Aspergillus fumigatus Lytic Polysaccharide Monooxygenase AfLPMO9D: Biochemical Properties and Photoactivation of a Multi-Domain AA9 Enzyme
Abstract
:1. Introduction
2. Material and Methods
2.1. Bioinformatic Analysis
2.2. AfLPMO9D Cloning and Expression in A. nidulans
2.3. AfLPMO9D Purification
2.4. Soluble Products Detection by HPAEC
2.5. Specificity to Different Substrates
2.6. Biochemical Characterization
2.7. Light-Driven Activation
2.8. Cellulose Hydrolysis
3. Results and Discussion
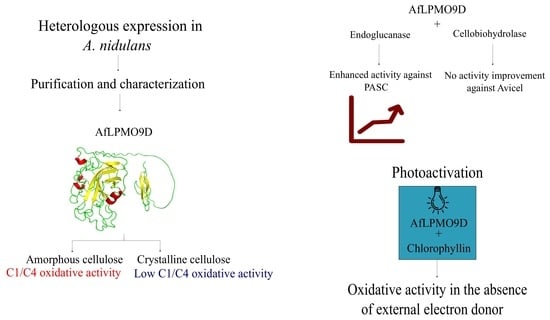
3.1. Protein Sequence Alignment and Heterologous Expression
3.2. AfLPMO9D Specificity for Different Cellulosic Substrates
3.3. AfLPMO9D Biochemical Properties
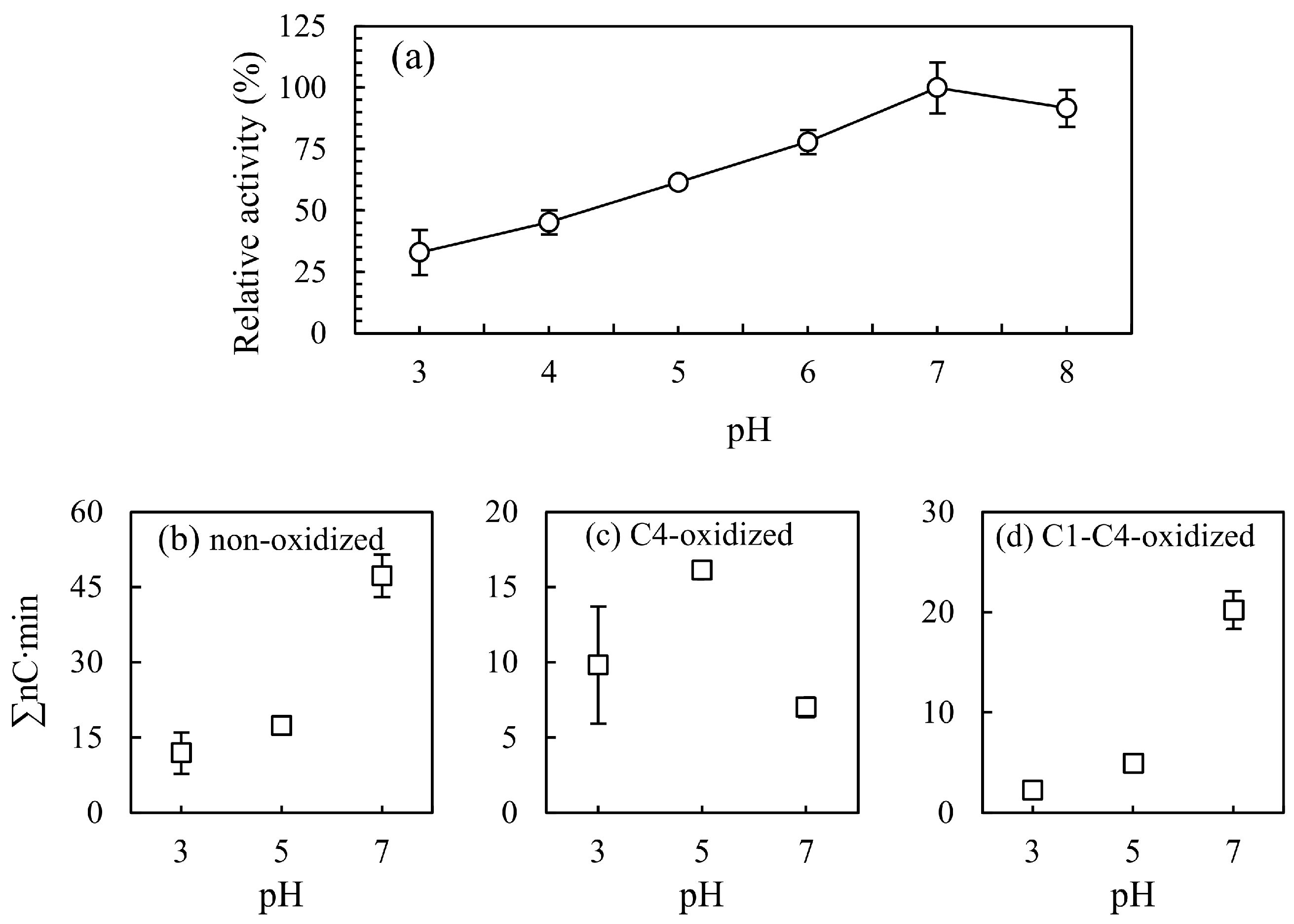
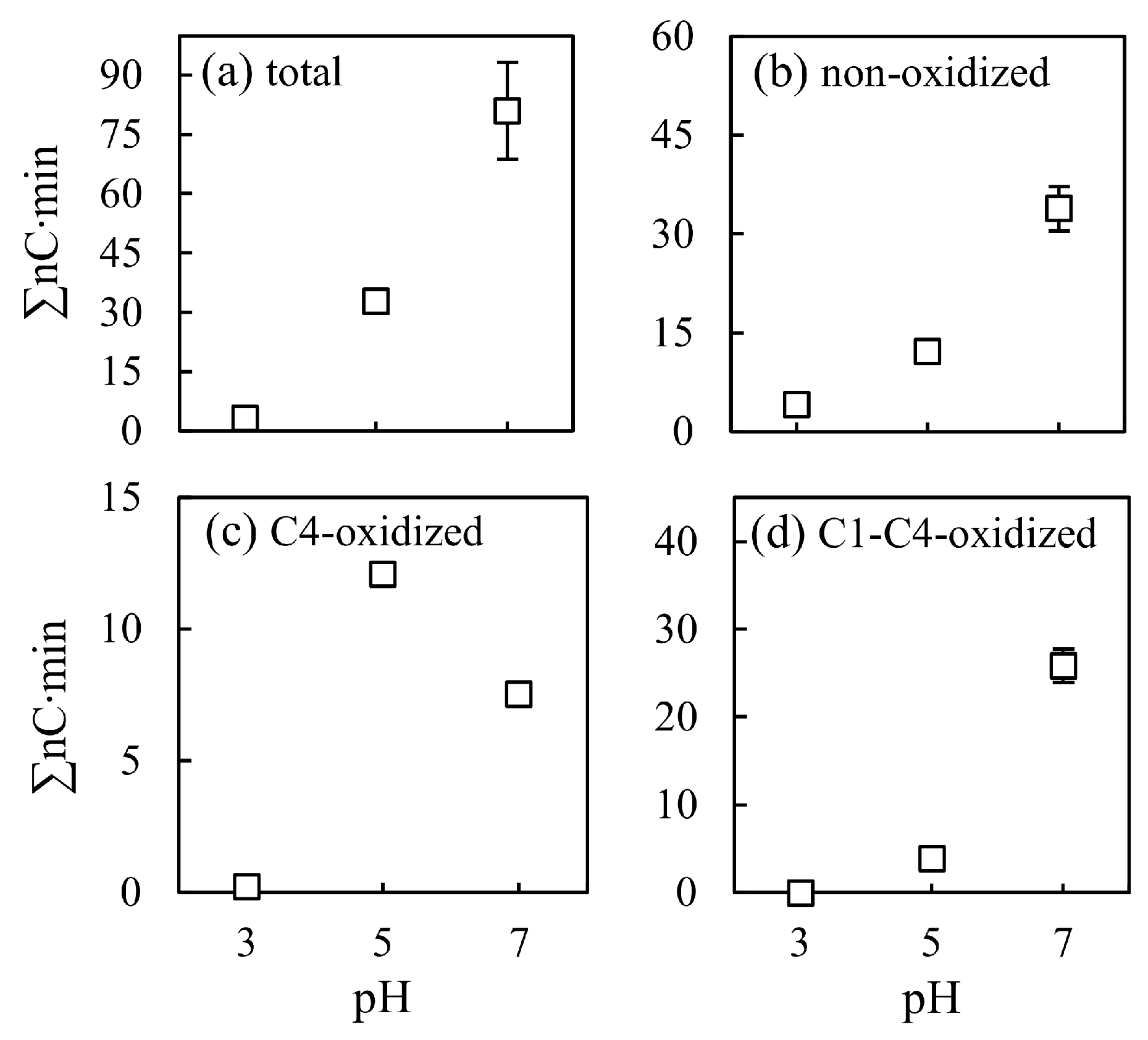
3.3.1. pH Effect
3.3.2. Effect of Temperature
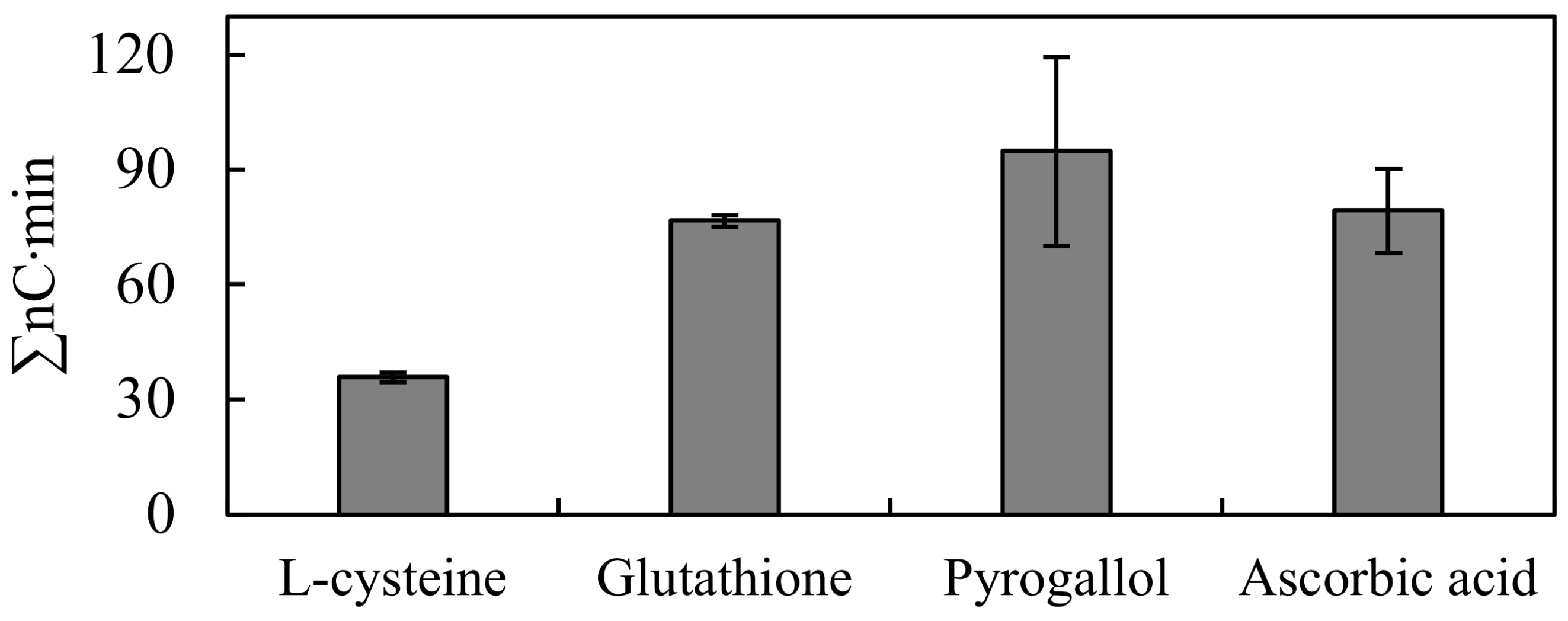
3.4. Effect of Different Reducing Agents
3.5. Light-Driven Activation of AfLPMO9D
3.6. Supplementation of Glycoside Hydrolases with AfLPMO9D
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Cherubini, F. The biorefinery concept: Using biomass instead of oil for producing energy and chemicals. Energy Convers. Manag. 2010, 51, 1412–1421. [Google Scholar] [CrossRef]
- Duarte, G.C.; Moreira, L.R.S.; Jaramillo, P.M.D.; Filho, E.X.F. Biomass-Derived Inhibitors of Holocellulases. BioEnergy Res. 2012, 5, 768–777. [Google Scholar] [CrossRef]
- Brodeur, G.; Yau, E.; Badal, K.; Collier, J.; Ramachandran, K.B.; Ramakrishnan, S. Chemical and Physicochemical Pretreatment of Lignocellulosic Biomass: A Review. Enzym. Res. 2011, 2011, e787532. [Google Scholar] [CrossRef]
- Kim, S.; Baek, S.-H.; Lee, K.; Hahn, J.-S. Cellulosic ethanol production using a yeast consortium displaying a minicellulosome and β-glucosidase. Microb. Cell Fact. 2013, 12, 14. [Google Scholar] [CrossRef] [PubMed]
- Bastawde, K.B. Xylan structure, microbial xylanases, and their mode of action. World J. Microbiol. Biotechnol. 1992, 8, 353–368. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.-H.P.; Lynd, L.R. Determination of the Number-Average Degree of Polymerization of Cellodextrins and Cellulose with Application to Enzymatic Hydrolysis. Biomacromolecules 2005, 6, 1510–1515. [Google Scholar] [CrossRef] [PubMed]
- Mansfield, S.D.; Meder, R. Cellulose hydrolysis—The role of monocomponent cellulases in crystalline cellulose degradation. Cellulose 2003, 10, 159–169. [Google Scholar] [CrossRef]
- Bischof, R.H.; Ramoni, J.; Seiboth, B. Cellulases and beyond: The first 70 years of the enzyme producer Trichoderma reesei. Microb. Cell Factories 2016, 15, 106. [Google Scholar] [CrossRef]
- Lopes, A.M.; Ferreira Filho, E.X.; Moreira, L.R.S. An update on enzymatic cocktails for lignocellulose breakdown. J. Appl. Microbiol. 2018, 125, 632–645. [Google Scholar] [CrossRef]
- Sharma, A.; Tewari, R.; Rana, S.S.; Soni, R.; Soni, S.K. Cellulases: Classification, Methods of Determination and Industrial Applications. Appl. Biochem. Biotechnol. 2016, 179, 1346–1380. [Google Scholar] [CrossRef]
- Morgenstern, I.; Powlowski, J.; Tsang, A. Fungal cellulose degradation by oxidative enzymes: From dysfunctional GH61 family to powerful lytic polysaccharide monooxygenase family. Briefings Funct. Genom. 2014, 13, 471–481. [Google Scholar] [CrossRef] [PubMed]
- Terrapon, N.; Lombard, V.; Drula, E.; Coutinho, P.M.; Henrissat, B. The CAZy Database/the Carbohydrate-Active Enzyme (CAZy) Database: Principles and Usage Guidelines. In A Practical Guide to Using Glycomics Databases; Springer: Tokyo, Japan, 2017; pp. 117–131. [Google Scholar] [CrossRef]
- Westereng, B.; Arntzen, M.; Aachmann, F.L.; Várnai, A.; Eijsink, V.G.; Agger, J.W. Simultaneous analysis of C1 and C4 oxidized oligosaccharides, the products of lytic polysaccharide monooxygenases acting on cellulose. J. Chromatogr. A 2016, 1445, 46–54. [Google Scholar] [CrossRef] [PubMed]
- Hamre, A.G.; Eide, K.B.; Wold, H.H.; Sørlie, M. Activation of enzymatic chitin degradation by a lytic polysaccharide monooxygenase. Carbohydr. Res. 2015, 407, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Frommhagen, M.; Sforza, S.; Westphal, A.H.; Visser, J.; A Hinz, S.W.; Koetsier, M.J.; van Berkel, W.J.H.; Gruppen, H.; A Kabel, M. Discovery of the combined oxidative cleavage of plant xylan and cellulose by a new fungal polysaccharide monooxygenase. Biotechnol. Biofuels 2015, 8, 101. [Google Scholar] [CrossRef]
- Agger, J.W.; Isaksen, T.; Várnai, A.; Vidal-Melgosa, S.; Willats, W.G.T.; Ludwig, R.; Horn, S.J.; Eijsink, V.G.H.; Westereng, B. Discovery of LPMO activity on hemicelluloses shows the importance of oxidative processes in plant cell wall degradation. Proc. Natl. Acad. Sci. USA 2014, 111, 6287–6292. [Google Scholar] [CrossRef] [PubMed]
- Latgé, J.-P. The pathobiology of Aspergillus fumigatus. Trends Microbiol. 2001, 9, 382–389. [Google Scholar] [CrossRef]
- Saroj, P.; Manasa, P.; Narasimhulu, K. Biochemical Characterization of Thermostable Carboxymethyl Cellulase and β-Glucosidase from Aspergillus fumigatus JCM 10253. Appl. Biochem. Biotechnol. 2022, 194, 2503–2527. [Google Scholar] [CrossRef]
- Velasco, J.; Pellegrini, V.d.O.A.; Sepulchro, A.G.V.; Kadowaki, M.A.S.; Santo, M.C.E.; Polikarpov, I.; Segato, F. Comparative analysis of two recombinant LPMOs from Aspergillus fumigatus and their effects on sugarcane bagasse saccharification. Enzym. Microb. Technol. 2021, 144, 109746. [Google Scholar] [CrossRef]
- Grigoriev, I.V.; Nikitin, R.; Haridas, S.; Kuo, A.; Ohm, R.; Otillar, R.; Riley, R.; Salamov, A.; Zhao, X.; Korzeniewski, F.; et al. MycoCosm portal: Gearing up for 1000 fungal genomes. Nucleic Acids Res. 2014, 42, D699–D704. [Google Scholar] [CrossRef]
- Nielsen, H. Predicting secretory proteins with SignalP. In Protein Function Prediction; Kihara, D., Ed.; Humana Press: New York, NY, USA, 2017; pp. 59–73. [Google Scholar] [CrossRef]
- E Hansen, J.; Lund, O.; Tolstrup, N.; A Gooley, A.; Williams, K.L.; Brunak, S. NetOglyc: Prediction of mucin type O-glycosylation sites based on sequence context and surface accessibility. Glycoconj. J. 1998, 15, 115–130. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Okonechnikov, K.; Golosova, O.; Fursov, M.; The UGENE Team. Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics 2012, 28, 1166–1167. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Schrödinger, LLC. The {PyMOL} Molecular Graphics System, Version~1.8; Schrödinger, LLC.: New York, NY, USA, 2015. [Google Scholar]
- Segato, F.; Damásio, A.R.; Gonçalves, T.A.; de Lucas, R.C.; Squina, F.M.; Decker, S.R.; Prade, R.A. High-yield secretion of multiple client proteins in Aspergillus. Enzym. Microb. Technol. 2012, 51, 100–106. [Google Scholar] [CrossRef] [PubMed]
- Higasi, P.M.; Velasco, J.A.; Pellegrini, V.O.; de Araújo, E.A.; França, B.A.; Keller, M.B.; Labate, C.A.; Blossom, B.M.; Segato, F.; Polikarpov, I. Light-stimulated T. thermophilus two-domain LPMO9H: Low-resolution SAXS model and synergy with cellulases. Carbohydr. Polym. 2021, 260, 117814. [Google Scholar] [CrossRef]
- Sepulchro, A.G.V.; Pellegrini, V.O.; Dias, L.D.; Kadowaki, M.A.; Cannella, D.; Polikarpov, I. Combining pieces: A thorough analysis of light activation boosting power and co-substrate preferences for the catalytic efficiency of lytic polysaccharide monooxygenase MtLPMO9A. Biofuel Res. J. 2021, 8, 1454–1464. [Google Scholar] [CrossRef]
- Keller, M.B.; Badino, S.F.; Blossom, B.M.; McBrayer, B.; Borch, K.; Westh, P. Promoting and Impeding Effects of Lytic Polysaccharide Monooxygenases on Glycoside Hydrolase Activity. ACS Sustain. Chem. Eng. 2020, 8, 14117–14126. [Google Scholar] [CrossRef]
- Schülein, M. Enzymatic properties of cellulases from Humicola insolens. J. Biotechnol. 1997, 57, 71–81. [Google Scholar] [CrossRef]
- Feng, X.; Ullah, N.; Wang, X.; Sun, X.; Li, C.; Bai, Y.; Chen, L.; Li, Z. Characterization of Bacterial Cellulose by Gluconacetobacter hansenii CGMCC 3917. J. Food Sci. 2015, 80, E2217–E2227. [Google Scholar] [CrossRef]
- Frommhagen, M.; Koetsier, M.J.; Westphal, A.H.; Visser, J.; Hinz, S.W.A.; Vincken, J.-P.; van Berkel, W.J.H.; Kabel, M.A.; Gruppen, H. Lytic polysaccharide monooxygenases from Myceliophthora thermophila C1 differ in substrate preference and reducing agent specificity. Biotechnol. Biofuels 2016, 9, 186. [Google Scholar] [CrossRef]
- Miller, G.L. Use of Dinitrosalicylic Acid Reagent for Determination of Reducing Sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Walton, P.H.; Davies, G.J. On the catalytic mechanisms of lytic polysaccharide monooxygenases. Curr. Opin. Chem. Biol. 2016, 31, 195–207. [Google Scholar] [CrossRef] [PubMed]
- Monclaro, A.V.; Filho, E.X.F. Fungal lytic polysaccharide monooxygenases from family AA9: Recent developments and application in lignocelullose breakdown. Int. J. Biol. Macromol. 2017, 102, 771–778. [Google Scholar] [CrossRef] [PubMed]
- Balajee, S.A.; Gribskov, J.L.; Hanley, E.; Nickle, D.; Marr, K.A. Aspergillus lentulus sp. nov., a New Sibling Species of A. fumigatus. Eukaryot. Cell 2005, 4, 625–632. [Google Scholar] [CrossRef] [PubMed]
- Rokas, A.; Mead, M.E.; Steenwyk, J.L.; Oberlies, N.H.; Goldman, G.H. Evolving moldy murderers: Aspergillus section Fumigati as a model for studying the repeated evolution of fungal pathogenicity. PLoS Pathog. 2020, 16, e1008315. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, K.B.; Macêdo, J.K.; Teixeira, T.; Barros, J.S.; Araújo, A.C.; Santos, F.P.; Quirino, B.F.; Brasil, B.S.; Salum, T.F.; Abdelnur, P.V.; et al. Recombinant expression of Thermobifida fusca E7 LPMO in Pichia pastoris and Escherichia coli and their functional characterization. Carbohydr. Res. 2017, 448, 175–181. [Google Scholar] [CrossRef]
- Petrović, D.M.; Bissaro, B.; Chylenski, P.; Skaugen, M.; Sørlie, M.; Jensen, M.S.; Aachmann, F.L.; Courtade, G.; Várnai, A.; Eijsink, V.G. Methylation of the N-terminal histidine protects a lytic polysaccharide monooxygenase from auto-oxidative inactivation. Protein Sci. 2018, 27, 1636–1650. [Google Scholar] [CrossRef]
- Uchiyama, T.; Uchihashi, T.; Ishida, T.; Nakamura, A.; Vermaas, J.V.; Crowley, M.F.; Samejima, M.; Beckham, G.T.; Igarashi, K. Lytic polysaccharide monooxygenase increases cellobiohydrolases activity by promoting decrystallization of cellulose surface. Sci. Adv. 2022, 8, eade5155. [Google Scholar] [CrossRef]
- Stepnov, A.A.; Christensen, I.A.; Forsberg, Z.; Aachmann, F.L.; Courtade, G.; Eijsink, V.G.H. The impact of reductants on the catalytic efficiency of a lytic polysaccharide monooxygenase and the special role of dehydroascorbic acid. FEBS Lett. 2021, 596, 53–70. [Google Scholar] [CrossRef]
- Tõlgo, M.; Hegnar, O.A.; Østby, H.; Várnai, A.; Vilaplana, F.; Eijsink, V.G.H.; Olsson, L. Comparison of Six Lytic Polysaccharide Monooxygenases from Thermothielavioides terrestris Shows That Functional Variation Underlies the Multiplicity of LPMO Genes in Filamentous Fungi. Appl. Environ. Microbiol. 2022, 88, e0009622. [Google Scholar] [CrossRef]
- Golten, O.; Ayuso-Fernández, I.; Hall, K.R.; Stepnov, A.A.; Sørlie, M.; Røhr, Å.K.; Eijsink, V.G.H. Reductants fuel lytic polysaccharide monooxygenase activity in a pH-dependent manner. FEBS Lett. 2023, 597, 1363–1374. [Google Scholar] [CrossRef]
- Shen, J.; Griffiths, P.T.; Campbell, S.J.; Utinger, B.; Kalberer, M.; Paulson, S.E. Ascorbate oxidation by iron, copper and reactive oxygen species: Review, model development, and derivation of key rate constants. Sci. Rep. 2021, 11, 7417. [Google Scholar] [CrossRef] [PubMed]
- Siegel, S.M.; Siegel, B.Z. Autoxidation of Pyrogallol: General Characteristics and Inhibition by Catalase. Nature 1958, 181, 1153–1154. [Google Scholar] [CrossRef] [PubMed]
- Akagawa, M.; Shigemitsu, T.; Suyama, K. Production of Hydrogen Peroxide by Polyphenols and Polyphenol-rich Beverages under Quasi-physiological Conditions. Biosci. Biotechnol. Biochem. 2003, 67, 2632–2640. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Beeson, W.T.; Phillips, C.M.; Marletta, M.A.; Cate, J.H. Structural basis for substrate targeting and catalysis by fungal polysaccharide monooxygenases. Structure 2012, 20, 1051–1061. [Google Scholar] [CrossRef]
- Liu, B.; Kognole, A.A.; Wu, M.; Westereng, B.; Crowley, M.F.; Kim, S.; Dimarogona, M.; Payne, C.M.; Sandgren, M. Structural and molecular dynamics studies of a C1-oxidizing lytic polysaccharide monooxygenase from Heterobasidion irregulare reveal amino acids important for substrate recognition. FEBS J. 2018, 285, 2225–2242. [Google Scholar] [CrossRef]
- Linder, M.; Nevanen, T.; Teeri, T.T. Design of a pH-dependent cellulose-binding domain. FEBS Lett. 1999, 447, 13–16. [Google Scholar] [CrossRef]
- Calderaro, F.; Keser, M.; Akeroyd, M.; Bevers, L.E.; Eijsink, V.G.H.; Várnai, A.; Berg, M.A.v.D. Characterization of an AA9 LPMO from Thielavia australiensis, TausLPMO9B, under industrially relevant lignocellulose saccharification conditions. Biotechnol. Biofuels 2020, 13, 195. [Google Scholar] [CrossRef]
- Alfani, F.; Gallifuoco, A.; Saporosi, A.; Spera, A.; Cantarella, M. Comparison of SHF and SSF processes for the bioconversion of steam-exploded wheat straw. J. Ind. Microbiol. Biotechnol. 2000, 25, 184–192. [Google Scholar] [CrossRef]
- Kerksick, C.; Willoughby, D. The Antioxidant Role of Glutathione and N-Acetyl-Cysteine Supplements and Exercise-Induced Oxidative Stress. J. Int. Soc. Sports Nutr. 2005, 2, 38–44. [Google Scholar] [CrossRef]
- Kommedal, E.G.; Angeltveit, C.F.; Klau, L.J.; Ayuso-Fernández, I.; Arstad, B.; Antonsen, S.G.; Stenstrøm, Y.; Ekeberg, D.; Gírio, F.; Carvalheiro, F.; et al. Visible light-exposed lignin facilitates cellulose solubilization by lytic polysaccharide monooxygenases. Nat. Commun. 2023, 14, 1063. [Google Scholar] [CrossRef] [PubMed]
- Müller, G.; Chylenski, P.; Bissaro, B.; Eijsink, V.G.H.; Horn, S.J. The impact of hydrogen peroxide supply on LPMO activity and overall saccharification efficiency of a commercial cellulase cocktail. Biotechnol. Biofuels 2018, 11, 209. [Google Scholar] [CrossRef] [PubMed]
- Bissaro, B.; Røhr, Å.K.; Müller, G.; Chylenski, P.; Skaugen, M.; Forsberg, Z.; Horn, S.J.; Vaaje-Kolstad, G.; Eijsink, V.G.H. Oxidative cleavage of polysaccharides by monocopper enzymes depends on H2O2. Nat. Chem. Biol. 2017, 13, 1123–1128. [Google Scholar] [CrossRef] [PubMed]
- Möllers, K.; Mikkelsen, H.; Simonsen, T.; Cannella, D.; Johansen, K.; Bjerrum, M.; Felby, C. On the formation and role of reactive oxygen species in light-driven LPMO oxidation of phosphoric acid swollen cellulose. Carbohydr. Res. 2017, 448, 182–186. [Google Scholar] [CrossRef] [PubMed]
- Blossom, B.M.; Russo, D.A.; Singh, R.K.; van Oort, B.; Keller, M.B.; Simonsen, T.I.; Perzon, A.; Gamon, L.F.; Davies, M.J.; Cannella, D.; et al. Photobiocatalysis by a Lytic Polysaccharide Monooxygenase Using Intermittent Illumination. ACS Sustain. Chem. Eng. 2020, 8, 9301–9310. [Google Scholar] [CrossRef]
- Bissaro, B.; Kommedal, E.; Røhr, Å.K.; Eijsink, V.G.H. Controlled depolymerization of cellulose by light-driven lytic polysaccharide oxygenases. Nat. Commun. 2020, 11, 890. [Google Scholar] [CrossRef]
- Velasco, J.; Sepulchro, A.G.V.; Higasi, P.M.R.; Pellegrini, V.O.A.; Cannella, D.; de Oliveira, L.C.; Polikarpov, I.; Segato, F. Light Boosts the Activity of Novel LPMO from Aspergillus fumigatus Leading to Oxidative Cleavage of Cellulose and Hemicellulose. ACS Sustain. Chem. Eng. 2022, 10, 16969–16984. [Google Scholar] [CrossRef]
- Kommedal, E.G.; Sæther, F.; Hahn, T.; Eijsink, V.G.H. Natural photoredox catalysts promote light-driven lytic polysaccharide monooxygenase reactions and enzymatic turnover of biomass. Proc. Natl. Acad. Sci. USA 2022, 119, e2204510119. [Google Scholar] [CrossRef]
- Tokin, R.; Ipsen, J.; Westh, P.; Johansen, K.S. The synergy between LPMOs and cellulases in enzymatic saccharification of cellulose is both enzyme- and substrate-dependent. Biotechnol. Lett. 2020, 42, 1975–1984. [Google Scholar] [CrossRef]
- Sagarika, M.S.; Parameswaran, C.; Senapati, A.; Barala, J.; Mitra, D.; Prabhukarthikeyan, S.; Kumar, A.; Nayak, A.K.; Panneerselvam, P. Lytic polysaccharide monooxygenases (LPMOs) producing microbes: A novel approach for rapid recycling of agricultural wastes. Sci. Total Environ. 2022, 806, 150451. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hamann, P.R.V.; Vacilotto, M.M.; Segato, F.; Polikarpov, I. Aspergillus fumigatus Lytic Polysaccharide Monooxygenase AfLPMO9D: Biochemical Properties and Photoactivation of a Multi-Domain AA9 Enzyme. Processes 2023, 11, 3230. https://doi.org/10.3390/pr11113230
Hamann PRV, Vacilotto MM, Segato F, Polikarpov I. Aspergillus fumigatus Lytic Polysaccharide Monooxygenase AfLPMO9D: Biochemical Properties and Photoactivation of a Multi-Domain AA9 Enzyme. Processes. 2023; 11(11):3230. https://doi.org/10.3390/pr11113230
Chicago/Turabian StyleHamann, Pedro Ricardo Vieira, Milena Moreira Vacilotto, Fernando Segato, and Igor Polikarpov. 2023. "Aspergillus fumigatus Lytic Polysaccharide Monooxygenase AfLPMO9D: Biochemical Properties and Photoactivation of a Multi-Domain AA9 Enzyme" Processes 11, no. 11: 3230. https://doi.org/10.3390/pr11113230
APA StyleHamann, P. R. V., Vacilotto, M. M., Segato, F., & Polikarpov, I. (2023). Aspergillus fumigatus Lytic Polysaccharide Monooxygenase AfLPMO9D: Biochemical Properties and Photoactivation of a Multi-Domain AA9 Enzyme. Processes, 11(11), 3230. https://doi.org/10.3390/pr11113230