Genetic Association of Diagnostic Traits of Metabolic Syndrome with Lysosomal Pathways: Insights from Target Gene Enrichment Analysis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Colocalization Analysis
2.2. Enrichment Analysis
3. Results
3.1. Colocalization Analysis
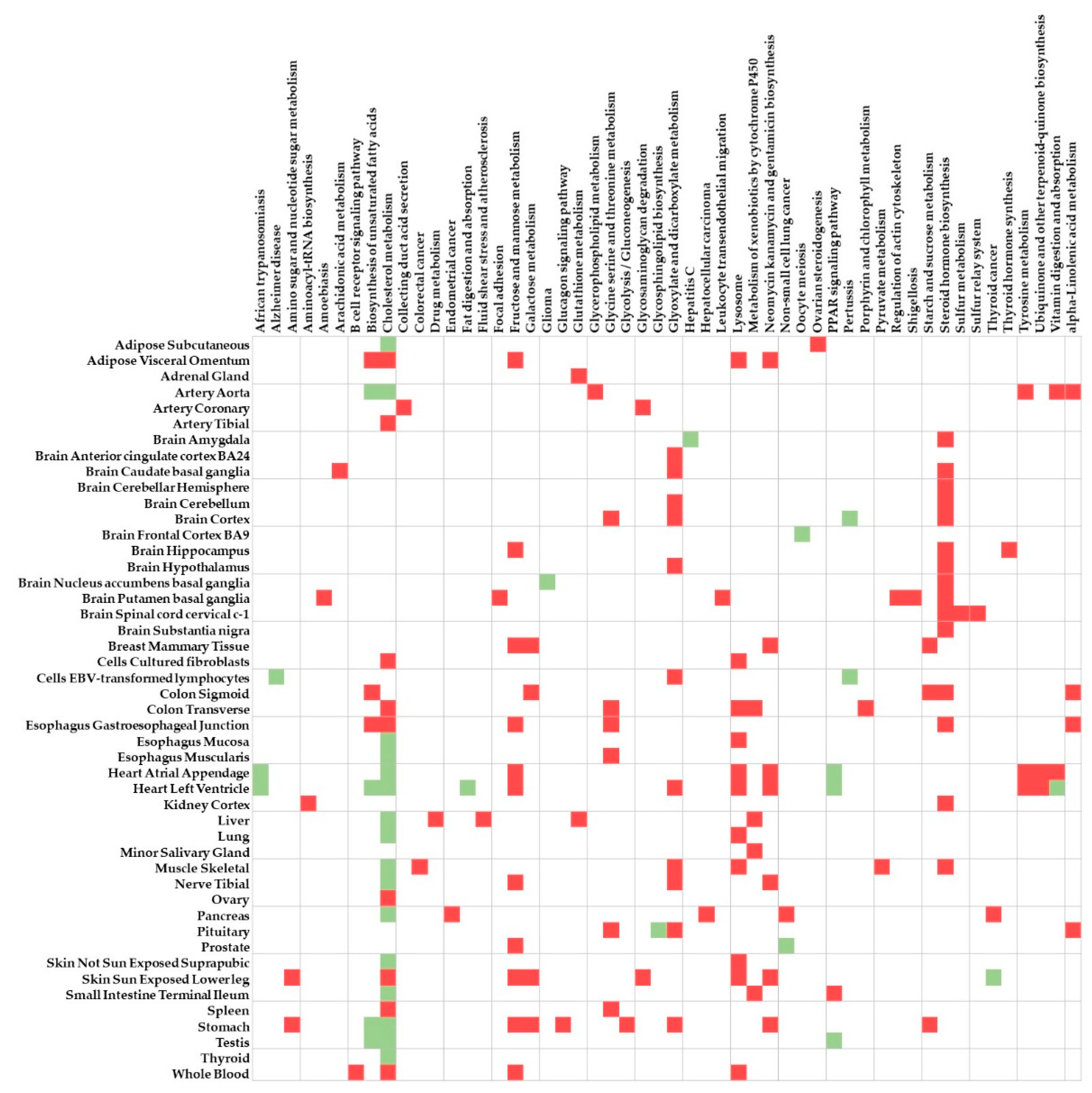
3.2. Enrichment Analysis
4. Discussion
4.1. Enrichment Analysis and Its Implications
4.2. Lysosomal Pathway and Diseases
4.3. Future Directions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dominguez, L.J.; Barbagallo, M. The biology of the metabolic syndrome and aging. Curr. Opin. Clin. Nutr. Metab. Care 2016, 19, 5–11. [Google Scholar] [CrossRef]
- Harrison, S.; Couture, P.; Lamarche, B. Diet quality, saturated fat and metabolic syndrome. Nutrients 2020, 12, 3232. [Google Scholar] [CrossRef]
- Borel, A.L. Sleep apnea and sleep habits: Relationships with metabolic syndrome. Nutrients 2019, 11, 2628. [Google Scholar] [CrossRef]
- Saladini, F.; Palatini, P. Arterial distensibility, physical activity, and the metabolic syndrome. Curr. Hypertens. Rep. 2018, 20, 39. [Google Scholar] [CrossRef]
- Wu, J.; Zhang, H.; Yang, L.; Shao, J.; Chen, D.; Cui, N.; Tang, L.; Fu, Y.; Xue, E.; Lai, C.; et al. Sedentary time and the risk of metabolic syndrome: A systematic review and dose–response meta-analysis. Obes. Rev. 2022, 23, e13510. [Google Scholar] [CrossRef] [PubMed]
- Åberg, F.; Byrne, C.D.; Pirola, C.J.; Männistö, V.; Sookoian, S. Alcohol consumption and metabolic syndrome: Clinical and epidemiological impact on liver disease. J. Hepatol. 2023, 78, 191–206. [Google Scholar] [CrossRef]
- Mazereel, V.; Detraux, J.; Vancampfort, D.; van Winkel, R.; De Hert, M. Impact of psychotropic medication effects on obesity and the metabolic syndrome in people with serious mental illness. Front. Endocrinol. 2020, 11, 573479. [Google Scholar] [CrossRef] [PubMed]
- Bellia, A.; Giardina, E.; Lauro, D.; Tesauro, M.; Di Fede, G.; Cusumano, G.; Federici, M.; Rini, G.B.; Novelli, G.; Lauro, R.; et al. ‘The Linosa Study’: Epidemiological and heritability data of the metabolic syndrome in a Caucasian genetic isolate. Nutr. Metab. Cardiovasc. Dis. 2009, 19, 455–461. [Google Scholar] [CrossRef]
- Zhang, S.; Liu, X.; Yu, Y.; Hong, X.; Christoffel, K.K.; Wang, B.; Tsai, H.-J.; Li, Z.; Liu, X.; Tang, G.; et al. Genetic and environmental contributions to phenotypic components of metabolic syndrome: A population-based twin study. Obesity 2009, 17, 1581–1587. [Google Scholar] [CrossRef]
- Oh, S.W.; Lee, J.E.; Shin, E.; Kwon, H.; Choe, E.K.; Choi, S.Y.; Rhee, H.; Choi, S.H. Genome-wide association study of metabolic syndrome in Korean populations. PLoS ONE 2020, 15, e0227357. [Google Scholar] [CrossRef]
- Fall, T.; Ingelsson, E. Genome-wide association studies of obesity and metabolic syndrome. Mol. Cell Endocrinol. 2014, 382, 740–757. [Google Scholar] [CrossRef] [PubMed]
- Wan, J.Y.; Goodman, D.L.; Willems, E.L.; Freedland, A.R.; Norden-Krichmar, T.M.; Santorico, S.A.; Edwards, K.L.; American Diabetes GENNID Study Group. Genome-wide association analysis of metabolic syndrome quantitative traits in the GENNID multiethnic family study. Diabetol. Metab. Syndr. 2021, 13, 59. [Google Scholar] [CrossRef]
- Lind, L. Genome-wide association study of the metabolic syndrome in UK Biobank. Metab. Syndr. Relat. Disord. 2019, 17, 505–511. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.M.; Kim, S.K.; Ahn, H.J.; Jeong, K.H. A pilot genome-wide association study identifies novel markers of metabolic syndrome in patients with psoriasis. Ann. Dermatol. 2023, 35, 285–292. [Google Scholar] [CrossRef]
- Prasad, G.; Bandesh, K.; Giri, A.K.; Kauser, Y.; Chanda, P.; Parekatt, V.; Mathur, S.; Madhu, S.V.; Venkatesh, P.; Bhansali, A.; et al. Genome-wide association study of metabolic syndrome reveals primary genetic variants at CETP locus in indians. Biomolecules 2019, 9, 321. [Google Scholar] [CrossRef]
- Tekola-Ayele, F.; Doumatey, A.P.; Shriner, D.; Bentley, A.R.; Chen, G.; Zhou, J.; Fasanmade, O.; Johnson, T.; Oli, J.; Okafor, G.; et al. Genome-wide association study identifies African-ancestry specific variants for metabolic syndrome. Mol. Genet. Metab. 2015, 116, 305–313. [Google Scholar] [CrossRef]
- Zabaneh, D.; Balding, D.J. A genome-wide association study of the metabolic syndrome in Indian Asian men. PLoS ONE 2010, 5, e11961. [Google Scholar] [CrossRef]
- Zhu, Y.; Zhang, D.; Zhou, D.; Li, Z.; Li, Z.; Fang, L.; Yang, M.; Shan, Z.; Li, H.; Chen, J.; et al. Susceptibility loci for metabolic syndrome and metabolic components identified in Han Chinese: A multi-stage genome-wide association study. J. Cell Mol. Med. 2017, 21, 1106–1116. [Google Scholar] [CrossRef]
- Willems, E.L.; Wan, J.Y.; Norden-Krichmar, T.M.; Edwards, K.L.; Santorico, S.A. Transethnic meta-analysis of metabolic syndrome in a multiethnic study. Genet. Epidemiol. 2020, 44, 16–25. [Google Scholar] [CrossRef]
- Zhang, X.; Ye, P.; Huang, H.; Wang, B.; Dong, F.; Ling, Q. TCF7L2 rs290487 C allele aberrantly enhances hepatic gluconeogenesis through allele-specific changes in transcription and chromatin binding. Aging 2020, 12, 13365–13387. [Google Scholar] [CrossRef]
- Carlson, J.C.; Weeks, D.E.; Hawley, N.L.; Sun, G.; Cheng, H.; Naseri, T.; Reupena, M.S.; Tuitele, J.; Deka, R.; McGarvey, S.T.; et al. Genome-wide association studies in Samoans give insight into the genetic architecture of fasting serum lipid levels. J. Hum. Genet. 2021, 66, 111–121. [Google Scholar] [CrossRef] [PubMed]
- Hindorff, L.A.; Sethupathy, P.; Junkins, H.A.; Ramos, E.M.; Mehta, J.P.; Collins, F.S.; Manolio, T.A. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl. Acad. Sci. USA 2009, 106, 9362–9367. [Google Scholar] [CrossRef] [PubMed]
- National Institutes of Health. Third Report of the National Cholesterol Education Program Expert Panel on detection, evaluation, and treatment of high blood cholesterol in adults (Adult Treatment Panel III). NIH Publ. 2011, 1, 3670. [Google Scholar]
- World Health Organization. Definition, Diagnosis and Classification of Diabetes Mellitus and Its Complications. Report of a WHO Consultation; World Health Organization: Geneva, Switzerland, 1999. [Google Scholar]
- Balkau, B.; Charles, M.A. Comment on the provisional report from the WHO consultation. European Group for the Study of Insulin Resistance (EGIR). Diabet. Med. 1999, 16, 442–443. [Google Scholar] [CrossRef] [PubMed]
- Alberti, K.G.; Zimmet, P.; Shaw, J. Metabolic syndrome—A new world-wide definition. A consensus statement from the International Diabetes Federation. Diabet. Med. 2006, 23, 469–480. [Google Scholar] [CrossRef]
- GTEx Consortium. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science 2020, 369, 1318–1330. [Google Scholar] [CrossRef]
- Ongen, H.; Buil, A.; Brown, A.A.; Dermitzakis, E.T.; Delaneau, O. Fast and efficient QTL mapper for thousands of molecular phenotypes. Bioinformatics 2016, 32, 1479–1485. [Google Scholar] [CrossRef]
- Kim, Y.; Lee, C. Enrichment of spatial eGenes colocalized with type 2 diabetes mellitus genome-wide association study signals in the lysosomal pathway. Appl. Sci. 2023, 13, 10447. [Google Scholar] [CrossRef]
- Xie, Z.; Bailey, A.; Kuleshov, M.V.; Clarke, D.J.B.; Evangelista, J.E.; Jenkins, S.L.; Lachmann, A.; Wojciechowicz, M.L.; Kropiwnicki, E.; Jagodnik, K.M.; et al. Gene set knowledge discovery with Enrichr. Curr. Protoc. 2021, 1, e90. [Google Scholar] [CrossRef]
- Storey, J.D.; Tibshirani, R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 2003, 100, 9440–9445. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Silveira Rossi, J.L.; Barbalho, S.M.; Reverete de Araujo, R.; Bechara, M.D.; Sloan, K.P.; Sloan, L.A. Metabolic syndrome and cardiovascular diseases: Going beyond traditional risk factors. Diabetes Metab. Res. Rev. 2022, 38, e3502. [Google Scholar] [CrossRef] [PubMed]
- Parapid, B.; Ostojic, M.C.; Lalic, N.M.; Micic, D.; Damjanovic, S.; Bubanja, D.; Simic, D.; Lalic, K.; Polovina, S.; Marinkovic, J.; et al. Risk factors clustering within the metabolic syndrome: A pattern or by chance? Hell. J. Cardiol. 2014, 55, 92–100. [Google Scholar]
- Marques, A.R.A.; Saftig, P. Lysosomal storage disorders—Challenges, concepts and avenues for therapy: Beyond rare diseases. J. Cell Sci. 2019, 132, jcs221739. [Google Scholar] [CrossRef]
- Platt, F.M. Emptying the stores: Lysosomal diseases and therapeutic strategies. Nat. Rev. Drug Discov. 2018, 17, 133–150. [Google Scholar] [CrossRef]
- Bassi, M.T.; Manzoni, M.; Monti, E.; Pizzo, M.T.; Ballabio, A.; Borsani, G. Cloning of the gene encoding a novel integral membrane protein, mucolipidin-and identification of the two major founder mutations causing mucolipidosis type IV. Am. J. Hum. Genet. 2000, 67, 1110–1120. [Google Scholar] [CrossRef]
- Cosma, M.P.; Pepe, S.; Annunziata, I.; Newbold, R.F.; Grompe, M.; Parenti, G.; Ballabio, A. The multiple sulfatase deficiency gene encodes an essential and limiting factor for the activity of sulfatases. Cell 2003, 113, 445–456. [Google Scholar] [CrossRef]
- Lee, J.H.; Yu, W.H.; Kumar, A.; Lee, S.; Mohan, P.S.; Peterhoff, C.M.; Wolfe, D.M.; Martinez-Vicente, M.; Massey, A.C.; Sovak, G.; et al. Lysosomal proteolysis and autophagy require presenilin 1 and are disrupted by Alzheimer-related PS1 mutations. Cell 2010, 141, 1146–1158. [Google Scholar] [CrossRef]
- Lee, J.H.; McBrayer, M.K.; Wolfe, D.M.; Haslett, L.J.; Kumar, A.; Sato, Y.; Lie, P.P.; Mohan, P.; Coffey, E.E.; Kompella, U.; et al. Presenilin 1 maintains lysosomal Ca(2+) homeostasis via TRPML1 by regulating vATPase-mediated lysosome acidification. Cell Rep. 2015, 12, 1430–1444. [Google Scholar] [CrossRef]
- Coen, K.; Flannagan, R.S.; Baron, S.; Carraro-Lacroix, L.R.; Wang, D.; Vermeire, W.; Michiels, C.; Munck, S.; Baert, V.; Sugita, S.; et al. Lysosomal calcium homeostasis defects, not proton pump defects, cause endo-lysosomal dysfunction in PSEN-deficient cells. J. Cell Biol. 2012, 198, 23–35. [Google Scholar] [CrossRef] [PubMed]
- Aflaki, E.; Borger, D.K.; Moaven, N.; Stubblefield, B.K.; Rogers, S.A.; Patnaik, S.; Schoenen, F.J.; Westbroek, W.; Zheng, W.; Sullivan, P.; et al. A new glucocerebrosidase chaperone reduces α-synuclein and glycolipid levels in iPSC-derived dopaminergic neurons from patients with Gaucher disease and parkinsonism. J. Neurosci. 2016, 36, 7441–7452. [Google Scholar] [CrossRef]
- Qu, X.; Yu, J.; Bhagat, G.; Furuya, N.; Hibshoosh, H.; Troxel, A.; Rosen, J.; Eskelinen, E.L.; Mizushima, N.; Ohsumi, Y.; et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J. Clin. Investig. 2003, 112, 1809–1820. [Google Scholar] [CrossRef]
- Tang, H.; Sebti, S.; Titone, R.; Zhou, Y.; Isidoro, C.; Ross, T.S.; Hibshoosh, H.; Xiao, G.; Packer, M.; Xie, Y.; et al. Decreased BECN1 mRNA expression in human breast cancer is associated with estrogen receptor-negative subtypes and poor prognosis. EBiomedicine 2015, 2, 255–263. [Google Scholar] [CrossRef]
- Xu, L.; Lin, D.C.; Yin, D.; Koeffler, H.P. An emerging role of PARK2 in cancer. J. Mol. Med. 2014, 92, 31–42. [Google Scholar] [CrossRef] [PubMed]
- Mészáros, G.; Pasquier, A.; Vivot, K.; Goginashvili, A.; Ricci, R. Lysosomes in nutrient signalling: A focus on pancreatic β-cells. Diabetes Obes. Metab. 2018, 20 (Suppl. S2), 104–115. [Google Scholar] [CrossRef]
- Gornicka, A.; Fettig, J.; Eguchi, A.; Berk, M.P.; Thapaliya, S.; Dixon, L.J.; Feldstein, A.E. Adipocyte hypertrophy is associated with lysosomal permeability both in vivo and in vitro: Role in adipose tissue inflammation. Am. J. Physiol. Endocrinol. Metab. 2012, 303, E597–E606. [Google Scholar] [CrossRef]
- Jaishy, B.; Abel, E.D. Lipids, lysosomes, and autophagy. J. Lipid Res. 2016, 57, 1619–1635. [Google Scholar] [CrossRef]
- Zhang, S.; Peng, X.; Yang, S.; Li, X.; Huang, M.; Wei, S.; Liu, J.; He, G.; Zheng, H.; Yang, L.; et al. The regulation, function, and role of lipophagy, a form of selective autophagy, in metabolic disorders. Cell Death Dis. 2022, 13, 132. [Google Scholar] [CrossRef]
- Lamri, A.; Pigeyre, M.; Garver, W.S.; Meyre, D. The Extending Spectrum of NPC1-Related Human Disorders: From Niemann-Pick C1 Disease to Obesity. Endocr. Rev. 2018, 39, 192–220. [Google Scholar] [CrossRef]
- Thelen, A.M.; Zoncu, R. Emerging Roles for the Lysosome in Lipid Metabolism. Trends Cell Biol. 2017, 27, 833–850. [Google Scholar] [CrossRef]
- Bhattacharya, A.; Mukherjee, R.; Kuncha, S.K.; Brunstein, M.E.; Rathore, R.; Junek, S.; Münch, C.; Dikic, I. A lysosome membrane regeneration pathway depends on TBC1D15 and autophagic lysosomal reformation proteins. Nat. Cell Biol. 2023, 25, 685–698. [Google Scholar] [CrossRef]
- Tan, J.X.; Finkel, T. A phosphoinositide signalling pathway mediates rapid lysosomal repair. Nature 2022, 609, 815–821. [Google Scholar] [CrossRef] [PubMed]
- Bonam, S.R.; Wang, F.; Muller, S. Lysosomes as a therapeutic target. Nat. Rev. Drug Discov. 2019, 18, 923–948. [Google Scholar] [CrossRef]
- Ryu, J.; Woo, J.; Shin, J.; Ryoo, H.; Kim, Y.; Lee, C. Profile of differential promoter activity by nucleotide substitution at GWAS signals for multiple sclerosis. Medicine 2014, 93, e281. [Google Scholar] [CrossRef]
- Lee, C. Towards the genetic architecture of complex gene expression traits: Challenges and prospects for eQTL mapping in humans. Genes 2022, 13, 235. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Lee, C. Antagonistic regulatory effects of a single cis-acting expression quantitative trait locus between transcription and translation of the MRPL43 gene. BMC Genom. Data 2022, 23, 42. [Google Scholar] [CrossRef]
- Lee, C. Genome-wide expression quantitative trait loci analysis using mixed models. Front. Genet. 2018, 9, 341. [Google Scholar] [CrossRef]
- Lee, C. Bayesian inference for mixed model-based genome-wide analysis of expression quantitative trait loci by Gibbs sampling. Front. Genet. 2019, 10, 199. [Google Scholar] [CrossRef] [PubMed]
- Lee, C. Heterogeneous genetic architecture by gender for precision medicine of cardiovascular disease. J. Geriatr. Cardiol. 2018, 15, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Lee, C. Analytical models for genetics of human traits influenced by sex. Curr. Genom. 2015, 17, 439–443. [Google Scholar] [CrossRef]
| MetS | WHR | FBG | GM | SBP | DBP | TG | HDL | |
|---|---|---|---|---|---|---|---|---|
| GWAS signal a | ||||||||
| No. of raw signals | 252 | 9919 | 538 | 2199 | 3674 | 2417 | 8414 | 10,774 |
| No. of unique signals | 225 | 4735 | 293 | 1499 | 2603 | 1626 | 3413 | 3718 |
| Colocalized eQTL-eGene b | ||||||||
| No. of eQTLs | 92 | 3220 | 55 | 433 | 929 | 650 | 1197 | 1519 |
| No. of eGenes+MHC | 72 | 566 | 40 | 229 | 470 | 381 | 454 | 522 |
| No. of eGenes | 67 | 545 | 40 | 224 | 464 | 374 | 439 | 507 |
| Biological Pathway | Tissue | p | PBH |
|---|---|---|---|
| Cholesterol metabolism | Heart left ventricle | 1.46 × 10−6 | 1.14 × 10−4 |
| Cholesterol metabolism | Skin—not sun-exposed suprapubic | 1.73 × 10−5 | 5.72 × 10−4 |
| Cholesterol metabolism | Thyroid | 6.15 × 10−6 | 9.16 × 10−4 |
| Cholesterol metabolism | Adipose—subcutaneous | 6.69 × 10−5 | 1.07 × 10−3 |
| Cholesterol metabolism | Muscle—skeletal | 2.57 × 10−5 | 1.14 × 10−3 |
| Cholesterol metabolism | Heart atrial appendage | 8.21 × 10−5 | 1.50 × 10−3 |
| Vitamin digestion and absorption | Heart left ventricle | 1.09 × 10−4 | 4.25 × 10−3 |
| Cholesterol metabolism | Esophagus—mucosa | 3.91 × 10−4 | 4.76 × 10−3 |
| PPAR signaling pathway | Testis | 7.41 × 10−4 | 6.07 × 10−3 |
| Biosynthesis of unsaturated fatty acids | Stomach | 2.77 × 10−3 | 8.03 × 10−3 |
| Cholesterol metabolism | Artery—aorta | 1.73 × 10−3 | 8.14 × 10−3 |
| Hepatitis C | Brain—amygdala | 8.63 × 10−3 | 1.51 × 10−2 |
| Cholesterol metabolism | Testis | 2.63 × 10−3 | 1.58 × 10−2 |
| Vitamin digestion and absorption | Brain—substantia nigra | 1.31 × 10−2 | 1.59 × 10−2 |
| Glycosphingolipid biosynthesis | Pituitary | 7.06 × 10−3 | 1.80 × 10−2 |
| PPAR signaling pathway | Heart left ventricle | 3.03 × 10−3 | 1.82 × 10−2 |
| Cholesterol metabolism | Liver | 4.95 × 10−3 | 1.94 × 10−2 |
| Cholesterol metabolism | Small intestine—terminal ileum | 3.27 × 10−3 | 1.95 × 10−2 |
| Oocyte meiosis | Brain—frontal cortex BA9 | 2.18 × 10−3 | 2.24 × 10−2 |
| African trypanosomiasis | Brain—substantia nigra | 2.02 × 10−2 | 2.34 × 10−2 |
| Biological Pathway | Tissue | p | PBH |
|---|---|---|---|
| Lysosome | Esophagus—mucosa | 1.58 × 10−6 | 5.52 × 10−5 |
| Steroid hormone biosynthesis | Brain—putamen basal ganglia | 5.23 × 10−5 | 9.57 × 10−4 |
| Lysosome | Whole blood | 4.27 × 10−5 | 1.31 × 10−3 |
| Glyoxylate and dicarboxylate metabolism | Brain—cortex | 6.00 × 10−5 | 1.50 × 10−3 |
| Steroid hormone biosynthesis | Brain—hippocampus | 1.45 × 10−4 | 1.84 × 10−3 |
| Steroid hormone biosynthesis | Brain—hypothalamus | 1.48 × 10−4 | 2.29 × 10−3 |
| Glyoxylate and dicarboxylate metabolism | Pituitary | 9.52 × 10−5 | 2.38 × 10−3 |
| Steroid hormone biosynthesis | Kidney—cortex | 6.67 × 10−4 | 2.85 × 10−3 |
| Glyoxylate and dicarboxylate metabolism | Brain—caudate basal ganglia | 3.49 × 10−4 | 4.92 × 10−3 |
| Steroid hormone biosynthesis | Brain—amygdala | 1.35 × 10−3 | 6.67 × 10−3 |
| Steroid hormone biosynthesis | Brain—cerebellar hemisphere | 6.73 × 10−4 | 8.04 × 10−3 |
| Glyoxylate and dicarboxylate metabolism | Stomach | 6.58 × 10−4 | 9.47 × 10−3 |
| Collecting duct acid secretion | Artery—coronary | 8.54 × 10−4 | 1.00 × 10−2 |
| Steroid hormone biosynthesis | Brain—cerebellum | 4.80 × 10−4 | 1.04 × 10−2 |
| Steroid hormone biosynthesis | Muscle—skeletal | 5.87 × 10−4 | 1.18 × 10−2 |
| Ovarian steroidogenesis | Adipose—subcutaneous | 4.71 × 10−4 | 1.40 × 10−2 |
| Steroid hormone biosynthesis | Brain—nucleus accumbens basal ganglia | 1.65 × 10−3 | 1.71 × 10−2 |
| Steroid hormone biosynthesis | Brain—caudate basal ganglia | 1.63 × 10−3 | 1.78 × 10−2 |
| Lysosome | Skin—sun-exposed lower leg | 8.70 × 10−4 | 1.94 × 10−2 |
| Lysosome | Heart atrial appendage | 1.20 × 10−3 | 2.33 × 10−2 |
| Biological Pathway | Tissue | p | PBH |
|---|---|---|---|
| Thyroid hormone synthesis | Brain—hippocampus | 2.06 × 10−4 | 1.85 × 10−3 |
| Fructose and mannose metabolism | Prostate | 5.44 × 10−4 | 9.79 × 10−3 |
| Biosynthesis of unsaturated fatty acids | Stomach | 6.03 × 10−4 | 1.07 × 10−2 |
| Galactose metabolism | Stomach | 7.97 × 10−4 | 1.07 × 10−2 |
| Starch and sucrose metabolism | Stomach | 1.07 × 10−3 | 1.07 × 10−2 |
| Biosynthesis of unsaturated fatty acids | Brain—hypothalamus | 9.41 × 10−3 | 1.88 × 10−2 |
| Arachidonic acid metabolism | Brain—caudate basal ganglia | 1.21 × 10−3 | 1.93 × 10−2 |
| PPAR signaling pathway | Small intestine—terminal ileum | 1.57 × 10−3 | 2.35 × 10−2 |
| alpha-Linolenic acid metabolism | Pituitary | 8.70 × 10−4 | 2.44 × 10−2 |
| Amoebiasis | Brain—putamen basal ganglia | 1.37 × 10−3 | 2.57 × 10−2 |
| Leukocyte transendothelial migration | Brain—putamen basal ganglia | 1.71 × 10−3 | 2.57 × 10−2 |
| alpha-Linolenic acid metabolism | Ovary | 9.96 × 10−3 | 2.69 × 10−2 |
| Biosynthesis of unsaturated fatty acids | Ovary | 1.08 × 10−2 | 2.69 × 10−2 |
| Biosynthesis of unsaturated fatty acids | Brain—spinal cord cervical c-1 | 5.39 × 10−3 | 2.69 × 10−2 |
| Glycolysis/Gluconeogenesis | Stomach | 3.68 × 10−3 | 2.76 × 10−2 |
| Cortisol synthesis and secretion | Brain—spinal cord cervical c-1 | 1.29 × 10−2 | 3.01 × 10−2 |
| Morphine addiction | Brain—spinal cord cervical c-1 | 1.81 × 10−2 | 3.01 × 10−2 |
| Cushing syndrome | Brain—spinal cord cervical c-1 | 3.06 × 10−2 | 3.06 × 10−2 |
| Purine metabolism | Brain—spinal cord cervical c-1 | 2.56 × 10−2 | 3.06 × 10−2 |
| Arachido + B2:F21nic acid metabolism | Brain—hippocampus | 1.82 × 10−2 | 3.27 × 10−2 |
| Biological Pathway | Tissue | p | PBH |
|---|---|---|---|
| Fructose and mannose metabolism | Skin—sun-exposed lower leg | 1.60 × 10−5 | 4.20 × 10−3 |
| Fructose and mannose metabolism | Whole blood | 4.96 × 10−5 | 1.24 × 10−2 |
| Neomycin, kanamycin, and gentamicin biosynthesis | Skin—sun-exposed lower leg | 1.58 × 10−4 | 1.38 × 10−2 |
| Fructose and mannose metabolism | Heart atrial appendage | 7.78 × 10−4 | 1.41 × 10−2 |
| Fructose and mannose metabolism | Heart left ventricle | 6.20 × 10−4 | 1.46 × 10−2 |
| Sulfur metabolism | Brain—spinal cord cervical c-1 | 4.05 × 10−4 | 2.05 × 10−2 |
| Sulfur relay system | Brain—spinal cord cervical c-1 | 2.53 × 10−4 | 2.05 × 10−2 |
| Neomycin, kanamycin, and gentamicin biosynthesis | Heart atrial appendage | 1.57 × 10−3 | 2.32 × 10−2 |
| Neomycin, kanamycin, and gentamicin biosynthesis | Heart left ventricle | 1.39 × 10−3 | 2.37 × 10−2 |
| Biosynthesis of unsaturated fatty acids | Adipose—visceral omentum | 8.44 × 10−4 | 2.50 × 10−2 |
| Neomycin, kanamycin, and gentamicin biosynthesis | Breast—mammary tissue | 1.58 × 10−3 | 2.59 × 10−2 |
| Fructose and mannose metabolism | Nerve—tibial | 2.18 × 10−4 | 2.69 × 10−2 |
| Neomycin, kanamycin, and gentamicin biosynthesis | Nerve—tibial | 1.85 × 10−4 | 2.69 × 10−2 |
| Fructose and mannose metabolism | Adipose—visceral omentum | 1.83 × 10−3 | 2.95 × 10−2 |
| Fructose and mannose metabolism | Stomach | 2.45 × 10−4 | 3.36 × 10−2 |
| Galactose metabolism | Skin—sun-exposed lower leg | 1.03 × 10−3 | 3.38 × 10−2 |
| Amino sugar and nucleotide sugar metabolism | Skin—sun-exposed lower leg | 1.31 × 10−3 | 3.44 × 10−2 |
| Neomycin, kanamycin, and gentamicin biosynthesis | Adipose—visceral omentum | 2.47 × 10−3 | 3.67 × 10−2 |
| Fructose and mannose metabolism | Brain—hippocampus | 3.69 × 10−4 | 3.76 × 10−2 |
| Lysosome | Whole blood | 1.31 × 10−3 | 4.10 × 10−2 |
| Biological Pathway | Tissue | p | PBH |
|---|---|---|---|
| Cholesterol metabolism | Esophagus—mucosa | 2.78 × 10−8 | 5.39 × 10−6 |
| Metabolism of xenobiotics by cytochrome P450 | Liver | 7.45 × 10−7 | 1.41 × 10−4 |
| Cholesterol metabolism | Skin—sun-exposed lower leg | 3.83 × 10−6 | 1.57 × 10−4 |
| Cholesterol metabolism | Nerve—tibial | 8.22 × 10−6 | 7.07 × 10−4 |
| Cholesterol metabolism | Heart atrial appendage | 3.85 × 10−5 | 1.12 × 10−3 |
| Cholesterol metabolism | Liver | 5.09 × 10−5 | 1.60 × 10−3 |
| Drug metabolism | Liver | 9.85 × 10−5 | 2.07 × 10−3 |
| Cholesterol metabolism | Thyroid | 6.13 × 10−5 | 2.20 × 10−3 |
| Cholesterol metabolism | Artery—tibial | 4.81 × 10−5 | 2.24 × 10−3 |
| Cholesterol metabolism | Esophagus—muscularis | 1.18 × 10−4 | 4.01 × 10−3 |
| Cholesterol metabolism | Testis | 1.26 × 10−4 | 5.76 × 10−3 |
| Cholesterol metabolism | Skin—not sun-exposed suprapubic | 1.96 × 10−4 | 7.89 × 10−3 |
| Cholesterol metabolism | Pancreas | 6.89 × 10−4 | 9.27 × 10−3 |
| Cholesterol metabolism | Adipose—subcutaneous | 4.12 × 10−4 | 1.40 × 10−2 |
| Cholesterol metabolism | Cells—cultured fibroblasts | 3.84 × 10−4 | 1.42 × 10−2 |
| Cholesterol metabolism | Lung | 3.68 × 10−4 | 1.45 × 10−2 |
| Cholesterol metabolism | Muscle—skeletal | 6.99 × 10−4 | 1.58 × 10−2 |
| Glutathione metabolism | Liver | 9.54 × 10−4 | 1.71 × 10−2 |
| Cholesterol metabolism | Esophagus—gastroesophageal junction | 1.17 × 10−3 | 2.60 × 10−2 |
| Cholesterol metabolism | Colon—transverse | 1.72 × 10−3 | 3.27 × 10−2 |
| Biological Pathway | Tissue | p | PBH |
|---|---|---|---|
| Cholesterol metabolism | Heart atrial appendage | 3.36 × 10−9 | 8.79 × 10−7 |
| Cholesterol metabolism | Skin—sun-exposed lower leg | 3.74 × 10−7 | 2.78 × 10−5 |
| Cholesterol metabolism | Esophagus—mucosa | 1.68 × 10−7 | 3.11 × 10−5 |
| Cholesterol metabolism | Esophagus—gastroesophageal junction | 9.60 × 10−7 | 4.10 × 10−5 |
| Cholesterol metabolism | Muscle—skeletal | 2.23 × 10−6 | 1.08 × 10−4 |
| Cholesterol metabolism | Esophagus—muscularis | 1.84 × 10−6 | 1.21 × 10−4 |
| Cholesterol metabolism | Skin—not sun-exposed suprapubic | 3.63 × 10−6 | 1.52 × 10−4 |
| Cholesterol metabolism | Testis | 9.81 × 10−7 | 2.64 × 10−4 |
| Cholesterol metabolism | Heart left ventricle | 7.61 × 10−6 | 2.95 × 10−4 |
| Cholesterol metabolism | Adipose—subcutaneous | 7.83 × 10−6 | 4.59 × 10−4 |
| Cholesterol metabolism | Spleen | 2.16 × 10−6 | 5.48 × 10−4 |
| Metabolism of xenobiotics by cytochrome P450 | Minor salivary gland | 4.61 × 10−5 | 1.48 × 10−3 |
| Cholesterol metabolism | Nerve—tibial | 5.21 × 10−6 | 1.54 × 10−3 |
| Cholesterol metabolism | Cells—cultured fibroblasts | 6.01 × 10−6 | 1.77 × 10−3 |
| Cholesterol metabolism | Lung | 2.57 × 10−5 | 1.81 × 10−3 |
| Cholesterol metabolism | Liver | 9.53 × 10−5 | 2.12 × 10−3 |
| Metabolism of xenobiotics by cytochrome P450 | Liver | 1.38 × 10−4 | 2.73 × 10−3 |
| Glycine serine and threonine metabolism | Colon—transverse | 1.38 × 10−4 | 4.31 × 10−3 |
| Cholesterol metabolism | Thyroid | 2.96 × 10−5 | 4.33 × 10−3 |
| Lysosome | Esophagus—mucosa | 1.61 × 10−4 | 5.18 × 10−3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
An, Y.; Seo, Y.; Lee, C. Genetic Association of Diagnostic Traits of Metabolic Syndrome with Lysosomal Pathways: Insights from Target Gene Enrichment Analysis. Processes 2023, 11, 3221. https://doi.org/10.3390/pr11113221
An Y, Seo Y, Lee C. Genetic Association of Diagnostic Traits of Metabolic Syndrome with Lysosomal Pathways: Insights from Target Gene Enrichment Analysis. Processes. 2023; 11(11):3221. https://doi.org/10.3390/pr11113221
Chicago/Turabian StyleAn, Yeeun, Yunji Seo, and Chaeyoung Lee. 2023. "Genetic Association of Diagnostic Traits of Metabolic Syndrome with Lysosomal Pathways: Insights from Target Gene Enrichment Analysis" Processes 11, no. 11: 3221. https://doi.org/10.3390/pr11113221
APA StyleAn, Y., Seo, Y., & Lee, C. (2023). Genetic Association of Diagnostic Traits of Metabolic Syndrome with Lysosomal Pathways: Insights from Target Gene Enrichment Analysis. Processes, 11(11), 3221. https://doi.org/10.3390/pr11113221







