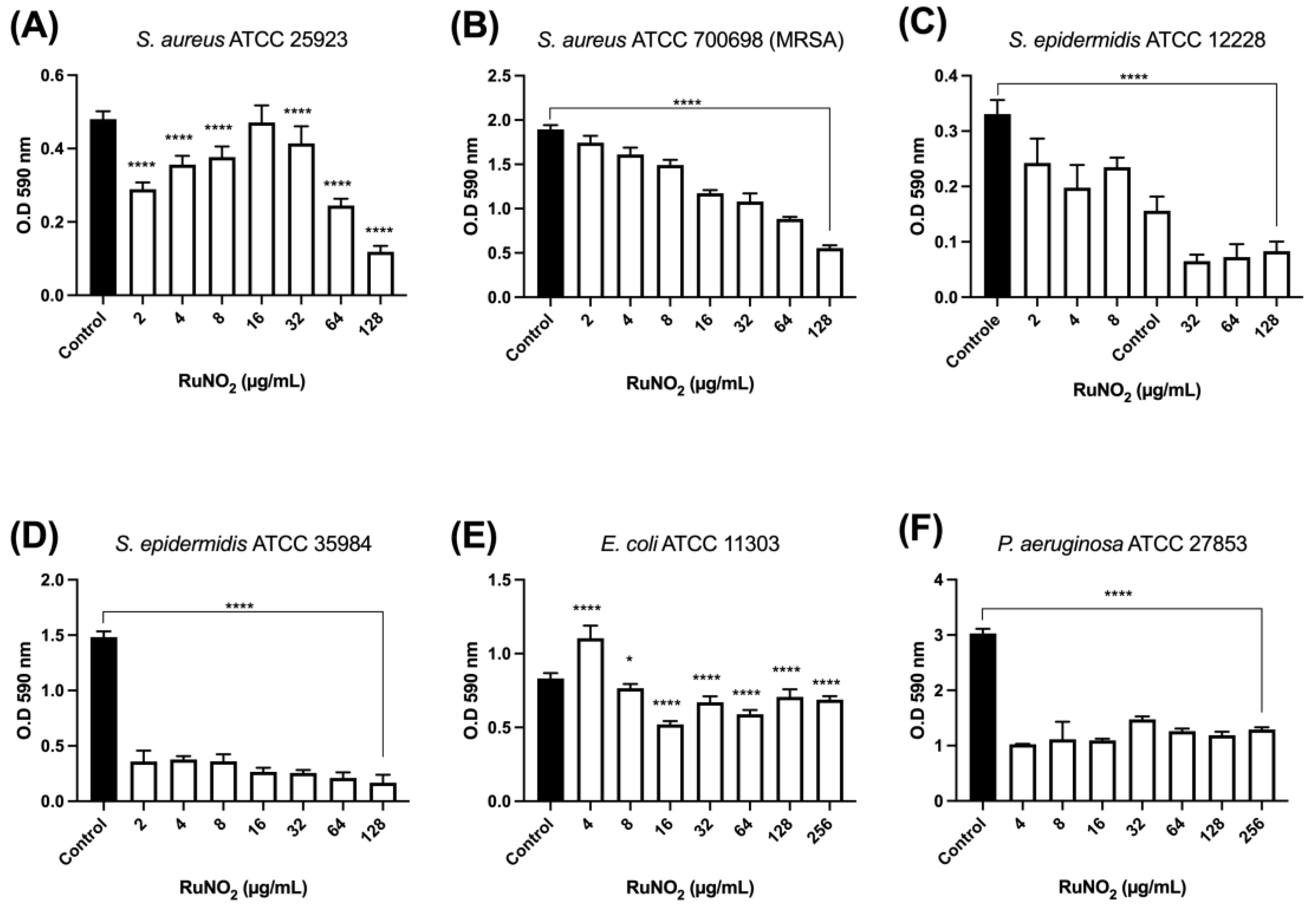
2.12. Effect of Complex I on Mature Biofilms
2.13. Scanning Electron Microscopy (SEM)
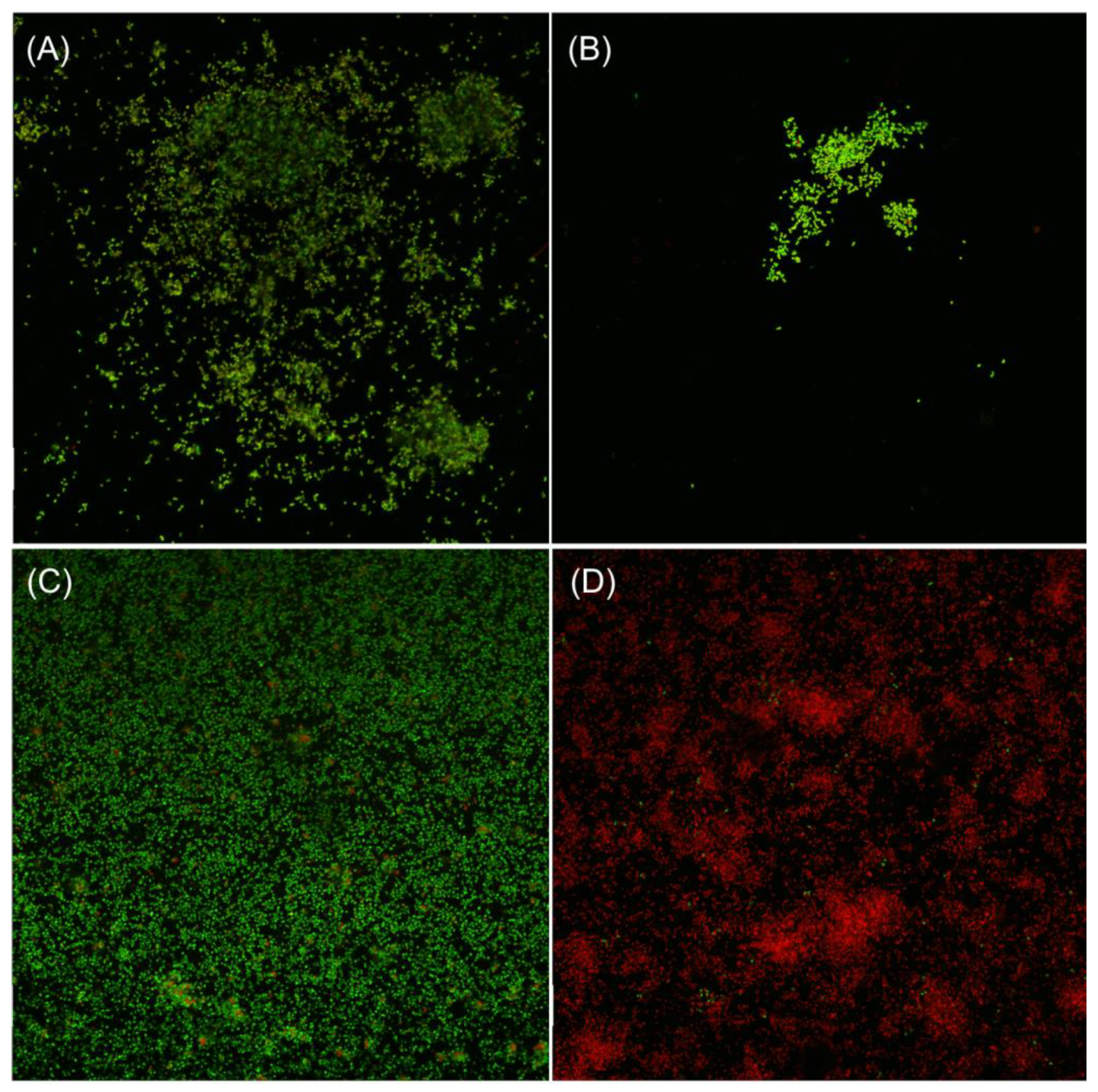
2.14. Confocal Laser Scanning Microscopy
2.15. Hemolytic Activity Assay
2.16. Cytotoxicity Activity
2.16.1. Cell Line and Culture Conditions
2.16.2. Cell Viability Assay
2.17. Statistical Analysis
3. Results
3.1. Synthesis of the Nitro Complex
3.2. Photochemistry
3.3. Carboxy-PTIO Reaction to Identify NO
3.4. DNA Photocleavage
3.5. Antibacterial Activity
3.6. Synergistic Effect of Complex I Combined with Antibiotics
3.7. Effect of Complex I on Biofilm Formation
3.8. Effect of Complex I on Mature Biofilm
3.9. Scanning Electron Microscopy (SEM)
3.10. Confocal Laser Scanning Microscopy Analyses
3.11. Hemolytic Activity
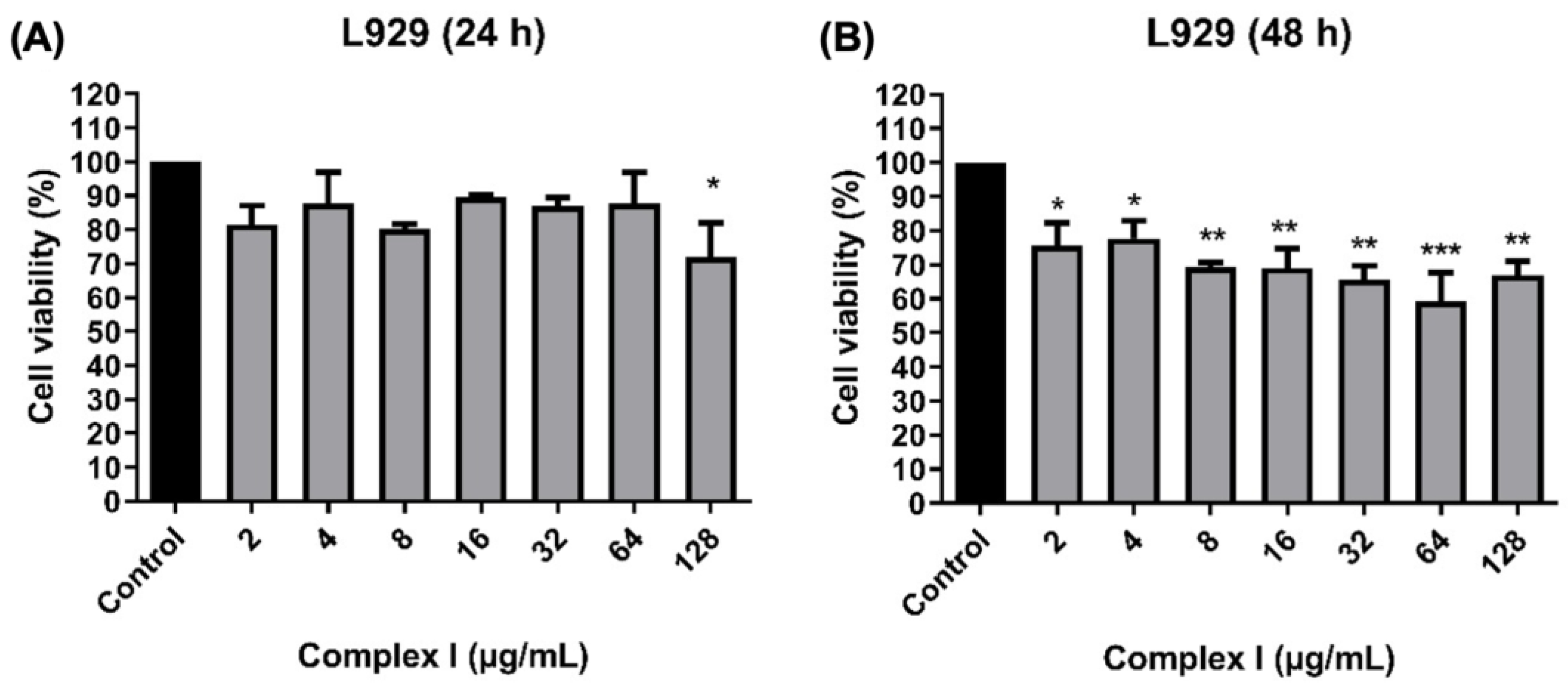
3.12. Cytotoxic Activity
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Blair, J.M.A. Introducing npj Antimicrobials and Resistance. npj Antimicrob. Resist. 2023, 1, 3. [Google Scholar] [CrossRef]
- Murray, C.J.L.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Robles Aguilar, G.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, J. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations: The Review on Antimicrobial Resistance; Wellcome Trust: London, UK, 2016. [Google Scholar]
- Ture, Z.; Güner, R.; Alp, E. Antimicrobial stewardship in the intensive care unit. J. Intensive Med. 2022, 3, 244–253. [Google Scholar] [CrossRef] [PubMed]
- WHO Regional Office for Europe/European Centre for Disease Prevention and Control. Antimicrobial Resistance Surveillance in Europe 2022—2020 Data; WHO Regional Office for Europe: Copenhagen, Denmark, 2022. [Google Scholar]
- Schilcher, K.; Horswill, A.R. Staphylococcal biofilm development: Structure, regulation, and treatment strategies. Microbiol. Mol. Biol. Rev. 2020, 84, e00026-19. [Google Scholar] [CrossRef] [PubMed]
- Flemming, H.C.; van Hullebusch, E.D.; Neu, T.R.; Nielsen, P.H.; Seviour, T.; Stoodley, P.; Wingender, J.; Wuertz, S. The biofilm matrix: Multitasking in a shared space. Nat. Rev. Microbiol. 2023, 21, 70–86. [Google Scholar] [CrossRef]
- Jamal, M.; Ahmad, W.; Andleeb, S.; Jalil, F.; Imran, M.; Nawaz, M.A.; Ussain, T.; Ali, M.; Rafiq, M.; Kamil, M.A. Bacterial biofilm and associated infections. J. Chin. Med. Assoc. 2018, 81, 7–11. [Google Scholar] [CrossRef]
- Kolpen, M.; Kragh, K.N.; Enciso, J.B.; Faurholt-Jepsen, D.; Lindegaard, B.; Egelund, G.B.; Jensen, A.V.; Ravn, P.; Mathiesen, I.H.M.; Gheorge, A.G.; et al. Bacterial biofilms predominate in both acute and chronic human lung infections. Thorax 2022, 77, 1015–1022. [Google Scholar] [CrossRef]
- Staats, A.; Li, D.; Sullivan, A.C.; Stoodley, P. Biofilm formation in periprosthetic joint infections. Ann. Jt. 2021, 6, 43. [Google Scholar] [CrossRef]
- Oliva, A.; Mascellino, M.T.; Nguyen, B.L.; De Angelis, M.; Cipolla, A.; Di Berardino, A.; Ciccaglioni, A.; Mastroianni, C.M.; Vullo, V. Detection of biofilm-associated implant pathogens in cardiac device infections: High sensitivity of sonication fluid culture even in the presence of antimicrobials. J. Glob. Infect. Dis. 2018, 10, 74–79. [Google Scholar] [CrossRef]
- Shrestha, L.B.; Baral, R.; Khanal, B. Comparative study of antimicrobial resistance and biofilm formation among Gram-positive uropathogens isolated from community acquired urinary tract infections and catheter-associated urinary tract infections. Infect. Drug Resist. 2019, 12, 957–963. [Google Scholar] [CrossRef]
- Ng, X.Y.; Fong, K.W.; Kiew, L.V.; Chung, P.Y.; Liew, Y.K.; Delsuc, N.; Zulkefeli, M.; Low, M.L. Ruthenium (II) polypyridyl complexes as emerging photosensitisers for antibacterial photodynamic therapy. J. Inorg. Biochem. 2024, 250, 112425. [Google Scholar] [CrossRef] [PubMed]
- Uruén, C.; Chopo-Escuin, G.; Tommassen, J.; Mainar-Jaime, R.C.; Arenas, J. Biofilms as Promoters of Bacterial Antibiotic Resistance and Tolerance. Antibiotics 2021, 10, 3. [Google Scholar] [CrossRef] [PubMed]
- Sousa, A.P.; Ellena, J.; Gondim, A.C.S.; Lopes, L.G.F.; Sousa, E.H.S.; Vasconcelos, M.A.; Teixeira, E.H.; Ford, P.C.; Holanda, A.K.M. Antimicrobial activity of cis -[Ru(bpy)2(L)(L’)]n+ complexes, where L = 4-(4-chlorobenzoyl)pyridine or 4-(benzoyl)pyridine and L’ = Cl− or CO. Polyhedron 2018, 144, 88–94. [Google Scholar] [CrossRef]
- Andrade, A.L.; Vasconcelos, M.A.; Arruda, F.V.S.; Nascimento Neto, L.G.; Carvalho, J.M.S.; Gondim, A.C.S.; Lopes, L.G.F.; Sousa, E.H.S.; Teixeira, E.H. Antimicrobial activity and antibiotic synergy of a biphosphinic ruthenium complex against clinically relevant bacteria. Biofouling 2020, 36, 442–454. [Google Scholar] [CrossRef]
- Sousa, A.P.; Gondim, A.C.S.; Sousa, E.H.S.; Lopes, L.G.F.; Martins, P.H.R.; Teixeira, E.H.; Vasconcelos, M.A.; Medeiros, E.J.T.; Batista, A.A.; Holanda, A.K.M. Biphosphinic ruthenium complexes as the promising antimicrobial agents. New J. Chem. 2020, 44, 21318–21325. [Google Scholar] [CrossRef]
- Sousa, A.P.; Gondim, A.C.S.; Sousa, E.H.S.; Vasconcelos, M.A.; Teixeira, E.H.; Bezerra, B.P.; Ayala, A.P.; Martins, P.H.R.; Lopes, L.G.F.; Holanda, A.K.M. An unusual bidentate methionine ruthenium(II) complex: Photo-uncaging and antimicrobial activity. J. Biol. Inorg. Chem. 2020, 25, 419–428. [Google Scholar] [CrossRef]
- Li, F.; Collins, J.G.; Keene, F.R. Ruthenium complexes as antimicrobial agents. Chem. Soc. Rev. 2015, 44, 2529–2542. [Google Scholar] [CrossRef]
- Southam, H.M.; Butler, J.A.; Chapman, J.A.; Poole, R.K. The Microbiology of Ruthenium Complexes. Adv. Microb. Physiol. 2017, 71, 1–96. [Google Scholar]
- Hamblin, M.R. Antimicrobial photodynamic inactivation: A bright new technique to kill resistant microbes. Curr. Opin. Microbiol. 2016, 33, 67–73. [Google Scholar] [CrossRef]
- Mendhi, J.; Asgari, M.; Ratheesha, G.; Prasadama, I.; Yanga, Y.; Xiao, Y. Dose controlled nitric oxide-based strategies for antibacterial property in biomedical devices. Appl. Mater. 2020, 19, 100562. [Google Scholar] [CrossRef]
- Hall, J.R.; Rouillard, K.R.; Suchyta, D.J.; Brown, M.D.; Ahonen, M.J.R.; Schoenfisc, M.H. Mode of nitric oxide delivery affects antibacterial action. ACS Biomater. Sci. Eng. 2020, 6, 433–441. [Google Scholar] [CrossRef] [PubMed]
- Sharma, B.; Shukla, S.; Rattan, R.; Fatima, M.; Goel, M.; Bhat, M.; Dutta, S.; Ranjan, R.K.; Sharma, M. Antimicrobial Agents Based on Metal Complexes: Present Situation and Future Prospects. Int. J. Biomater. 2022, 2022, 6819080. [Google Scholar] [CrossRef] [PubMed]
- Giri, B.; Saini, T.; Kumbhakar, S.; Karumban, K.S.; Muley, A.; Misra, A.; Maji, S. Near-IR light-induced photorelease of nitric oxide (NO) on ruthenium nitrosyl complexes: Formation, reactivity, and biological effects. Dalton Trans. 2020, 49, 10772–10785. [Google Scholar] [CrossRef] [PubMed]
- Bourassa, J.L.; Ford, P.C. Flash and continuous photolysis studies of Roussin’s red salt dianion Fe2S2(NO)42− in solution. Coord. Chem. Rev. 2000, 200, 887–900. [Google Scholar] [CrossRef]
- Wecksler, S.R.; Mikhailovsky, A.; Korystov, D.; Buller, F.; Kannan, R.; Tan, L.-S.; Ford, P.C. Single- and two-photon properties of a dye-derivatized Roussin’s red salt ester (Fe2(μ-RS)2(NO)4) with a large TPA cross section. Inorg. Chem. 2007, 46, 395–402. [Google Scholar] [CrossRef]
- Lima, R.G.; Sauaia, M.G.; Bonaventura, D.; Tedesco, A.C.; Lopez, R.F.V.; Bendhack, L.M.; Silva, R.S. Controlled nitric oxide photo-release from nitro ruthenium complexes: The vasodilator response produced by UV light irradiation. Inorg. Chim. Acta 2005, 358, 2643–2650. [Google Scholar] [CrossRef]
- Silva, H.V.R.; Dias, J.S.M.; Ferreira-Silva, G.Á.; Vegas, L.C.; Ionta, M.; Corrêa, C.C.; Batista, A.A.; Barbosa, M.I.F.; Doriguetto, A.C. Phosphine/diimine ruthenium complexes with Cl−, CO, NO+, NO2−, NO3− and pyridine ligands: Proapoptotic activity on triple-negative breast cancer cells and DNA/HSA interactions. Polyhedron 2018, 144, 55–65. [Google Scholar] [CrossRef]
- Agilent. CrysAlis PRO; Agilent Technologies Ltd.: Oxfordshire, UK, 2013; pp. 1–60. [Google Scholar]
- Sheldrick, G.M. Crystal Structure Refinement with SHELXL. Acta Cryst. C 2015, 71, 3–8. [Google Scholar] [CrossRef]
- Dolomanov, O.V.; Bourhis, L.J.; Gildea, R.J.; Howard, J.A.K.; Puschmann, H. OLEX2: A Complete Structure Solution, Refinement and Analysis Program. J. Appl. Cryst. 2009, 42, 339–341. [Google Scholar] [CrossRef]
- Sheldrick, G.M. TWINABS; University of Göttingen: Göttingen, Germany, 2009. [Google Scholar]
- Macrae, C.F.; Edgington, P.R.; McCabe, P.; Pidcock, E.; Shields, G.P.; Taylor, R.; Towler, M.; van de Streek, J. Mercury: Visualization and Analysis of Crystal Structures. J. Appl. Cryst. 2006, 39, 453–457. [Google Scholar] [CrossRef]
- M07-A10; Method for Dilution Antibacterial Susceptibility Tests for Bacteria That Grow Aerobically: Approved Standard, 10th ed. Clinical and Laboratory Standards Institute: Pennsylvania, PA, USA, 2015.
- Das, B.; Mandal, D.; Dash, S.K.; Chattopadhyay, S.; Tripathy, S.; Dolai, D.P.; Dey, S.K.; Roy, S. Eugenol Provokes ROS-Mediated Membrane Damage-Associated Antibacterial Activity Against Clinically Isolated Multidrug-Resistant Staphylococcus aureus Strains. Infect. Dis. 2016, 9, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Odds, F.C. Synergy, antagonism, and what the chequerboard puts between them. J. Antimicrob. Chemother. 2003, 52, 1. [Google Scholar] [CrossRef] [PubMed]
- Castelo-Branco, D.S.; Riello, G.B.; Vasconcelos, D.C.; Guedes, G.M.; Serpa, R.; Bandeira, T.J.; Monteiro, A.J.; Cordeiro, R.A.; Rocha, M.F.; Sidrim, J.J.; et al. Farnesol increases the susceptibility of Burkholderia pseudomallei biofilm to antimicrobials used to treat melioidosis. J. Appl. Microbiol. 2016, 120, 600–606. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Ma, Y.; Xiong, Y.; Tan, Y.; Duan, X.; Liao, X.; Wang, J. Ruthenium polypyridine complexes with triphenylamine groups as antibacterial agents against Staphylococcus aureus with membrane-disruptive mechanism. Front. Chem. 2022, 10, 1035741. [Google Scholar] [CrossRef]
- Holanda, A.K.M.; da Silva, F.O.N.; Sousa, J.R.; Diogenes, I.C.N.; Carvalho, I.M.M.; Moreira, I.S.; Clarke, M.J.; Lopes, L.G.F. Photochemical NO release from nitrosyl Ru-II complexes with C-bound imidazoles. Inorganica Chim. Acta 2008, 361, 2929–2933. [Google Scholar] [CrossRef]
- da Silveira Carvalho, J.M.; Batista, A.H.M.; Nogueira, N.A.P.; Holanda, A.K.M.; Sousa, J.R.; Zampiere, D.; Bezerra, M.J.B.; Barreto, F.S.; Moraes, M.O.; Batista, A.A.; et al. A biphosphinic ruthenium complex with potent anti-bacterial and anti-cancer activity. New J. Chem. 2017, 41, 13085–13095. [Google Scholar] [CrossRef]
- Hernando-Amado, S.; Coque, T.M.; Baquero, F.; Martínez, J.L. Defining and combating antibiotic resistance from One Health and Global Health perspectives. Nat. Rev. Microbiol. 2019, 4, 1432–1442. [Google Scholar] [CrossRef]
- Ghosh, S.; Amariei, G.; Mosquera, M.E.G.; Rosal, R. Polymeric ruthenium precursor as a photoactivated antimicrobial agent. J. Hazard. Mater. 2021, 402, 123788. [Google Scholar] [CrossRef]
- Burnham, C.; Leeds, J.; Nordmann, P.; O’Grady, J.; Patel, J. Diagnosing antimicrobial resistance. Nat. Rev. Microbiol. 2017, 15, 697–703. [Google Scholar] [CrossRef]
- Ghosh, T.; Biswas, M.K.; Chatterjee, S.; Roy, P. In-vitro study on the hemolytic activity of different extracts of Indian medicinal plant Croton bonplandianum with phytochemical estimation: A new era in drug development. J. Drug Deliv. Ther. 2018, 8, 155–160. [Google Scholar] [CrossRef]
- Kaviyarasu, K.; Geetha, N.; Kanimozhi, K.; Magdalane, C.M.; Sivaranjani, S.; Ayeshamariam, A.; Kennedy, J.; Maaza, M. In vitro cytotoxicity effect and antibacterial performance of human lung epithelial cells A549 activity of Zinc oxide doped TiO2 nanocrystals: Investigation of bio-medical application by chemical method. Mater. Sci. Eng. 2017, 74, 325–333. [Google Scholar] [CrossRef] [PubMed]
- Amariei, G.; Kokol, V.; Boltes, K.; Letón, P.; Rosal, R. Incorporation of antimicrobial peptides on electrospun nanofibres for biomedical applications. RSC Adv. 2018, 8, 28013–28023. [Google Scholar] [CrossRef] [PubMed]
- Magdalane, C.M.; Kaviyarasu, K.; Raja, A.; Arularasu, M.V.; Mola, G.T.; Isaev, A.B.; Aldhabi, N.A.; Arasu, M.V.; Jeyaraj, B.; Kennedy, J.; et al. Photocatalytic decomposition effect of erbium doped cerium oxide nanostructures driven by visible light irradiation: Investigation of cytotoxicity, antibacterial growth inhibition using catalysts. J. Photochem. Photobiol. B. 2018, 185, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Conti, L.; Mengoni, A.; Giacomazzo, G.E.; Mari, L.; Perfetti, M.; Fagorzi, C.; Sorace, L.; Valtancoli, B.; Giorgi, C. Exploring the potential of highly charged Ru(II)- and heteronuclear Ru(II)/Cu(II)-polypyridyl complexes as antimicrobial agents. J. Inorg. Biochem. 2021, 220, 11467. [Google Scholar] [CrossRef]
- Kay, M.I.; Frazer, B.C. A neutron diffraction refinement of the low temperature phase of NaNO2. Acta Crystallogr. 1961, 14, 56–57. [Google Scholar] [CrossRef]
- Bastos, T.M.; Barbosa, M.I.F.; Silva, M.M.; Júnior, J.W.C.; Meira, C.S.; Guimarães, E.T.; Ellena, J.; Moreira, D.R.M.; Batista, A.A.; Soares, M.B.P. Nitro/nitrosyl ruthenium complexes are potent and selective anti-Trypanosoma cruzi agents causing autophagy and necrotic parasite death. Antimicrob. Agents Chemother. 2014, 58, 6044–6055. [Google Scholar] [CrossRef]
- Freedman, D.A.; Kruger, S.; Roosa, C.; Wymer, C. Synthesis, characterization, and reactivity of [Ru(bpy)(CH3CN)3(NO2)]PF6, a synthon for [Ru(bpy)(L3)NO2] complexes. Inorg. Chem. 2006, 45, 9558–9568. [Google Scholar] [CrossRef]
- Sousa, A.P.; Fernandes, A.F.; Paz, I.A.; Nascimento, N.R.F.; Ellena, J.; Sousa, E.H.S.; Lopes, I.G.F.; Holanda, A.K.M. A Potential Visible-Light NO Releaser: Synthesis, Reactivity and Vasodilator Properties. J. Braz. Chem. Soc. 2017, 28, 2117–2129. [Google Scholar] [CrossRef]
- Honorato, J.; Colina-Vegas, L.; Correa, R.S.; Guedes, A.P.M.; Miyata, M.; Pavan, F.R.; Ellena, J.; Batista, A.A. Esterification of the free carboxylic group from the lutidinic acid ligand as a tool to improve the cytotoxicity of Ru(ii) complexes. Inorg. Chem. Front. 2019, 6, 376–390. [Google Scholar] [CrossRef]
- Bogdan, C.; Röllinghoff, M.; Diefenbach, A. The role of nitric oxide in innate immunity. Immunol. Rev. 2000, 173, 17–26. [Google Scholar] [CrossRef]
- Abreu, F.D.; Paulo, T.F.; Gehlen, M.H.; Ando, R.A.; Lopes, L.G.F.; Gondim, A.C.S.; Vasconcelos, M.A.; Teixeira, E.H.; Sousa, E.H.S.; Carvalho, I.M.M. Aryl-Substituted Ruthenium(II) Complexes: A Strategy for Enhanced Photocleavage and Efficient DNA Binding. Inorg. Chem. 2017, 56, 9084–9096. [Google Scholar] [CrossRef] [PubMed]
- Sharma, D.; Misba, L.; Khan, A.U. Antibiotics versus Biofilm: An Emerging Battleground in Microbial Communities. Antimicrob. Resist. Infect. Control 2019, 8, 76. [Google Scholar] [CrossRef] [PubMed]
- Parvin, F.; Hu, H.; Whiteley, G.S.; Glasbey, T.; Vickery, K. Difficulty in Removing Biofilm from Dry Surfaces. J. Hosp. Infect. 2019, 103, 465–467. [Google Scholar] [CrossRef] [PubMed]
- Fotinos, N.; Convert, M.; Piffaretti, J.C.; Gurny, R.; Lange, N. Effects on Gram-Negative and Gram-Positive Bacteria Mediated by 5-Aminolevulinic Acid and 5-Aminolevulinic Acid Derivatives. Antimicrob. Agents Chemother. 2008, 52, 1366–1373. [Google Scholar] [CrossRef]
- Otter, J.A.; Vickery, K.; Walker, J.T.; De Lancey Pulcini, E.; Stoodley, P.; Goldenberg, S.D.; Salkeld, J.A.G.; Chewins, J.; Yezli, S.; Edgeworth, J.D. Surface-Attached Cells, Biofilms and Biocide Susceptibility: Implications for Hospital Cleaning and Disinfection. J. Hosp. Infect. 2015, 89, 16–27. [Google Scholar] [CrossRef]
- Sharma, A.; Gupta, S.; Sarethy, I.P.; Dang, S.; Gabrani, R. Green tea extract: Possible mechanism and antibacterial activity on skin pathogens. Food Chem. 2012, 135, 672–675. [Google Scholar] [CrossRef]
- Flemming, H.C.; Wingender, J. The biofilm matrix. Nat. Rev. Microbiol. 2010, 8, 623–633. [Google Scholar] [CrossRef]
- Sun, W.Z.; Jian, Y.; Zhou, M.X.; Yao, Y.S.; Tian, N.; Li, C.; Chen, J.; Wang, X.S.; Zhou, Q.X. Selective and efficient photoinactivation of intracellular Staphylococcus aureus and MRSA with little accumulation of drug resistance: Application of a Ru(II) complex with photolabile ligands. J. Med. Chem. 2021, 64, 7359–7370. [Google Scholar] [CrossRef]
- Yan, J.; Bassler, B.L. Surviving as a community: Antibiotic tolerance and persistence in bacterial biofilms. Cell Host Microbe 2019, 26, 15–21. [Google Scholar] [CrossRef]
- Gafur, A.; Sukamdani, G.Y.; Kristi, N.; Maruf, A.; Xu, J.; Chen, X.; Wang, G.; Ye, Z. From bulk to nano-delivery of essential phytochemicals: Recent progress and strategies for antibacterial resistance. J. Mater. Chem. B 2020, 8, 9825–9835. [Google Scholar] [CrossRef]
- Wang, L.L.; Battini, N.; Bheemanaboina, R.R.Y.; Ansari, M.F.; Chen, J.P.; Xie, Y.P.; Cai, G.X.; Zhang, S.L.; Zhou, C.H. A new exploration towards aminothiazolquinolone oximes as potentially multi-targeting antibacterial agents: Design, synthesis and evaluation acting on microbes, DNA, HSA and topoisomerase IV. Eur. J. Med. Chem. 2019, 179, 166–181. [Google Scholar] [CrossRef] [PubMed]
- Rhoads, D.D.; Wolcott, R.D.; Percival, S.L. Biofilms in wounds: Management strategies. J. Wound Care 2008, 17, 502–508. [Google Scholar] [CrossRef] [PubMed]
- Anderson, G.G.; O’Toole, G.A. Innate and induced resistance mechanisms of bacterial biofilms. In Bacterial Biofilms; Romeo, T., Ed.; Springer: Berlin/Heidelberg, Germany, 2008; Volume 322, pp. 85–105. [Google Scholar]
- Sun, D.; Zhang, W.; Yang, E.; Li, N.; Liu, H.; Wang, W. Investigation of antibacterial activity and related mechanism of a ruthenium(II) polypyridyl complex. Inorg. Chem. Commun. 2015, 56, 17–21. [Google Scholar] [CrossRef]
- Jin, H.; Yang, L.; Ahonen, M.J.R.; Schoenfisch, M.H. Nitric Oxide-Releasing Cyclodextrins. J. Am. Chem. Soc. 2018, 140, 14178–14184. [Google Scholar] [CrossRef]
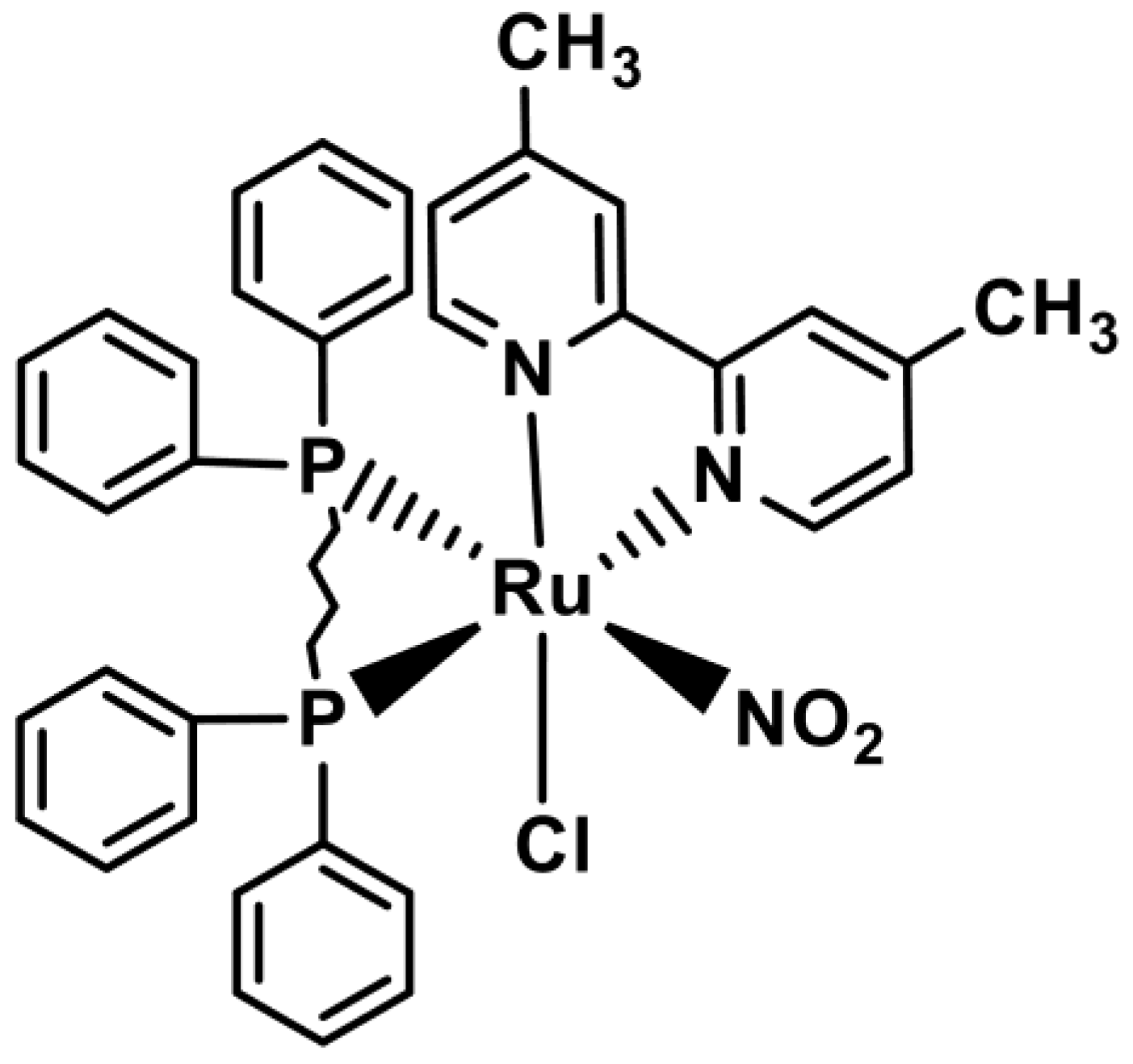
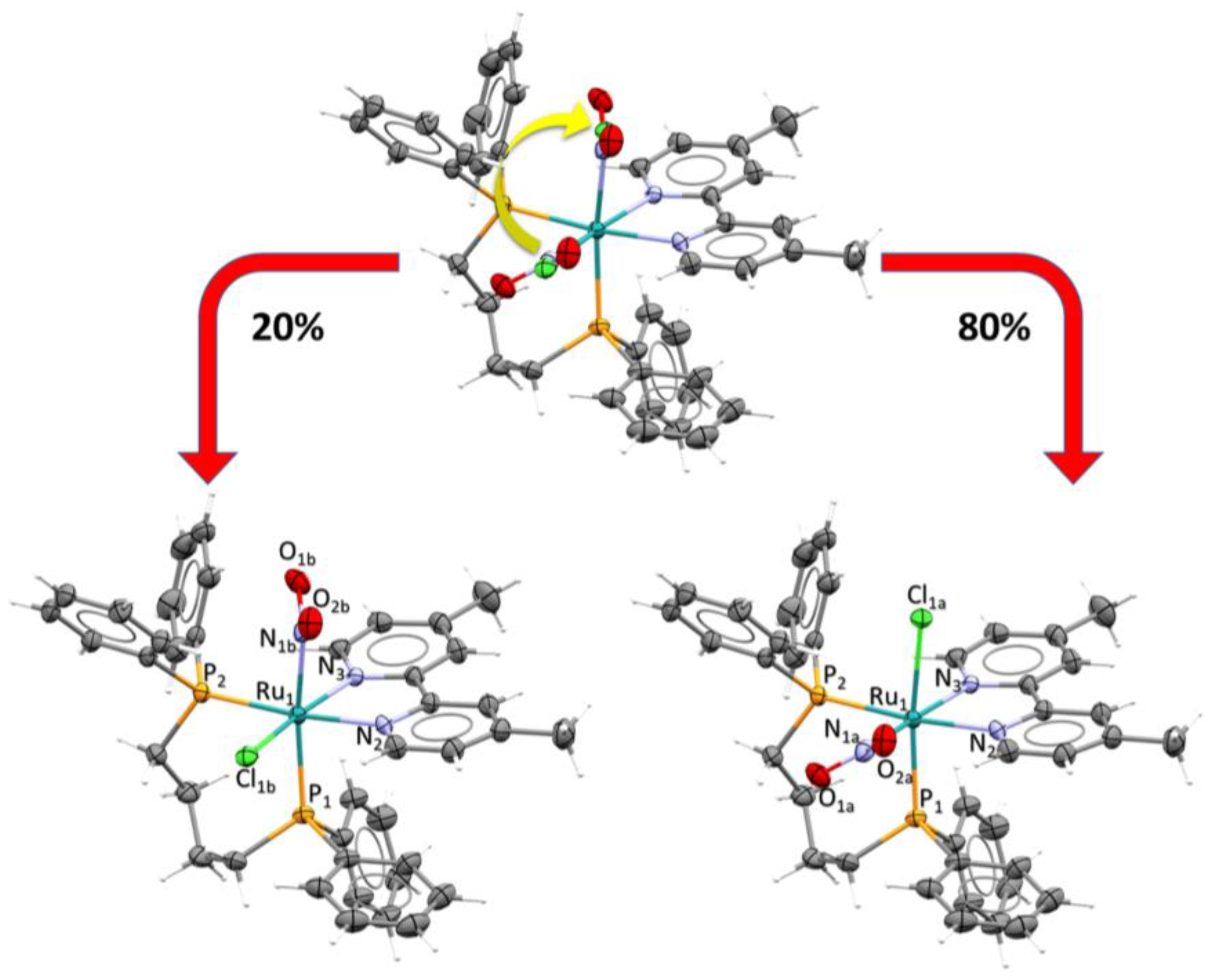





| Empirical formula | Ru1C42H48ClN3O4P2 |
| Formula weight | 857.29 |
| Temperature/K | 293 (2) |
| Crystal system | Monoclinic |
| Space group | P21/n |
| a/Å | 12.3962 (2) |
| b/Å | 19.0198 (3) |
| c/Å | 16.9362 (3) |
| α/° | 90 |
| β/° | 92.645 (2) |
| γ/° | 90 |
| Volume/Å3 | 3988.85 (11) |
| Z | 4 |
| ρcalcg/cm3 | 1.428 |
| μ/mm−1 | 0.586 |
| F(000) | 1776.0 |
| Crystal size/mm3 | 0.545 × 0.218 × 0.208 |
| Radiation | MoKα (λ = 0.71073) |
| 2Θ range for data collection/° | 5.27 to 53.998 |
| Index ranges | −15 ≤ h ≤ 15, −24 ≤ k ≤ 24, −21 ≤ l ≤ 21 |
| Reflections collected | 52,577 |
| Independent reflections | 8703 [Rint = 0.0256, Rsigma = 0.0164] |
| Data/restraints/parameters | 8703/0/502 |
| Goodness-of-fit on F2 | 1.199 |
| Final R indexes [I > =2σ (I)] | R1 = 0.0329, wR2 = 0.0739 |
| Final R indexes [all data] | R1 = 0.0449, wR2 = 0.0865 |
| Largest diff. peak/hole/e Å−3 | 0.60/−0.45 |
| Bond Angles (°) | Bond Lengths (Å) | ||
|---|---|---|---|
| P1-Ru1-P2 | 97.70 (2) | Ru1-P2 | 2.3540 (6) |
| N2-Ru1-N3 | 76.58 (8) | Ru1-P1 | 2.3269 (6) |
| P2-Ru1-N2 | 173.49 (6) | Ru1-N2 | 2.121 (2) |
| N1b-Ru1-Cl1b | 91.8 (7) | Ru1-N3 | 2.115 (2) |
| N1a-Ru1-Cl1a | 93.7 (4) | Ru1-N1a | 2.084 (2) |
| O1a-N1a-O2a | 120.2 (10) | Ru1-N1b | 2.085 (2) |
| O1b-N1b-O2b | 111.6 (17) | Ru1-Cl1a | 2.453 (1) |
| - | - | Ru1-Cl1b | 2.372 (2) |
| N1a-O1a | 1.197 (17) | ||
| N1a-O2a | 1.225 (15) | ||
| N1b-O2b | 1.25 (2) | ||
| N1b-O1b | 1.23 (3) |
| Complex I | Ampicillin | Tetracycline | ||||||
|---|---|---|---|---|---|---|---|---|
| LED OFF | LED ON | |||||||
| MIC | MBC | MIC | MBC | MIC | MBC | MIC | MBC | |
| S. aureus (ATCC 25923) | 8 | 16 | 4 | 4 | 0.25 S | 0.5 | 0.25 S | 4 |
| S. aureus (ATCC 700698) | 16 | 32 | 8 | 16 | 64 R | 64 | 128 R | 128 |
| S. epidermidis (ATCC 12228) | 8 | 8 | 4 | 4 | 2 S | 2 | 0.25 S | 4 |
| S. epidermidis (ATCC 35984) | 16 | 32 | 4 | 8 | 1024 R | 1024 | 1 S | 4 |
| E. coli (ATCC 11303) | 256 | ND | 256 | ND | 1 S | 2 | 1 S | 4 |
| P. aeruginosa (ATCC 27853) | 256 | ND | 256 | ND | 128 R | ND | 64 R | 128 |
| Complex I/Ampicillin | Complex I/Tetracycline | |||||
|---|---|---|---|---|---|---|
| MIC a | MIC b | FICI | MIC a | MIC b | FICI | |
| S. aureus (ATCC 25923) | 0.24 | 0.09 | 0.311 (S) | 1.9 | 0.04 | 0.625 (NI) |
| S. aureus (ATCC 700698) | 0.98 | 12.5 | 0.375 (S) | 0.48 | 50 | 0.562 (NI) |
| S. epidermidis (ATCC 12228) | 0.24 | 0.39 | 0.311 (S) | 1.9 | 0.04 | 0.625 (NI) |
| S. epidermidis (ATCC 35984) | 0.24 | 125 | 0.186 (S) | 3.9 | 0.04 | 1.063 (NI) |
| E. coli (ATCC 11303) | 250 | 0.02 | 1.063 (NI) | 15.6 | 0.19 | 0.311 (S) |
| P. aeruginosa (ATCC 27853) | 500 | 252 | 4.0 (A) | 15.6 | 16 | 0.311 (S) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Andrade, A.L.; Pinheiro, A.A.; Malveira, E.A.; Azevedo, I.M.F.; Gondim, A.C.S.; Batista, A.A.; Neto, J.H.A.; Castellano, E.E.; Holanda, A.K.M.; Vasconcelos, M.A.; et al. The Effect of a Photoactivated Ruthenium Nitrocomplex [RuCl(NO2)(dppb)(4,4-2 Mebipy)] on the Viability of Eukaryotic and Prokaryotic Cells, Including Bacterial Biofilms. Processes 2024, 12, 2841. https://doi.org/10.3390/pr12122841
Andrade AL, Pinheiro AA, Malveira EA, Azevedo IMF, Gondim ACS, Batista AA, Neto JHA, Castellano EE, Holanda AKM, Vasconcelos MA, et al. The Effect of a Photoactivated Ruthenium Nitrocomplex [RuCl(NO2)(dppb)(4,4-2 Mebipy)] on the Viability of Eukaryotic and Prokaryotic Cells, Including Bacterial Biofilms. Processes. 2024; 12(12):2841. https://doi.org/10.3390/pr12122841
Chicago/Turabian StyleAndrade, Alexandre L., Aryane A. Pinheiro, Ellen A. Malveira, Isa M. F. Azevedo, Ana C. S. Gondim, Alzir A. Batista, João H. A. Neto, Eduardo E. Castellano, Alda K. M. Holanda, Mayron A. Vasconcelos, and et al. 2024. "The Effect of a Photoactivated Ruthenium Nitrocomplex [RuCl(NO2)(dppb)(4,4-2 Mebipy)] on the Viability of Eukaryotic and Prokaryotic Cells, Including Bacterial Biofilms" Processes 12, no. 12: 2841. https://doi.org/10.3390/pr12122841
APA StyleAndrade, A. L., Pinheiro, A. A., Malveira, E. A., Azevedo, I. M. F., Gondim, A. C. S., Batista, A. A., Neto, J. H. A., Castellano, E. E., Holanda, A. K. M., Vasconcelos, M. A., & Teixeira, E. H. (2024). The Effect of a Photoactivated Ruthenium Nitrocomplex [RuCl(NO2)(dppb)(4,4-2 Mebipy)] on the Viability of Eukaryotic and Prokaryotic Cells, Including Bacterial Biofilms. Processes, 12(12), 2841. https://doi.org/10.3390/pr12122841








