Abstract
Recent studies have demonstrated that cancer cells can elude immune cells by creating a sanctuary within the tumor’s microenvironment. Large amounts of immune-suppressing signaling proteins can be expressed by cancer cells. One of the most important mechanisms in this system is immune suppression caused by tumors and the modulation of the immune checkpoint. The immune checkpoint is modulated by both the programmed cell death protein 1 (PD-1) and its ligands, programmed death ligand 1 (PD-L1) and PD-L2. Non-coding RNAs (ncRNA), including the more well-known microRNAs (miRNAs), long non-coding RNAs (lncRNAs), and circular RNAs (circRNAs), all play roles in the regulation of biological processes and extensive diseases such as cancer. Thus, the focus of this study is on the interactions between the programmed death protein and its ligands with miRNAs, lncRNAs, and circRNAs during tumorigenesis and tumor progression. Furthermore, some FDA-approved drugs for the treatment of various cancers were based on their interactions with PD-1, PD-Ls, and ncRNAs. This promising strategy is still in the production stages, with additional results and clinical trials being processed.
1. Introduction
The immune system plays a pivotal role in halting the progression of cancer. Cancer is a condition in which the immune system fails to recognize and eliminate abnormal cells and tissues. Tumor cells can evade the immune system in part because of the immunosuppressive qualities of the tumor microenvironment. It is commonly assumed that immunological checkpoints are negative modulators of the immune response due to their primary role in preventing tissue damage [1]. These checkpoints are frequently enhanced in response to the continuous activation of immune cells. When this checkpoint is engaged, the immune cell is prevented from launching a cytotoxic response. Therefore, the immunity cells are unable to kill other cells. To maintain the stability of the autoimmunity/self-tolerance balance, these checkpoint proteins are important. However, tumors may benefit from this network as they attempt to spread and grow.
Tumor cells may be able to avoid being destroyed by the immune system because of the immunosuppressive conditions brought about by immune checkpoint signaling [2]. Checkpoints are inhibitive in nature and comprise the cytotoxic T-lymphocyte-associated molecule-4, PD-1, and PD-L1 [3]. PD-1 is a costimulatory molecule that has a repressive function in adjusting T-cell stimulation in the periphery. PD-L1 and PD-L2 are ligands for PD-1, and both can bind to PD-1. PD-L1 is a type 1 transmembrane protein that plays a role in dampening the immune response to infection to prevent persistent tissue damage [4]. Because of its role, PD-L1 is extensively distributed throughout the body. PD-L2 is a ligand of PD-1 that has received very little research attention. It is mostly found on activated dendritic cells and macrophages, both of which participate in the suppression of the immune response against tumors [5].
In recent years, ncRNAs have been established as crucial participants in epigenetic gene regulation. It has been hypothesized that ncRNAs promote epigenetic modifications by recruiting chromatin-remodeling complexes that control transcription [6]. It has also been proven that a great number of ncRNAs act as negative regulators of transcription as well as transcription cofactors [7]. ncRNAs are also thought to have a role in the control of post-transcriptional processes such the splicing, transport, translation, and destruction of mRNAs [8]. In particular, miRNAs, lncRNAs, and circRNAs are the focus of several studies due to the likelihood that they play a role in the regulation of a wide range of cellular procedures, both normally and pathologically. In the milieu of tumor immunity, a substantial number of miRNAs and, to a lesser degree, lncRNAs and circRNAs, have been discovered as powerful regulators of tumor immunity through the direct control of genes that govern the equilibrium between immune stimulation and repression [9]. Using the current literature, we explored interactions between ncRNAs and PD-L1/-L2 within the context of human neoplasms. Additionally, we highlight existing and new notions within the fields of immunology and ncRNA research.
2. Immune Checkpoints and Modulators
An organism’s immune system acts as one of its three lines of defense, protecting it from potentially harmful outside agents. The skin and mucous membranes, which serve as a natural barrier, make up the first line of defense. Natural bactericidal substances, which serve as the “vanguard” of the immune response, make up the second line of defense. Finally, the immune organs and cells, which are responsible for generating specific immune responses, make up the third line of defense. The T-cell receptor (TCR) and the peptide–major histocompatibility complex (pMHC) on the surface of antigen-presenting cells (APCs) interact to produce the first signal, and an antigen-independent co-signaling molecule is required for the second signal [4].
T-cells must receive two different signals to fully activate. The interaction of the antigenic peptide with MHC on the surface of APCs produces signal one. It is significant to highlight the immunological checkpoints that can operate as co-stimulators or co-inhibitors and tightly modulate the stimulating T-cells. If the binding of costimulatory receptors, such as CD28, occurs concurrently with the binding of the antigen/MHC and TCR, then T-lymphocytes may multiply and move toward a particular antigen. On the other hand, the activation of T-cells would be prevented if coinhibitory receptors such as CTLA-4 were activated concurrently with the binding of antigen/MHC and TCR [10,11]. CTLA-4 is not present in naive T-cells; however, it is quickly increased when T-cells are activated. Its primary function is to govern the increase in T-cells through the early priming phase in lymphoid organs. Naive T-cells do not have CTLA-4. When CTLA-4 connects to B7 proteins, it interferes with the costimulatory signals that are being sent out by CD28, which ultimately helps to inhibit overactive immunity. This co-inhibitor aids in the fight against unwanted autoimmunity and limits damage to healthy tissues [12,13].
PD-1 is an essential component in the process of keeping peripheral tolerance intact. After Src homology-2 (SH2) domain-containing phosphatases 1/2 (SHP1/2) are recruited, the TCR-mediated regulation of T-cell proliferation and cytokine production can occur [14]. This happens because the engagement of PD-1 by its ligands causes the recruitment of SHP1/2. It has been shown that an increased expression of PD-L1 on tumors has a strong correlation with an advanced disease state and an unfavorable prognosis in several kinds of cancer.
Immune checkpoint medicines do not directly eliminate cancer cells; rather, they make use of the capacity of the immune system of the host to re-enhance the body’s natural ability to fight tumors [15,16]. The development of immunotherapy with the use of immune checkpoint inhibitors (ICIs) has meaningfully enhanced the overall survival rate in a sizeable proportion of society [17], which has resulted in a revolution in the treatment of cancer. Since their discovery, ICIs have assumed the role of primary treatment for a variety of cancers. In general, the method by which ICIs exert their impact is to remove the inhibitory brakes on T-cells, resulting in an increased immunological response. Although T-cells constitute the foundation of an ICI-mediated immune response, ICIs also activate innate, adaptive, and peripheral immunological responses [18]. Because of the careful coordination of these processes, the anti-tumor immune response is much improved overall when mediated by ICIs.
The FDA has authorized three ICIs to date. The first antibody to be discovered was ipilimumab, which was an antagonist antibody against CTLA-4. It was given to people who had advanced melanoma. The second type of antibody acts by reducing the amount of PD-1 generated by T-cells. The FDA has approved pembrolizumab and nivolumab as PD-1 inhibitors (Figure 1). These drugs were approved for the treatment of melanoma; however, they are currently used to treat a variety of other cancers. The third class of pharmacological medications works by inhibiting PD-L1 expression in cancer cells. Drugs in this class include atezolizumab, durvalumab, and avelumab. These medications were originally given to treat advanced-stage melanomas; however, they are now approved for the treatment of other cancers [19,20].
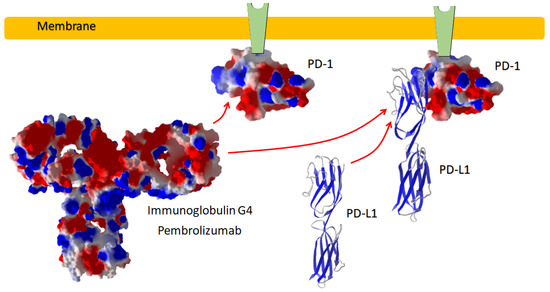
Figure 1.
PD-1 inhibitor antibody. Pembroliczumab binds with the surface of the ectodomain of PD-1, preventing the immune-suppressive reaction by the binding of PD-L1 to its site on PD-1.
3. PD-L1 and PD-L2 in Cancer
In 1992, Tasuku Honjo and coworkers identified PD-1, and this molecule was shown to be involved with apoptosis (Figure 2) [21]. This finding emerged when the PD-1 gene was cloned from dying immune cell lines. By studying the immune responses of PD-1-deficient mice, scientists were able to demonstrate that PD-1 is synthesized for use as a negative modulator for immune responses [22]. Antigen-experienced memory T-cells in peripheral tissues express proteins at a higher level than B-cells, activated monocytes, dendritic cells (DC), or natural killer (NK) cells [23]. PD-1 is a member of the type II CD28 family of protein receptors. The extracellular region is structurally and functionally similar to immunoglobulin V and includes a transmembrane region, an intracellular region consisting of about 95 residues, and two phosphorylation sites located in the immunoreceptor tyrosine-based inhibitory motif (ITIM) and the immunoreceptor tyrosine-based switch motif (IgV). Src homology phosphatase-1 (SHP-1) and SHP-22 are phosphorylated, which can suppress TCR signaling [24,25].
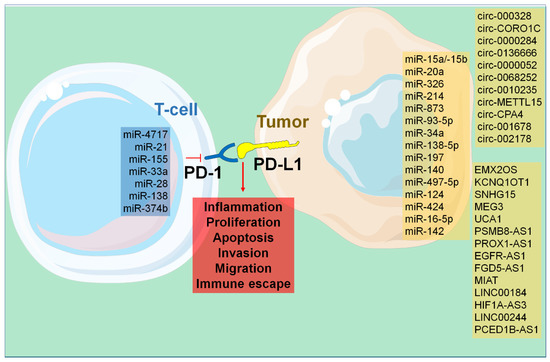
Figure 2.
Cellular processes governed by the activation of PD-1/-L1 and -L2.
PD-L1 and PD-L2 are the two PD-1 ligands; they belong to the B7 family of transmembrane proteins and function as type I receptors. The human B7-H1 gene was identified and cloned in 1999 by Lieping Chen and colleagues, who also found that it suppressed T-cells via generating IL-10 [26]. The finding that PD-1 interacted with B7-H1 led to the molecule being renamed PD-L1. The CD274 gene, located on human chromosome 9, encodes the 290-amino-acid protein receptor known as PD-L1 [27]. It consists of a transmembrane region, an intracellular region, and two extracellular regions that are structurally comparable to those of IgV and IgC. With only 30 amino acids, the PD-L1 intracellular domain is extremely short and seemingly useless. Multiple cell types, such as antigen-presenting cells, T-cells, B-cells, monocytes, and epithelial cells, might exhibit this immunological checkpoint. The protein is also upregulated in some cells in response to stimulation by pro-inflammatory cytokines, such as interferon-gamma (IFN-γ) and interleukin 4 (IL-4), by the transcription factors signal transducer and activator of transcription 1 (STAT1) and IFN regulatory factor 1 (IRF1) [28,29,30].
PD-L2 is encoded by the Pdcd1lg2 gene, which is located 42 kilobase pairs (kb) from the Cd274 gene in the human genome. It is composed of 7 exons and 273 amino acid residues split between cytoplasmic, transmembrane, IgV-like, and IgC-like domains. Bone-marrow-derived DCs, macrophages, and mast cells do not naturally produce PD-L2 but can be induced to do so [31,32].
4. NcRNAs and Cancer
For several decades, the relatively small section of the genome that is responsible for protein coding was the primary focus of medical research. The decoding of the human genome revealed that fewer than 2% of our genes eventually code for proteins, and many scientists concluded that the other 98% of our genome was nothing more than non-functional “junk” [33,34]. Regardless, the ENCODE research discovered that the section of the genome that does not code for proteins is replicated in hundreds of RNA molecules. These RNA molecules appear to have an essential role in the whole spectrum of human illness, including cancer, besides modulating essential biological processes such as growth, development, and organ function [35,36]. The overall length of an ncRNA may be used to categorize it into one of many distinct types. The miRNAs, transfer RNA-derived small RNAs (tsRNAs), and Piwi-interacting RNA (piRNAs) are examples of the tiny ncRNAs that play an essential role in cancer. On the other end of the size spectrum are the lncRNAs, which are untranslated RNAs that are longer than 200 nucleotides and comprise subclasses such as pseudogenes and circRNAs [37].
4.1. miRNAs
The family of ncRNAs known as microRNAs (miRNAs, miR) has been researched the most, since it is responsible for negatively influencing the expression of up to 60% of protein-coding genes [38]. One of the most significant characteristics of miRNAs is their ability to target several mRNA molecules at once. For example, a single miRNA may target up to 200 distinct mRNAs, and many miRNAs can modify the same mRNA target [39]. The synthesis of miRNA starts in the nucleus with the transcription of a long hairpin transcript called pri-miRNA (Figure 3).
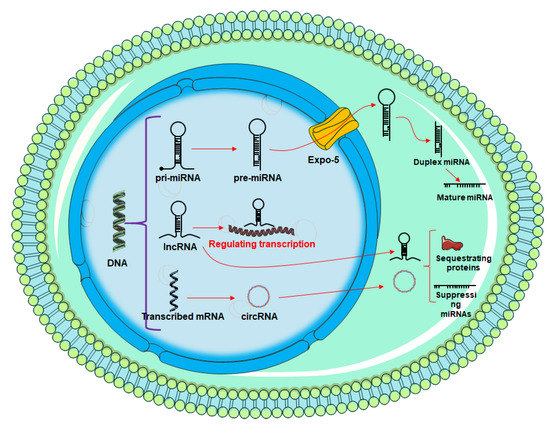
Figure 3.
Overview of miRNA and circRNA production. The process is initiated in the nucleus, then exported to the cytoplasm for downstream activities.
The length of the pri-miRNA transcript can range from hundreds to thousands of nucleotides. In addition, RNA polymerase III Drosha and DGCR8 catalyze an enzymatic mechanism that converts pri-miRNA to pre-miRNA, a shorter transcript (DiGeorge syndrome critical region 8). Approximately 70 nucleotides make up this pre-miRNA transcript. Once pre-miRNA has been transported to the cytoplasm by nuclear receptor exportin 5, it is further processed by the Dicer complex, which first forms a mature miRNA duplex of around 22 nucleotide lengths before finally reducing it to a single-stranded miRNA. As a result of sequence complementarity, typically in the 3′ untranslated region (UTR), mature miRNA loaded onto AGO2 and the RNA-induced silencing complex (RISC) will direct the targeting of certain mRNA transcripts, resulting in translational repression or mRNA destruction [40].
Changes in the expression of miRNA caused by physiological processes are critical components in the regulation of complex genomic networks and, as a consequence, cellular signaling cascades. As part of the pathological cellular alterations that can occur as a result of various diseases [41], reformed miRNA expression has a vital impact on the modification of protein expression in many disease situations. These short RNAs offer therapeutic potential for the focused modification of cell processes that are critical to a disease phenotype [42], in addition to the diagnostic potential of changed miRNA expression levels.
4.2. LncRNAs
A transcript that is longer than 200 nucleotides is referred to as a lncRNA. There are around 16,000 lncRNA genes in the human genome [43]. lncRNAs are classified into one of five different types, each of which is determined by the relative position of the protein gene and the lncRNA within the genome [44]. Sense lncRNAs, antisense lncRNAs, bidirectional lncRNAs, intronic lncRNAs, and intergenic lncRNAs all fall into this category (also known as long intergenic ncRNAs [lincRNAs]). lncRNAs have recently been found to play an important role in a wide variety of biological processes through regulating gene expression in several distinct types of cancer. Furthermore, invasion, metastasis, and chemotherapy resistance have all been linked to the abnormal expression of lncRNAs [45,46].
4.3. CircRNAs
CircRNAs are a specific kind of ncRNA that are single-stranded and are found in very large quantities in human cells. Back-splicing is one of the processes that can lead to exon shuffling, and it is the mechanism that is responsible for the production of the vast majority of circRNAs. Because of their covalently-closed loop structure, lack of open terminals, and resistance to digestion by exonucleases and RNase R [47], circRNAs are more stable and better-conserved over evolutionary time than linear RNAs. Since circRNAs do not have any free terminals, they are not easily broken down by enzymes such as RNase R or exonucleases. Many different functions of circRNAs have been discovered in recent years. These functions include scavenging for miRNAs, regulating transcription, connecting proteins, and facilitating translation [48].
5. Regulation of PD-L1 and PD-L2 in Cancer by ncRNAs
Below is the information about the ability of ncRNAs to modify the activity of PD-L1/L2 in cancers (Table 1). There is a special focus on the interaction of miRNAs and PD-L1, which is shown in Figure 4.
Figure 4.
An overview of miRNAs with the ability to target PD-1/PD-L1 in human malignancies.
5.1. miRNAs
According to a recent study, miR-15a/miR-15b is responsible for the degradation of PD-L1 mRNA. The RNA-induced silencing complex and the 3′-UTR play a role in this phenomenon. Additional evidence suggests that miR-15a/miR-15b promotes neuroblastoma-specific CD8+T-cell activation and cytotoxicity in vitro. In addition, the injection of miR-15a-overexpressing murine cells reduced tumor growth and vasculature and enhanced CD8+T and NK cell activation and infiltration into tumors in vivo. The anti-PD-L1 antibody has also been demonstrated to interfere with miR-15a/miR-15b-induced CD8+T and NK cell-regulated anti-cancer responses [49]. NSCLC cells were more likely to proliferate when miR-20a was present because it reduced the amount of PTEN that was expressed and increased the amount of PD-L1 [50].
Shao and colleagues found that the expression of miR-326 in LUAD tissue had a negative correlation with PD-L1/B7-H3. The change in the cytokine profile of CD8+ T-cells and the reduced migratory capacity of tumor cells are both brought about as a result of the inhibition of PD-L1 and B7-H3 expression, due to the overexpression of miR-326. At the same time, the downregulation of miR-326 facilitated the migration of tumor cells. In addition, inhibiting PD-L1 and B7-H3 had a dampening impact on the effect that the miR-326 inhibitor had on stimulating tumor growth. After miR-326 was overexpressed in tumor-bearing mice, there was a considerable increase in the infiltration of CD8+ T-cells, as well as a significant enhancement in the production of both TNF-α and IFN-γ, both of which contributed to the growth of the tumor [51].
In diffuse large B-cell lymphoma (DLBCL) tissues and cell lines, miR-214 was found to be downregulated, whilst PD-L1 was found to be increased. This was in comparison with normal neighboring tissues or normal B-cells. This suggested that there was an inverse relationship between the expression levels. The overexpression of miR-214 in OCI-Ly3 cells led to the triggering of apoptosis, as well as the inhibition of cell viability and invasion. In addition to this, it was discovered that miR-214 targets PD-L1 mRNA by binding to the 3′-untranslated region. OCI-Ly3 cells had a less malignant behavior after PD-L1 expression was knocked down. Targeting PD-L1 in vivo with an overexpression of miR-214 was shown to limit the development of tumors [52].
When PD-L1 was overexpressed, it activated the PI3K/Akt and ERK1/2 pathways, which in turn caused breast cancer cells to become resistant to chemotherapy and exhibit stem cell characteristics. A correlation between PD-L1 expression and stemness marker expression provided proof of this. Reduced PD-L1 expression and PI3K/Akt/ERK1/2 signaling [53] were responsible for miR-873′s effects on breast cancer stemness and chemoresistance. Mechanistically, miR-873 bound to the 3′-UTR of PD-L1 and inhibited its expression, hence repressing its production. MiR-93-5p has been definitively linked to tumor growth and immune regulation in breast cancer, and it directly targets PD-L1 and CCND1 to accomplish its goals [54]. Wang et al. analyzed miR-34a and PD-L1 expression in 44 AML specimens (AML). An unfavorable correlation between PD-L1 and miR-34a expression was observed. The overexpression of miR-34a inhibited PD-L1 expression and reduced surface PD-L1 expression in HL-60 and Kasumi-1 cells. It was observed that miR-34a was a possible binding partner for the PD-L1-3′UTR region. In a similar vein, miR-34a was able to reverse the increase in PD-L1 surface expression that had been induced by chemotherapeutic medications, and was also able to reduce PD-L1-specific T-cell death after transfection. In addition, it was found that PD-L1 expression and AKT activation form a positive feedback loop [55].
MiR-138-5p, a tumor suppressor in CRC, works by reducing levels of the protein PD-L1. PD-L1 and miR-138-5p may be used as CRC biomarkers for optimum treatment selection and clinical outcome prediction [56], as patients with low miR-138-5p and high PD-L1 had shorter overall survival. In their study, Fujita et al. reported that PD-L1 is a possible biomarker of the miR-197-mediated CKS1B/STAT3 axis, which, in turn, regulates the expression of several oncogenic genes such as Bcl-2, c-Myc, and cyclin D1. Increased chemoresistance, tumorigenicity, and pulmonary metastasis are seen both in vitro and in vivo when miR-197 is downregulated in platinum-resistant NSCLC [57]. Furthermore, Xie and coworkers found that, whereas NSCLC cells expressed high amounts of PD-L1 and cyclin E, they expressed low levels of miR-140. In A549 and NCI-H1650 cells, miR-140 overexpression suppressed cell growth, and this effect was correlated with decreased cyclin E expression. The direct binding of miR-140 to the 3′ untranslated region of PD-L1 was responsible for its downregulation (UTR). Inhibiting PD-L1 had a comparable effect on cyclin E expression, resulting in a decrease in its levels independent of miR-140 levels [58]. It was discovered that an increase in PD-L1 expression was connected to a decrease in the level of miRNA-497-5p. The downregulation of miRNA-497-5p was connected not only with an increase in PD-L1 expression but also with a decreased chance of survival. In two different cell lines derived from renal cell carcinoma, PD-L1 served as a direct target for miRNA-497-5p. In addition, this miRNA increased apoptosis while inhibiting cell proliferation, clone formation, and migration in in vitro experiments, and this was the case despite the fact that it suppressed cell proliferation [59].
It was also shown that miR-124 expression is drastically lower in CRC tissues compared with almost normal samples, and that this reduction is correlated negatively with PD-L1. Additional research showed that transfecting HT29 and SW480 cells with miR-124 mimics significantly reduced PD-L1 mRNA, protein, and cell surface expression. Interleukin (IL)-10, IL-2, tumor necrosis factor (TNF), and transforming growth factor (TGF) beta expression were all downregulated in coculture models, with the additional effect of suppressing Tregs [60]. Based on research into the underlying mechanisms, it was revealed that miR-424(322) suppressed the production of PD-L1 and CD80. The inhibition of the PD-L1 immunological checkpoint is observed concurrently with the reversal of chemoresistance that results from the restoration of miR-424(322) expression. This synergistic impact of chemotherapy and immunotherapy is linked to the proliferation of functional cytotoxic CD8+ T-cells, as well as the suppression of myeloid-derived suppressive cells (MDSCs) and Tregs [61]. In addition to this, it was shown that the expression of miR-224-5p was considerably elevated in the urine extracellular vesicles (EVs) of renal cell carcinoma (RCC) patients in comparison with healthy volunteers. The overexpression of miR-224-5p was found to be responsible for the inhibition of RCC cell growth as well as the induction of cell cycle arrest. Through a combination of prediction and confirmation, the cyclin D1-encoding gene, CCND1, was found to be a direct target of the miRNA known as miR-224-5p. In addition to this, the expression of miR-224-5p in RCC cells led to an increase in both their invasiveness and their capacity to metastasize. It is interesting to note that, by reducing CCND1, miR-224-5p was also able to boost the stability of the PD-L1 protein. This impact had the potential to be conveyed by EVs and further increased the resistance of RCC cells to toxicity dependent on T-cells [62].
The results of a recent study suggest that M1 macrophages had a negative association with the advancement of gastric cancer (GC), which shed light on the anti-tumor role that M1 macrophages play. Exosomal miR-16-5p can be produced by M1 macrophages, and this miR-16-5p can precisely target and inhibit PDL1 expression on GC cells. Blocking the PD1/PDL1 checkpoint may result in T-cell activation and the suppression of GC proliferation [63]. Furthermore, Zhao et al. found that miRNA-3127-5p promotes STAT3 phosphorylation by restricting autophagy; autophagy may preserve pSTAT3 into the nucleus in miRNA-3127-5p knockdown cells; and immune escape driven by high doses of PD-L1 leads to lung cancer chemoresistance [64]. Increasing miR-142-5p levels has been shown to slow the development of pancreatic cancer in animal and human studies. MiR-142-5p overexpression on tumor cells downregulates PD-L1 expression, which in turn increases the number of CD4+ and CD8+ T lymphocytes, decreases the percentage of PD-1+ T lymphocytes, and boosts the amount of IIFN-γ and TNF-α in tumor microenvironments. Consequently, inhibiting the PD-L1/PD-1 pathway is one mechanism through which upregulating miR-142-5p expression can boost anti-tumor immunity [65]. Over-expression of the let-7 family was also found to inhibit immune evasion in head and neck squamous cell carcinoma (HNSCC) through suppressing TCF-4 expression, -catenin/STT3-mediated PD-L1 glycosylation, and PD-L1 stability, while simultaneously helping PD-L1 ubiquitination and degradation, as well as the T-cell recognition of HNSCC cells [66]. Furthermore, miR-140-3p has been demonstrated to be a tumor suppressor in colorectal cancer via directly reducing PD-L1 and deactivating the PI3K/AKT pathway, indicating that miR-140-3p may be a promising marker for CRC diagnostics and therapy [67].
5.2. lncRNAs
The existence of lncRNAs affects a wide range of epigenetic regulatory mechanisms. The bulk of carcinogenesis and progression cannot be studied in isolation from the function that lncRNAs play in their regulation. When EMX2OS was knocked out, almost all of the cancerous characteristics of ovarian cancer cells were hindered; however, when EMX2OS was overexpressed, the same activities were promoted. In an in vivo xenograft model of human ovarian cancer, EMX2OS hastened tumor development. AKT3, a direct target of miR-654, was overexpressed due to the fact that EMX2OS may independently bind to miR-654 and suppresses its production. The overexpression of miR-654 or knockdown of EMX2OS was found to negatively affect cell proliferation, invasion, and sphere formation; however, restoring AKT3 was proven to rescue the cells from this fate. Moreover, PD-L1 was identified as the key oncogenic component in ovarian cancer cells, where it operated downstream of AKT3. The ectopic expression of PD-L1 prevented the anti-cancer effects of EMX2OS knockdown and AKT3 silencing in ovarian cancer cells [68]. Additionally, lncRNA KCNQ1OT1 acts as a sponge for miR-15a, which then allows it to enhance immune evasion and the advancement of malignant prostate cancer by up-regulating PD-L1 [69].
Dang and colleagues illustrated that SNHG15 and PD-L1 could be positively associated with one another. The upregulation of SNHG15 in gastric cells led to a reduction in the expression of miR-141, an inhibitor of PD-L1. This led to an increase in PD-L1 expression, which in turn enabled gastric cancer cells to be resistant to immune responses [70]. In addition, Xu et al. discovered that the amount of PD-L1 protein in invasive endometrial cancer cells might be controlled via the activity of miR-216a, which directly reduced PD-L1. They also discovered a mechanism by which the lncRNA MEG3 represses the production of miR-216a, which ultimately results in enhanced PD-L1 expression and a considerable reduction in cell motility and invasion [71].
The levels of expression of the lncRNAs UCA1 and PD-L1 were shown to be higher in anaplastic thyroid cancer (ATC) tissues and cells. The knocking down of UCA1 and PD-L1 led to an increase in the cytotoxic CD8+ T-cells’ ability to destroy ATC cells. UCA1 acted as a negative regulator of the production of miR-148a, which in turn caused miR-148a to target PD-L1 and reduce the expression of that gene. In addition, it was shown that UCA1 inhibited the killing function of cytotoxic CD8+ T-cells and decreased the release of cytokines by working in conjunction with PD-L1 and miR-148a. Restoring the reduction in the lethal impact of CD8+ T-cells in vivo was achieved by silencing either UCA1 or PD-L1 in ATC cells [72]. Furthermore, it has been shown that the lncRNA PSMB8-AS1 promotes the proliferation and metastasis of pancreatic cancer (PC) cells by sponging up miR-382-3p, thereby increasing STAT1 expression. One plausible therapeutic target for PC is the PSMB8-AS1/miR-382-3p/STAT1/PD-L1 axis [73]. PC may benefit from targeting this axis with therapy because STAT1 transcriptionally upregulated PD-L1 and its stability.
Recent research found that the GC cell ectopic expression of miR-877-5p suppressed cell growth, reduced invasion, and induced cell death. Furthermore, it was shown that PD-L1, which is a target of miR-877-direct 5p, was negatively regulated. PD-L1 serves as a target for miR-877-5p. Several tests showed that miR-877-5p has a direct correlation with PD-L1, and several related experiments were carried out. It has been demonstrated that knocking down the lncRNA PROX1-AS1 reduced cell proliferation and that an increase in invasion ability was brought about by an inhibitor of miR-877-5p. However, a consequence of this inhibitor of miR-877-5p was later recovered by depleting the PD-L1 protein [74]. In addition, it was revealed that EGFR-AS1 increased PD-L1 expression in leiomyosarcoma cells by way of the EGFR/MYC pathway. This, in turn, suppresses T-cell infiltration and contributes to immunological escape [75]. Moreover, Zhang et al. [76] showed that silencing lncRNA FGD5-AS1 increased PD-L1 via sponging miR-497-5p, hence reducing malignant behaviors in colon cancer cells in vitro and in vivo. Proliferation, viability, invasion, migration, and EMT are all facets that fall under this category. Furthermore, it was found that MIAT was responsible for controlling the expression of PD-L1, and that both MIAT and PD-L1 were significantly upregulated in HCC tissues. T-cell cytotoxicity toward HCC cells increased after MIAT was inhibited. At the transcriptional level, MIAT was able to reduce miR-411-5p expression, increase STAT3 activity, and then increase PD-L1 expression [77].
Docetaxel resistance and a poor prognosis in prostate cancer were both discovered to be connected with the gene LINC00184. In prostate cancer cells, this gene influenced docetaxel resistance as well as the immunological response mediated by T-cells. The adsorption of miR-105-5p led to the induction of LINC00184, which then negatively regulated miR-105-5p and, as a consequence, decreased the expression level of PD-L1 [78]. HIF1A-AS2 is an oncogene that regulates the miR-429/PD-L1 axis to promote the proliferation and metastasis of GC cells; it is also a strong predictor of malignancy and prognosis in GC [79]. In addition, HIF1A-AS3 predicts the likelihood of survival in GC patients. A recent study found that HOXA-AS3 was significantly upregulated in HCC cells, and that it controlled PD-L1 expression by sponging miR-455-5p. The miR-455-5p/PD-L1 axis was also targeted by HOXA-AS3 overexpression, contributing to HCC cell invasion and proliferation [80]. Through a process known as the downregulation of PD-L1 expression, LINC00244 was able to reduce the proliferation, invasion, and metastasis of HCC. In addition, the decreased expression of LINC00244 triggered pathways that are involved in the epithelial–mesenchymal transition (EMT), which accelerated the fast development and metastasis of HCC cells [81]. In addition, PCED1B-AS1 and miR-194-5p were increased in head and neck cancer. In this cancer, PCED1B-AS1 had a positive correlation with PD-Ls (PD-L1 and PD-L2); however, it had a negative correlation with miR-194-5p. PCED1B-AS1 had an interaction with mir-194-5p, which resulted in the suppressed expression of PD-Ls. Through the process of sponging mir-194-5p, PCED1B-AS1 was able to increase the expression of PD-Ls. PCED1B-AS1 is responsible for the immunosuppression mediated by PD-Ls in co-cultured T-cells. Exosomes containing PCED1B-AS1 were produced by HCC cells, and these exosomal PCED1B-AS1 molecules boosted the expression of PD-Ls in receipt head and neck cancer cells while inhibiting the expression of PD-Ls in receipt T-cells and macrophages. PCED1B-AS1 suppressed apoptosis while simultaneously promoting cancerous features and in vivo tumor development in nude mice that had been xenografted with human tumor cells [82].
5.3. circRNAs
Growing amounts of data suggest that circRNAs have functional roles in cellular procedures such as miRNA sponging, transcription regulation, and gene expression regulation. It has been proven that hsa-circ-0003288 increased EMT and invasion of HCC by way of the PI3K/Akt pathway via the hsa-circ-0003288/miR-145/PD-L1 axis [83]. Wu et al. found a new circRNA, which they named circCORO1C. HCC patients and cell lines were discovered to have significantly higher expression of this circRNA. The proliferation and metastasis of hepatocellular carcinoma are mediated by CircCORO1C [84]. This is accomplished by increasing the expression of PD-L1 via the NF-kB pathway. Furthermore, circ 0000284 was shown to be upregulated in NSCLC, and can be used to indicate a bad prognosis for individuals with NSCLC. The downregulation of circ-0000284 inhibited the development of NSCLC tumors in both in vitro and in vivo experimental settings. In addition, circ-0000284 was a miRNA target of miR-377-3p, which targeted PD-L1 [85].
When compared with a normal colon epithelial cell line, the findings of a recent study revealed that the levels of PD-L1 and circ-0136666 were up in colorectal cancer cells; however, miR-497 was shown to be at a lower level in colorectal cancer cells. It was determined that circ-0136666 specifically targets miR-497, which controls PD-L1 by binding to its 3′UTR. Circ-0136666 was able to control cell proliferation and death by targeting miR-497 and modulating the expression of PD-L1. It is noteworthy that circ-0136666 can activate Treg cells via the miR-497/PD-L1 axis and its downstream signal pathway in Treg cells [86]. When it comes to regulating cell proliferation, migration, invasion, and clonogenicity in HNSCC, the overexpression of PD-L1 is essential on both the clinical and fundamental biological levels. Signaling via IFN-γ/JAK2/STAT1 plays a significant role in the process that leads to the induction of PD-L1 overexpression.
It has been established that increased circ-0000052 is responsible for PD-L1 overexpression. It does this by binding competitively to miR-382-3p and reducing the suppressive impact that miR-382-3p has on PD-L1 [87]. Additionally, it was shown that a high level of circ-0068252 was associated with a poor prognosis of non-small cell lung cancer as well as cisplatin resistance. It is possible that knocking down the circ-0068252 gene will make cisplatin more effective in treating cisplatin-resistant NSCLC cells. In addition, knocking down circ-0068252 was able to regulate the immunological microenvironment, and this regulation was mediated by CD8+ T-cells. By absorbing miR-1304-5p, circ 0068252 has the potential to stimulate the expression of PD-L1 [88]. Circ-0010235 was elevated in lung cancer, and via sponging miR-636 and upregulating PD-L1, circ-0010235 accelerated lung cancer development and immune escape [89]. In addition, inhibiting Circ-METTL15 activity in lung cancer cells led to an upsurge in the rate of apoptosis, as well as a reduction in some cancerous characteristics, including immune evasion. Because it acted as a sponge for miR-1299, Circ-METTL15 was able to exert a regulatory influence on lung cancer via miR-1299. In addition, PD-L1 was shown to be a functional target of miR-1299, and research found that miR-1299 prevented the growth of lung cancer cells by lowering PD-L1 expression [76].
A recent study discovered that inhibiting circ-CPA4 using a knock-down strategy reduced levels of PD-L1 both inside and outside of cells via targeting let-7 miRNA. On the one hand, PD-L1 acted as a regulator of proliferation, motility, stemness, and chemoresistant properties in NSCLC cells that were treated with cisplatin. On the other hand, PD-L1 secreted inactivated CD8+ T-cells to promote immune evasion. This was accomplished by triggering extracellular and intracellular pathways that facilitated death in impaired cells [90]. Circ-001678 might act as a sponge for miR-326 and boost ZEB1 expression. Treatment with an anti-PD-L1/PD-1 antibody combination might counteract the effects of circ-001678 on tumor growth. The stimulation of the PD-1/PD-L1 pathway by circ-001678 led to the promotion of CD8+ T-cell death, which resulted in the induction of NSCLC cell immune escape via control of the miR-326/ZEB1 axis. These results suggest that circ-001678 functions as a sponge for miR-326, which then upregulates ZEB1 expression and induces the PD-1/PD-L1 pathway-dependent immune escape, hence aiding the formation of malignant tumors in NSCLC [78]. In addition, Wang and colleagues discovered that circ-002178 was considerably elevated in lung adenocarcinoma tissues, the cells and exosomes of cancer cells, and blood from patients. Through the process of sponging miR-34 in cancer cells, circ-002178 has the potential to increase PD-L1 expression. Additionally, circ-002178 might be transported into T-cells through the exosomes that are released by cancer cells in order to enhance PD1 expression. This would be accomplished by sequestering miR-28-5p [91].

Table 1.
ncRNAs and their mechanisms to regulate PD-Ls.
Table 1.
ncRNAs and their mechanisms to regulate PD-Ls.
| ncRNA | Name | Target | Role | Reference |
|---|---|---|---|---|
| miRNA | miR-15a/-15b | PD-L1 | Induces activation and cytotoxicity in CD8+T and NK cells against neuroblastoma | [49] |
| miR-20a | PD-L1 | Reduces the amount of PTEN and PD-L1 | [50] | |
| miR-326 | PD-L1 | Change in the cytokine profile of CD8+ T-cells and the reduced migratory capacity of tumor cells | [51] | |
| miR-214 | PD-L1 | Induction of apoptosis as well as repressing cell viability and migration | [52] | |
| miR-873 | PD-L1 | Chemoresistance and stemness-like features in breast cancer cells via activating PI3K/Akt and ERK1/2 pathways | [53] | |
| miR-93-5p | PD-L1 | Affects tumor growth and immune modulation in breast cancer | [54] | |
| miR-34a | PD-L1 | Inhibited PD-L1 expression | [55] | |
| miR-138-5p | PD-L1 | A tumor suppressor in CRC | [56] | |
| miR-197 | PD-L1 | Mediates CKS1B/STAT3 axis tumor growth | [57] | |
| miR-140 | PD-L1 | Represses cellular proliferation, related to lower expression of cyclin E and inactivating the PI3K/AKT pathway | [58] | |
| miRNA-497-5p | PD-L1 | Increases apoptosis while inhibiting cell proliferation, clone formation, and migration | [59] | |
| miR-124 | PD-L1 | Suppresses Tregs by altering IL-10, IL-2, TNF, and TGF-β | [60] | |
| miR-424 | PD-L1 | Increase the proliferation of functional cytotoxic CD8+ T-cells, MDSCs and Tregs | [61] | |
| miR-16-5p | PD-L1 | T-cell stimulation and suppression of tumor proliferation | [62] | |
| miR-142-5p | PD-L1 | Growth in the number of CD4+ and CD8+ T lymphocytes, a reduction in the percentage of PD-1+ T lymphocytes, and an upsurge in the quantity of IFN-γ and TNF-α in TME | [63] | |
| lncRNAs | EMX2OS | PD-L1 | Directly interacts with miR-654 and suppresses its expression | [68] |
| KCNQ1OT1 | PD-L1 | Sponging miR-15a | [69] | |
| SNHG15 | PD-L1 | Reduction in the expression of miR-141, an inhibitor of PD-L1. | [70] | |
| MEG3 | PD-L1 | Represses the production of miR-216a, which ultimately results in enhanced PD-L1 expression | [71] | |
| UCA1 | PD-L1 | Sponging miR-148a which could target PD-L1 | [72] | |
| PSMB8-AS1 | PD-L1 | Sponging miR-382–3p in order to increase STAT1 expression | [73] | |
| PROX1-AS1 | PD-L1 | Inhibits miR-877-5p, which could target PD-L1 | [74] | |
| EGFR-AS1 | PD-L1 | Suppresses T-cell infiltration and contributes to immunological escape | [75] | |
| FGD5-AS1 | PD-L1 | Elevates PD-L1 via sponging miR-497-5p | [76] | |
| MIAT | PD-L1 | Suppresses the expression of miR-411-5p, boosts the activity of STAT3, and finally raises the level of PD-L1 expression | [77] | |
| LINC00184 | PD-L1 | Negatively regulates miR-105-5p and, as a consequence, decreases the expression level of PD-L1 | [78] | |
| HIF1A-AS3 | PD-L1 | Sponges miR-455-5p | [79] | |
| LINC00244 | PD-L1 | Represses PD-L1 expression | [81] | |
| PCED1B-AS1 | PD-L1/-L2 | Sponges mir-194-5p | [82] | |
| circRNAs | circ-0003288 | PD-L1 | Increases EMT and invasion of HCC by way of the PI3K/Akt pathway | [83] |
| circCORO1C | PD-L1 | Increasing the expression of PD-L1 via the NF-kB pathway | [84] | |
| circ-0000284 | PD-L1 | Is a miRNA target of miR-377-3p, which targets PD-L1 | [85] | |
| circ-0136666 | PD-L1 | Directly targets miR-497, which also regulates PD-L1 | [86] | |
| circ-0000052 | PD-L1 | Signaling via IFN-γ/JAK2/STAT1 | [87] | |
| circ-0068252 | PD-L1 | By absorbing miR-1304-5p, this circRNA has the potential to stimulate the expression of PD-L1 | [88] | |
| circ-0010235 | PD-L1 | Sponging miR-636 and upregulating PD-L1 | [89] | |
| circ-METTL15 | PD-L1 | Acts as a sponge for miR-1299 | [76] | |
| circ-CPA4 | PD-L1 | Targeting let-7 miRNA | [90] | |
| circ-001678 | PD-L1 | Sponge for miR-326 and boosts ZEB1 expression | [78] | |
| circ-002178 | PD-L1 | Sequesters miR-28-5p | [91] |
6. FDA-Approved Drugs
FDA-approved immunotherapies targeting PD-1/PD-L1 signaling might help stimulate long-lasting anti-tumor immune responses in certain patients with advanced malignancies [92,93,94]. This has led to immunotherapies becoming the first-line treatment for specific kinds of neoplasms. There is a correlation between the overexpression of PD-L1 and PD-1 on tumor cells and tumor-infiltrating lymphocytes, respectively, and a poor prognosis of illness in some human malignancies. Certain cancer cells have the potential to produce inhibitory ligands, which have the capability of binding co-inhibitory receptor molecules. Consequently, normal anti-tumor immune responses are suppressed, which contributes to immune evasion. Therefore, blocking these immunological targets might cause the patient’s immune system to mount an attack against the tumor.
Three anti-PD-L1 antibodies have been granted FDA approval: atezolizumab (IgG4 mAb), durvalumab (IgG1 mAb), and avelumab (IgG1 mAb) [95]. Patients with locally advanced or metastatic urothelial carcinoma have access to atezolizumab, the first PD-LI inhibitor licensed by the FDA [96]. In addition, patients with metastatic non-small-cell lung cancer (NSCLC), whose illness progressed while they were receiving platinum-containing medications for chemotherapy, have been approved to utilize this antibody [97]. Furthermore, the FDA has authorized the use of the combination of the antiangiogenic medications atezolizumab and bevacizumab for the treatment of persons with unresectable or metastatic hepatocellular carcinoma (HCC) [98]. BRAF V600E mutation-positive unresectable or metastatic melanoma patients may utilize atezolizumab in conjunction with cobimetinib, an inhibitor of the mitogen-activated extracellular kinase (MEK), and vemurafenib, a repressor of the B-Raf enzyme [99]. Locally progressed or metastatic urothelial carcinoma and Merkel cell carcinoma (MCC), a rare but lethal form of skin cancer, were the initial indications for the use of durvalumab [100,101]. Patients with advanced NSCLC may benefit from first therapy with durvalumab in addition to etoposide and carboplatin or cisplatin [102]. This treatment schedule has been given the green light. In 2017, the FDA approved avelumab for the treatment of MCC and metastatic urothelial carcinoma [101,103]. FDA approval was received in 2019 for the use of avelumab in combination with the tyrosine kinase inhibitor axitinib as a first-line therapy for patients with advanced renal cell carcinoma (RCC) [104].
7. Conclusions and Future Perspectives
NcRNA and PD-L1/PD-L2 interactions determine how well patients respond to anti-cancer medications. The therapeutic techniques employed may also have an impact on these connections. The most surprising discovery of this research was the presence of several ncRNAs connected to PD-L1/L2 in exosomes discharged into the environment by tumor cells. This finding lends credence to the idea that these transcripts could be used as biomarkers for the early identification of neoplasia using biofluids, and it also draws attention to the crucial roles played by these lnRNAs during tumor development and in the regulation of the tumor microenvironment. Due to the promising outcomes seen in recent years from clinical trials with anti-PD-L1/-L2 medicines, scientists are now looking into alternative therapeutic approaches. Considering their potential role as modulators of PD-L1/PD-L2 expression and activity, ncRNAs could be exploited as therapeutic targets in this context. Additionally, ncRNAs associated with PD-L1/-L2 can be used to predict a patient’s response to PD-L1/-L2 inhibitors. The apparent lack of response to this medication may be due to the expression levels of PD-L1/-L2-associated ncRNAs. As a result, antisense oligonucleotides or siRNA can be used to modulate the synthesis of these transcripts to enhance the therapeutic response to anti-PD-L1/-L2 treatment. High-throughput sequencing tools and a bioinformatics approach can be used to achieve this. This objective, however, cannot be achieved without further investigation into the idea of tissue-specific panels of ncRNAs.
Author Contributions
Conceptualization, M.K. and D.A.; methodology, M.K.; software, M.K.; validation, M.K., I.M.E.-S., S.S. and Y.H.; formal analysis, M.K.; investigation, M.K., A.M.A.M.; resources, M.K.; data curation, M.K., M.K.Z. and D.A.; writing—original draft preparation, M.K.; writing—review and editing, K.M.Z., N.E. and A.H., E.-A.M.E.-H.; visualization, M.G.E. and A.E.; supervision, I.A.; project administration, F.I. and M.S.; funding acquisition, M.K. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Deanship of Scientific Research, Vice Presidency for Graduate Studies and Scientific Research, King Faisal University, Saudi Arabia (Grant No. Grant2615).
Data Availability Statement
All data can be found within the manuscript.
Acknowledgments
The authors would like to acknowledge the support from the KFU mentors program. We appreciate the financial support from the Deanship of Scientific Research, Vice Presidency for Graduate Studies and Scientific Research, King Faisal University, Saudi Arabia (Grant No. Grant2615).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Mittal, D.; Gubin, M.M.; Schreiber, R.D.; Smyth, M.J. New insights into cancer immunoediting and its three component phases—Elimination, equilibrium and escape. Curr. Opin. Immunol. 2014, 27, 16–25. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Xu, C. Immune checkpoint signaling and cancer immunotherapy. Cell Res. 2020, 30, 660–669. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Siddiqui, B.A.; Anandhan, S.; Yadav, S.S.; Subudhi, S.K.; Gao, J.; Goswami, S.; Allison, J.P. The next decade of immune checkpoint therapy. Cancer Discov. 2021, 11, 838–857. [Google Scholar] [CrossRef]
- Pardoll, D.M. The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer 2012, 12, 252–264. [Google Scholar] [CrossRef] [PubMed]
- Yearley, J.H.; Gibson, C.; Yu, N.; Moon, C.; Murphy, E.; Juco, J.; Lunceford, J.; Cheng, J.; Chow, L.Q.; Seiwert, T.Y. PD-L2 expression in human tumors: Relevance to anti-PD-1 therapy in cancer. Clin. Cancer Res. 2017, 23, 3158–3167. [Google Scholar] [CrossRef] [PubMed]
- Ponting, C.P.; Oliver, P.L.; Reik, W. Evolution and functions of long noncoding RNAs. Cell 2009, 136, 629–641. [Google Scholar] [CrossRef]
- Feng, J.; Bi, C.; Clark, B.S.; Mady, R.; Shah, P.; Kohtz, J.D. The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev. 2006, 20, 1470–1484. [Google Scholar] [CrossRef]
- Siomi, H.; Siomi, M.C. On the road to reading the RNA-interference code. Nature 2009, 457, 396–404. [Google Scholar] [CrossRef]
- Liu, L.; Wang, Q.; Qiu, Z.; Kang, Y.; Liu, J.; Ning, S.; Yin, Y.; Pang, D.; Xu, S. Noncoding RNAs: The shot callers in tumor immune escape. Signal Transduct. Target. Ther. 2020, 5, 1–24. [Google Scholar] [CrossRef]
- Dyck, L.; Mills, K.H. Immune checkpoints and their inhibition in cancer and infectious diseases. Eur. J. Immunol. 2017, 47, 765–779. [Google Scholar] [CrossRef]
- Torphy, R.J.; Schulick, R.D.; Zhu, Y. Newly emerging immune checkpoints: Promises for future cancer therapy. Int. J. Mol. Sci. 2017, 18, 2642. [Google Scholar] [CrossRef]
- Chikuma, S. CTLA-4, an essential immune-checkpoint for T-cell activation. In Emerging Concepts Targeting Immune Checkpoints in Cancer and Autoimmunity; Springer: Berlin/Heidelberg, Germany, 2017; pp. 99–126. [Google Scholar]
- Zam, W.; Ali, L. Immune checkpoint inhibitors in the treatment of cancer. Curr. Rev. Clin. Exp. Pharmacol. Former. Curr. Clin. Pharmacol. 2022, 17, 103–113. [Google Scholar]
- Dermani, F.K.; Samadi, P.; Rahmani, G.; Kohlan, A.K.; Najafi, R. PD-1/PD-L1 immune checkpoint: Potential target for cancer therapy. J. Cell. Physiol. 2019, 234, 1313–1325. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, H.; Liu, C.; Wang, Z.; Wu, W.; Zhang, N.; Zhang, L.; Hu, J.; Luo, P.; Zhang, J. Immune checkpoint modulators in cancer immunotherapy: Recent advances and emerging concepts. J. Hematol. Oncol. 2022, 15, 1–53. [Google Scholar] [CrossRef] [PubMed]
- Chowell, D.; Yoo, S.-K.; Valero, C.; Pastore, A.; Krishna, C.; Lee, M.; Hoen, D.; Shi, H.; Kelly, D.W.; Patel, N. Improved prediction of immune checkpoint blockade efficacy across multiple cancer types. Nat. Biotechnol. 2022, 40, 499–506. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.X.; Kurra, V.; Gainor, J.F.; Sullivan, R.J.; Flaherty, K.T.; Lee, S.I.; Fintelmann, F.J. Immune checkpoint inhibitor cancer therapy: Spectrum of imaging findings. Radiographics 2017, 37, 2132–2144. [Google Scholar] [CrossRef]
- Thallinger, C.; Füreder, T.; Preusser, M.; Heller, G.; Müllauer, L.; Höller, C.; Prosch, H.; Frank, N.; Swierzewski, R.; Berger, W. Review of cancer treatment with immune checkpoint inhibitors. Wien. Klin. Wochenschr. 2018, 130, 85–91. [Google Scholar] [CrossRef]
- Ohaegbulam, K.C.; Assal, A.; Lazar-Molnar, E.; Yao, Y.; Zang, X. Human cancer immunotherapy with antibodies to the PD-1 and PD-L1 pathway. Trends Mol. Med. 2015, 21, 24–33. [Google Scholar] [CrossRef]
- Pennock, G.K.; Chow, L.Q. The evolving role of immune checkpoint inhibitors in cancer treatment. Oncologist 2015, 20, 812–822. [Google Scholar] [CrossRef]
- Ishida, Y.; Agata, Y.; Shibahara, K.; Honjo, T. Induced expression of PD-1, a novel member of the immunoglobulin gene superfamily, upon programmed cell death. EMBO J. 1992, 11, 3887–3895. [Google Scholar] [CrossRef]
- Nishimura, H.; Nose, M.; Hiai, H.; Minato, N.; Honjo, T. Development of lupus-like autoimmune diseases by disruption of the PD-1 gene encoding an ITIM motif-carrying immunoreceptor. Immunity 1999, 11, 141–151. [Google Scholar] [CrossRef]
- Keir, M.E.; Butte, M.J.; Freeman, G.J.; Sharpe, A.H. PD-1 and its ligands in tolerance and immunity. Annu. Rev. Immunol. 2008, 26, 677–704. [Google Scholar] [CrossRef]
- Konstantinidou, M.; Zarganes-Tzitzikas, T.; Magiera-Mularz, K.; Holak, T.A.; Dömling, A. Immune checkpoint PD-1/PD-L1: Is there life beyond antibodies? Angew. Chem. Int. Ed. 2018, 57, 4840–4848. [Google Scholar] [CrossRef] [PubMed]
- Tang, Q.; Chen, Y.; Li, X.; Long, S.; Shi, Y.; Yu, Y.; Wu, W.; Han, L.; Wang, S. The role of PD-1/PD-L1 and application of immune-checkpoint inhibitors in human cancers. Front. Immunol. 2022, 13, 964442. [Google Scholar] [CrossRef]
- Dong, H.; Zhu, G.; Tamada, K.; Chen, L. B7-H1, a third member of the B7 family, co-stimulates T-cell proliferation and interleukin-10 secretion. Nat. Med. 1999, 5, 1365–1369. [Google Scholar] [CrossRef] [PubMed]
- Doroshow, D.B.; Bhalla, S.; Beasley, M.B.; Sholl, L.M.; Kerr, K.M.; Gnjatic, S.; Wistuba, I.I.; Rimm, D.L.; Tsao, M.S.; Hirsch, F.R. PD-L1 as a biomarker of response to immune-checkpoint inhibitors. Nat. Rev. Clin. Oncol. 2021, 18, 345–362. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Gilligan, B.M.; Yuan, J.; Li, T. Current status and perspectives in translational biomarker research for PD-1/PD-L1 immune checkpoint blockade therapy. J. Hematol. Oncol. 2016, 9, 1–21. [Google Scholar] [CrossRef]
- Zak, K.M.; Grudnik, P.; Magiera, K.; Dömling, A.; Dubin, G.; Holak, T.A. Structural biology of the immune checkpoint receptor PD-1 and its ligands PD-L1/PD-L2. Structure 2017, 25, 1163–1174. [Google Scholar] [CrossRef]
- Conroy, J.M.; Pabla, S.; Nesline, M.K.; Glenn, S.T.; Papanicolau-Sengos, A.; Burgher, B.; Andreas, J.; Giamo, V.; Wang, Y.; Lenzo, F.L. Next generation sequencing of PD-L1 for predicting response to immune checkpoint inhibitors. J. Immunother. Cancer 2019, 7, 18. [Google Scholar] [CrossRef] [PubMed]
- Akinleye, A.; Rasool, Z. Immune checkpoint inhibitors of PD-L1 as cancer therapeutics. J. Hematol. Oncol. 2019, 12, 92. [Google Scholar] [CrossRef]
- Miao, Y.R.; Thakkar, K.N.; Qian, J.; Kariolis, M.S.; Huang, W.; Nandagopal, S.; Yang, T.T.C.; Diep, A.N.; Cherf, G.M.; Xu, Y. Neutralization of PD-L2 is essential for overcoming immune checkpoint blockade resistance in ovarian cancer. Clin. Cancer Res. 2021, 27, 4435–4448. [Google Scholar] [CrossRef]
- Mattick, J.S.; Makunin, I.V. Non-coding RNA. Hum. Mol. Genet. 2006, 15, R17–R29. [Google Scholar] [CrossRef]
- Slack, F.J. Regulatory RNAs and the demise of ‘junk’ DNA. Genome Biol. 2006, 7, 328. [Google Scholar] [CrossRef]
- Adams, B.D.; Parsons, C.; Walker, L.; Zhang, W.C.; Slack, F.J. Targeting noncoding RNAs in disease. J. Clin. Investig. 2017, 127, 761–771. [Google Scholar] [CrossRef]
- Deveson, I.W.; Hardwick, S.A.; Mercer, T.R.; Mattick, J.S. The dimensions, dynamics, and relevance of the mammalian noncoding transcriptome. Trends Genet. 2017, 33, 464–478. [Google Scholar] [CrossRef] [PubMed]
- Slack, F.J.; Chinnaiyan, A.M. The role of non-coding RNAs in oncology. Cell 2019, 179, 1033–1055. [Google Scholar] [CrossRef] [PubMed]
- Lewis, B.P.; Burge, C.B.; Bartel, D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005, 120, 15–20. [Google Scholar] [CrossRef]
- Krek, A.; Grün, D.; Poy, M.N.; Wolf, R.; Rosenberg, L.; Epstein, E.J.; MacMenamin, P.; Da Piedade, I.; Gunsalus, K.C.; Stoffel, M. Combinatorial microRNA target predictions. Nat. Genet. 2005, 37, 495–500. [Google Scholar] [CrossRef] [PubMed]
- Diener, C.; Keller, A.; Meese, E. Emerging concepts of miRNA therapeutics: From cells to clinic. Trends Genet. 2022, 38, 613–626. [Google Scholar] [CrossRef]
- Subramanian, S.; Steer, C.J. MicroRNA regulation in health and disease. Genes 2019, 10, 457. [Google Scholar] [CrossRef]
- Huang, W. MicroRNAs: Biomarkers, diagnostics, and therapeutics. In Bioinformatics in MicroRNA Research; Springer: Berlin/Heidelberg, Germany, 2017; pp. 57–67. [Google Scholar]
- Sun, Q.; Hao, Q.; Prasanth, K.V. Nuclear long noncoding RNAs: Key regulators of gene expression. Trends Genet. 2018, 34, 142–157. [Google Scholar] [CrossRef] [PubMed]
- Kong, Y.; Lu, Z.; Liu, P.; Liu, Y.; Wang, F.; Liang, E.Y.; Hou, F.F.; Liang, M. Long noncoding RNA: Genomics and relevance to physiology. Compr. Physiol. 2011, 9, 933–946. [Google Scholar]
- Bhan, A.; Soleimani, M.; Mandal, S.S. Long noncoding RNA and cancer: A new paradigm. Cancer Res. 2017, 77, 3965–3981. [Google Scholar] [CrossRef] [PubMed]
- Safarzadeh, E.; Asadzadeh, Z.; Safaei, S.; Hatefi, A.; Derakhshani, A.; Giovannelli, F.; Brunetti, O.; Silvestris, N.; Baradaran, B. MicroRNAs and lncRNAs—A new layer of myeloid-derived suppressor cells regulation. Front. Immunol. 2020, 11, 572323. [Google Scholar] [CrossRef]
- Patop, I.L.; Kadener, S. circRNAs in Cancer. Curr. Opin. Genet. Dev. 2018, 48, 121–127. [Google Scholar] [CrossRef]
- Yu, T.; Wang, Y.; Fan, Y.; Fang, N.; Wang, T.; Xu, T.; Shu, Y. CircRNAs in cancer metabolism: A review. J. Hematol. Oncol. 2019, 12, 1–10. [Google Scholar] [CrossRef]
- Pathania, A.S.; Prathipati, P.; Olwenyi, O.A.; Chava, S.; Smith, O.V.; Gupta, S.C.; Chaturvedi, N.K.; Byrareddy, S.N.; Coulter, D.W.; Challagundla, K.B. miR-15a and miR-15b modulate natural killer and CD8+ T-cell activation and anti-tumor immune response by targeting PD-L1 in neuroblastoma. Mol. Ther.-Oncolytics 2022, 25, 308–329. [Google Scholar] [CrossRef]
- Gong, J.; Shen, Y.; Jiang, F.; Wang, Y.; Chu, L.; Sun, J.; Shen, P.; Chen, M. MicroRNA-20a promotes non-small cell lung cancer proliferation by upregulating PD-L1 by targeting PTEN. Oncol. Lett. 2022, 23, 148. [Google Scholar] [CrossRef]
- Shao, L.; He, Q.; Wang, J.; He, F.; Lin, S.; Wu, L.; Gao, Y.; Ma, W.; Dong, J.; Yang, X. MicroRNA-326 attenuates immune escape and prevents metastasis in lung adenocarcinoma by targeting PD-L1 and B7-H3. Cell Death Discov. 2021, 7, 145. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.-R.; Zhang, X.; Zhang, Y. MiR-214 prevents the progression of diffuse large B-cell lymphoma by targeting PD-L1. Cell. Mol. Biol. Lett. 2019, 24, 68. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Guo, Q.; Li, X.; Yang, X.; Ni, H.; Wang, T.; Zhao, Q.; Liu, H.; Xing, Y.; Xi, T. MiR-873/PD-L1 axis regulates the stemness of breast cancer cells. EBioMedicine 2019, 41, 395–407. [Google Scholar] [CrossRef]
- Yang, M.; Xiao, R.; Wang, X.; Xiong, Y.; Duan, Z.; Li, D.; Kan, Q. MiR-93-5p regulates tumorigenesis and tumor immunity by targeting PD-L1/CCND1 in breast cancer. Ann. Transl. Med. 2022, 10, 203. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, J.; Dong, K.; Lin, F.; Long, M.; Ouyang, Y.; Wei, J.; Chen, X.; Weng, Y.; He, T. Tumor suppressor miR-34a targets PD-L1 and functions as a potential immunotherapeutic target in acute myeloid leukemia. Cell. Signal. 2015, 27, 443–452. [Google Scholar] [CrossRef]
- Zhao, L.; Yu, H.; Yi, S.; Peng, X.; Su, P.; Xiao, Z.; Liu, R.; Tang, A.; Li, X.; Liu, F. The tumor suppressor miR-138-5p targets PD-L1 in colorectal cancer. Oncotarget 2016, 7, 45370. [Google Scholar] [CrossRef] [PubMed]
- Fujita, Y.; Yagishita, S.; Hagiwara, K.; Yoshioka, Y.; Kosaka, N.; Takeshita, F.; Fujiwara, T.; Tsuta, K.; Nokihara, H.; Tamura, T. The clinical relevance of the miR-197/CKS1B/STAT3-mediated PD-L1 network in chemoresistant non-small-cell lung cancer. Mol. Ther. 2015, 23, 717–727. [Google Scholar] [CrossRef] [PubMed]
- Xie, W.-B.; Liang, L.-H.; Wu, K.-G.; Wang, L.-X.; He, X.; Song, C.; Wang, Y.-Q.; Li, Y.-H. MiR-140 expression regulates cell proliferation and targets PD-L1 in NSCLC. Cell. Physiol. Biochem. 2018, 46, 654–663. [Google Scholar] [CrossRef]
- Qu, F.; Ye, J.; Pan, X.; Wang, J.; Gan, S.; Chu, C.; Chu, J.; Zhang, X.; Liu, M.; He, H. MicroRNA-497-5p down-regulation increases PD-L1 expression in clear cell renal cell carcinoma. J. Drug Target. 2019, 27, 67–74. [Google Scholar] [CrossRef]
- Roshani Asl, E.; Rasmi, Y.; Baradaran, B. MicroRNA-124-3p suppresses PD-L1 expression and inhibits tumorigenesis of colorectal cancer cells via modulating STAT3 signaling. J. Cell. Physiol. 2021, 236, 7071–7087. [Google Scholar] [CrossRef]
- Xu, S.; Tao, Z.; Hai, B.; Liang, H.; Shi, Y.; Wang, T.; Song, W.; Chen, Y.; OuYang, J.; Chen, J. miR-424 (322) reverses chemoresistance via T-cell immune response activation by blocking the PD-L1 immune checkpoint. Nat. Commun. 2016, 7, 11406. [Google Scholar] [CrossRef]
- Qin, Z.; Hu, H.; Sun, W.; Chen, L.; Jin, S.; Xu, Q.; Liu, Y.; Yu, L.; Zeng, S. miR-224-5p contained in urinary extracellular vesicles regulates PD-L1 expression by inhibiting cyclin D1 in renal cell carcinoma cells. Cancers 2021, 13, 618. [Google Scholar] [CrossRef]
- Li, Z.; Suo, B.; Long, G.; Gao, Y.; Song, J.; Zhang, M.; Feng, B.; Shang, C.; Wang, D. Exosomal miRNA-16-5p derived from M1 macrophages enhances T cell-dependent immune response by regulating PD-L1 in gastric cancer. Front. Cell Dev. Biol. 2020, 8, 572689. [Google Scholar] [CrossRef] [PubMed]
- Tang, D.; Zhao, D.; Wu, Y.; Yao, R.; Zhou, L.; Lu, L.; Gao, W.; Sun, Y. The miR-3127-5p/p-STAT 3 axis up-regulates PD-L1 inducing chemoresistance in non-small-cell lung cancer. J. Cell. Mol. Med. 2018, 22, 3847–3856. [Google Scholar] [CrossRef]
- Jia, L.; Xi, Q.; Wang, H.; Zhang, Z.; Liu, H.; Cheng, Y.; Guo, X.; Zhang, J.; Zhang, Q.; Zhang, L. miR-142-5p regulates tumor cell PD-L1 expression and enhances anti-tumor immunity. Biochem. Biophys. Res. Commun. 2017, 488, 425–431. [Google Scholar] [CrossRef]
- Yu, D.; Liu, X.; Han, G.; Liu, Y.; Zhao, X.; Wang, D.; Bian, X.; Gu, T.; Wen, L. The let-7 family of microRNAs suppresses immune evasion in head and neck squamous cell carcinoma by promoting PD-L1 degradation. Cell Commun. Signal. 2019, 17, 173. [Google Scholar] [CrossRef]
- Jiang, W.; Li, T.; Wang, J.; Jiao, R.; Shi, X.; Huang, X.; Ji, G. miR-140-3p suppresses cell growth and induces apoptosis in colorectal cancer by targeting PD-L1. OncoTargets Ther. 2019, 12, 10275. [Google Scholar] [CrossRef]
- Duan, M.; Fang, M.; Wang, C.; Wang, H.; Li, M. LncRNA EMX2OS induces proliferation, invasion and sphere formation of ovarian cancer cells via regulating the miR-654-3p/AKT3/PD-L1 axis. Cancer Manag. Res. 2020, 12, 2141. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.-H.; Li, B.; Liu, D.-G.; Zhang, B.; Yang, X.; Tu, Y.-L. LncRNA KCNQ1OT1 sponges miR-15a to promote immune evasion and malignant progression of prostate cancer via up-regulating PD-L1. Cancer Cell Int. 2020, 20, 394. [Google Scholar] [CrossRef]
- Dang, S.; Malik, A.; Chen, J.; Qu, J.; Yin, K.; Cui, L.; Gu, M. LncRNA SNHG15 contributes to immuno-escape of gastric cancer through targeting miR141/PD-L1. OncoTargets Ther. 2020, 13, 8547. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Dong, P.; Xiong, Y.; Chen, R.; Konno, Y.; Ihira, K.; Yue, J.; Watari, H. PD-L1 is a tumor suppressor in aggressive endometrial cancer cells and its expression is regulated by miR-216a and lncRNA MEG3. Front. Cell Dev. Biol. 2020, 8, 598205. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, Y.; Zheng, J.; Yao, C.; Lu, X. LncRNA UCA1 attenuated the killing effect of cytotoxic CD8+ T cells on anaplastic thyroid carcinoma via miR-148a/PD-L1 pathway. Cancer Immunol. Immunother. 2021, 70, 2235–2245. [Google Scholar] [CrossRef]
- Zhang, H.; Zhu, C.; He, Z.; Chen, S.; Li, L.; Sun, C. LncRNA PSMB8-AS1 contributes to pancreatic cancer progression via modulating miR-382-3p/STAT1/PD-L1 axis. J. Exp. Clin. Cancer Res. 2020, 39, 179. [Google Scholar] [CrossRef]
- Guo, T.; Wang, W.; Ji, Y.; Zhang, M.; Xu, G.; Lin, S. LncRNA PROX1-AS1 facilitates gastric cancer progression via miR-877-5p/PD-L1 Axis. Cancer Manag. Res. 2021, 13, 2669. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.-L.; Fan, L.; Huang, G.-R.; Sun, Z.-F. lncRNA EGFR-AS1 facilitates leiomyosarcoma progression and immune escape via the EGFR–MYC–PD-L1 axis. Int. Immunol. 2022, 34, 365–377. [Google Scholar] [CrossRef]
- Zhang, R.; Shang, L.; Nan, J.; Niu, K.; Dai, J.; Jin, X.; Zhang, X. Circ-METTL15 contributes to the proliferation, metastasis, immune escape and restrains apoptosis in lung cancer by regulating miR-1299/PDL1 axis. Autoimmunity 2022, 55, 8–20. [Google Scholar] [CrossRef]
- Zhang, X.; Pan, B.; Qiu, J.; Ke, X.; Shen, S.; Wang, X.; Tang, N. lncRNA MIAT targets miR-411-5p/STAT3/PD-L1 axis mediating hepatocellular carcinoma immune response. Int. J. Exp. Pathol. 2022, 103, 102–111. [Google Scholar] [CrossRef] [PubMed]
- Tian, Q.; Wu, T.; Zhang, X.; Xu, K.; Yin, X.; Wang, X.; Shi, S.; Wang, P.; Gao, L.; Xu, S. Immunomodulatory functions of the circ_001678/miRNA-326/ZEB1 axis in non-small cell lung cancer via the regulation of PD-1/PD-L1 pathway. Hum. Mol. Genet. 2022, 31, 4094–4106. [Google Scholar] [CrossRef] [PubMed]
- Mu, L.; Wang, Y.; Su, H.; Lin, Y.; Sui, W.; Yu, X.; Lv, Z. HIF1A-AS2 promotes the proliferation and metastasis of gastric cancer cells through miR-429/PD-L1 axis. Dig. Dis. Sci. 2021, 66, 4314–4325. [Google Scholar] [CrossRef]
- Zeng, C.; Ye, S.; Chen, Y.; Zhang, Q.; Luo, Y.; Gai, L.; Luo, B. HOXA-AS3 Promotes Proliferation and Migration of Hepatocellular Carcinoma Cells via the miR-455-5p/PD-L1 Axis. J. Immunol. Res. 2021, 2021, 9289719. [Google Scholar] [CrossRef]
- Sun, Z.; Xue, C.; Li, J.; Zhao, H.; Du, Y.; Du, N. LINC00244 suppresses cell growth and metastasis in hepatocellular carcinoma by downregulating programmed cell death ligand 1. Bioengineered 2022, 13, 7635–7647. [Google Scholar] [CrossRef]
- Fan, F.; Chen, K.; Lu, X.; Li, A.; Liu, C.; Wu, B. Dual targeting of PD-L1 and PD-L2 by PCED1B-AS1 via sponging hsa-miR-194-5p induces immunosuppression in hepatocellular carcinoma. Hepatol. Int. 2021, 15, 444–458. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Zhang, P.; Liang, H.; Xu, Y.; Shen, J.; Wang, W.; Li, M.; Huang, J.; Ni, C.; Zhang, X. Circular RNA hsa_circ_0003288 induces EMT and invasion by regulating hsa_circ_0003288/miR-145/PD-L1 axis in hepatocellular carcinoma. Cancer Cell Int. 2021, 21, 212. [Google Scholar] [CrossRef]
- Wu, F.; Sun, G.; Zheng, W.; Tang, W.; Cheng, Y.; Wu, L.; Li, X.; Tao, J.; Ma, S.; Cao, H. circCORO1C promotes the proliferation and metastasis of hepatocellular carcinoma by enhancing the expression of PD-L1 through NF-κB pathway. J. Clin. Lab. Anal. 2021, 35, e24003. [Google Scholar] [CrossRef]
- Li, L.; Zhang, Q.; Lian, K. Circular RNA circ_0000284 plays an oncogenic role in the progression of non-small cell lung cancer through the miR-377-3p-mediated PD-L1 promotion. Cancer Cell Int. 2020, 20, 247. [Google Scholar] [CrossRef]
- Xu, Y.-J.; Zhao, J.-M.; Gao, C.; Ni, X.-F.; Wang, W.; Hu, W.-W.; Wu, C.-P. Hsa_circ_0136666 activates Treg-mediated immune escape of colorectal cancer via miR-497/PD-L1 pathway. Cell. Signal. 2021, 86, 110095. [Google Scholar] [CrossRef]
- Zhang, D.J.; Fu, Z.M.; Guo, Y.Y.; Guo, F.; Wan, Y.N.; Guan, G.F. Circ_0000052/miR-382-3p axis induces PD-L1 expression and regulates cell proliferation and immune evasion in head and neck squamous cell carcinoma. J. Cell. Mol. Med. 2022, 27, 113–126. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Xu, J.; Wu, G.; Ren, Y.; Wang, X.; Zhang, Q. Circular RNA hsa_circ_0068252 functions in cisplatin resistance and immune response via miR-1304-5p/PD-L1 axis in non-small cell lung cancer. Chemotherapy 2022, 67, 223–233. [Google Scholar] [CrossRef]
- Zhao, J.; Yan, W.; Huang, W.; Li, Y. Circ_0010235 facilitates lung cancer development and immune escape by regulating miR-636/PDL1 axis. Thorac. Cancer 2022, 13, 965–976. [Google Scholar] [CrossRef] [PubMed]
- Hong, W.; Xue, M.; Jiang, J.; Zhang, Y.; Gao, X. Circular RNA circ-CPA4/let-7 miRNA/PD-L1 axis regulates cell growth, stemness, drug resistance and immune evasion in non-small cell lung cancer (NSCLC). J. Exp. Clin. Cancer Res. 2020, 39, 149. [Google Scholar] [CrossRef]
- Wang, J.; Zhao, X.; Wang, Y.; Ren, F.; Sun, D.; Yan, Y.; Kong, X.; Bu, J.; Liu, M.; Xu, S. circRNA-002178 act as a ceRNA to promote PDL1/PD1 expression in lung adenocarcinoma. Cell Death Dis. 2020, 11, 32. [Google Scholar] [CrossRef] [PubMed]
- Fehrenbacher, L.; Spira, A.; Ballinger, M.; Kowanetz, M.; Vansteenkiste, J.; Mazieres, J.; Park, K.; Smith, D.; Artal-Cortes, A.; Lewanski, C. Atezolizumab versus docetaxel for patients with previously treated non-small-cell lung cancer (POPLAR): A multicentre, open-label, phase 2 randomised controlled trial. Lancet 2016, 387, 1837–1846. [Google Scholar] [CrossRef]
- Rosenberg, J.E.; Hoffman-Censits, J.; Powles, T.; Van Der Heijden, M.S.; Balar, A.V.; Necchi, A.; Dawson, N.; O’Donnell, P.H.; Balmanoukian, A.; Loriot, Y. Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: A single-arm, multicentre, phase 2 trial. Lancet 2016, 387, 1909–1920. [Google Scholar] [CrossRef] [PubMed]
- Balar, A.V.; Castellano, D.; O’Donnell, P.H.; Grivas, P.; Vuky, J.; Powles, T.; Plimack, E.R.; Hahn, N.M.; de Wit, R.; Pang, L. First-line pembrolizumab in cisplatin-ineligible patients with locally advanced and unresectable or metastatic urothelial cancer (KEYNOTE-052): A multicentre, single-arm, phase 2 study. Lancet Oncol. 2017, 18, 1483–1492. [Google Scholar] [CrossRef]
- Philips, G.K.; Atkins, M. Therapeutic uses of anti-PD-1 and anti-PD-L1 antibodies. Int. Immunol. 2015, 27, 39–46. [Google Scholar] [CrossRef]
- Balar, A.V.; Galsky, M.D.; Rosenberg, J.E.; Powles, T.; Petrylak, D.P.; Bellmunt, J.; Loriot, Y.; Necchi, A.; Hoffman-Censits, J.; Perez-Gracia, J.L. Atezolizumab as first-line treatment in cisplatin-ineligible patients with locally advanced and metastatic urothelial carcinoma: A single-arm, multicentre, phase 2 trial. Lancet 2017, 389, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Socinski, M.A.; Jotte, R.M.; Cappuzzo, F.; Orlandi, F.; Stroyakovskiy, D.; Nogami, N.; Rodríguez-Abreu, D.; Moro-Sibilot, D.; Thomas, C.A.; Barlesi, F. Atezolizumab for first-line treatment of metastatic nonsquamous NSCLC. N. Engl. J. Med. 2018, 378, 2288–2301. [Google Scholar] [CrossRef] [PubMed]
- Casak, S.J.; Donoghue, M.; Fashoyin-Aje, L.; Jiang, X.; Rodriguez, L.; Shen, Y.-L.; Xu, Y.; Jiang, X.; Liu, J.; Zhao, H. FDA Approval Summary: Atezolizumab Plus Bevacizumab for the Treatment of Patients with Advanced Unresectable or Metastatic Hepatocellular CarcinomaFDA Approval: Atezolizumab plus Bevacizumab in HCC. Clin. Cancer Res. 2021, 27, 1836–1841. [Google Scholar] [CrossRef]
- Gutzmer, R.; Stroyakovskiy, D.; Gogas, H.; Robert, C.; Lewis, K.; Protsenko, S.; Pereira, R.P.; Eigentler, T.; Rutkowski, P.; Demidov, L. Atezolizumab, vemurafenib, and cobimetinib as first-line treatment for unresectable advanced BRAFV600 mutation-positive melanoma (IMspire150): Primary analysis of the randomised, double-blind, placebo-controlled, phase 3 trial. Lancet 2020, 395, 1835–1844. [Google Scholar] [CrossRef]
- Syed, Y.Y. Durvalumab: First global approval. Drugs 2017, 77, 1369–1376. [Google Scholar] [CrossRef]
- Baker, M.; Cordes, L.; Brownell, I. Avelumab: A new standard for treating metastatic Merkel cell carcinoma. Expert Rev. Anticancer Ther. 2018, 18, 319–326. [Google Scholar] [CrossRef]
- Paz-Ares, L.; Dvorkin, M.; Chen, Y.; Reinmuth, N.; Hotta, K.; Trukhin, D.; Statsenko, G.; Hochmair, M.J.; Özgüroğlu, M.; Ji, J.H. Durvalumab plus platinum–etoposide versus platinum–etoposide in first-line treatment of extensive-stage small-cell lung cancer (CASPIAN): A randomised, controlled, open-label, phase 3 trial. Lancet 2019, 394, 1929–1939. [Google Scholar] [CrossRef]
- Powles, T.; Park, S.H.; Voog, E.; Caserta, C.; Valderrama, B.P.; Gurney, H.; Kalofonos, H.; Radulović, S.; Demey, W.; Ullén, A. Avelumab maintenance therapy for advanced or metastatic urothelial carcinoma. N. Engl. J. Med. 2020, 383, 1218–1230. [Google Scholar] [CrossRef] [PubMed]
- Choueiri, T.K.; Larkin, J.; Oya, M.; Thistlethwaite, F.; Martignoni, M.; Nathan, P.; Powles, T.; McDermott, D.; Robbins, P.B.; Chism, D.D. Preliminary results for avelumab plus axitinib as first-line therapy in patients with advanced clear-cell renal-cell carcinoma (JAVELIN Renal 100): An open-label, dose-finding and dose-expansion, phase 1b trial. Lancet Oncol. 2018, 19, 451–460. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).